A Quantitative Survey of Bacterial Persistence in the Presence of Antibiotics: Towards Antipersister Antimicrobial Discovery
Abstract
1. Introduction
2. Results
2.1. Data Summary and Praxis
2.2. Bactericidal vs. Bacteriostatic Antibiotics
2.3. Growth Phase and Gram Staining Were Related to Significantly Different Levels of Bacterial Persisters to Antibiotics
2.4. Trends of Time–Kill in Relation to Growth Phase, Media Type, and Gram Staining
2.5. The Paradoxical Effect of Concentration on Bacterial Killing by Some Antibiotics
2.6. Top Six Bacteria
2.7. Cheminformatics of Antibiotics Represented in This Survey and Fluorophores Transported by E. coli
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hobby, G.L.; Meyer, K.; Chaffee, E. Observations on the mechanism of action of penicillin. Proc. Soc. Exp. Biol. Med. 1942, 50, 281–285. [Google Scholar] [CrossRef]
- Bigger, J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Greenwood, D.; O’Grady, F. Trimodal response of Escherichia coli and Proteus mirabilis to penicillins. Nature 1970, 228, 457–458. [Google Scholar] [CrossRef]
- Greenwood, D.; O’Grady, F. The effect of osmolality on the response of Escherichia coli and Proteus mirabilis to penicillins. Br. J. Exp. Pathol. 1972, 53, 457–464. [Google Scholar] [PubMed]
- Greenwood, D.; O’Grady, F. FL 1060: A new beta-lactam antibiotic with novel properties. J. Clin. Pathol. 1973, 26, 1–6. [Google Scholar] [CrossRef]
- Greenwood, D. Effect of osmolality on the response of Escherichia coli to mecillinam. Antimicrob. Agents Chemother. 1976, 10, 824–826. [Google Scholar] [CrossRef]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Sebastian, J.; Swaminath, S.; Nair, R.R.; Jakkala, K.; Pradhan, A.; Ajitkumar, P. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Michiels, J.E.; Van den Bergh, B.; Verstraeten, N.; Fauvart, M.; Michiels, J. In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob. Agents Chemother. 2016, 60, 4630–4637. [Google Scholar] [CrossRef] [PubMed]
- Van den Bergh, B.; Fauvart, M.; Michiels, J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 2017, 41, 219–251. [Google Scholar] [CrossRef] [PubMed]
- Defraine, V.; Fauvart, M.; Michiels, J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resist. Updates 2018, 38, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Hannan, T.J.; Totsika, M.; Mansfield, K.J.; Moore, K.H.; Schembri, M.A.; Hultgren, S.J. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 2012, 36, 616–648. [Google Scholar] [CrossRef]
- Blango, M.G.; Ott, E.M.; Erman, A.; Veranic, P.; Mulvey, M.A. Forced resurgence and targeting of intracellular uropathogenic Escherichia coli reservoirs. PLoS ONE 2014, 9, e93327. [Google Scholar] [CrossRef]
- Leatham-Jensen, M.P.; Mokszycki, M.E.; Rowley, D.C.; Deering, R.; Camberg, J.L.; Sokurenko, E.V.; Tchesnokova, V.L.; Frimodt-Moller, J.; Krogfelt, K.A.; Nielsen, K.L.; et al. Uropathogenic Escherichia coli metabolite-dependent quiescence and persistence may explain antibiotic tolerance during urinary tract infection. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Benavides-Montano, J.A.; Vadyvaloo, V. Yersinia pestis resists predation by Acanthamoeba castellanii and exhibits prolonged intracellular survival. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Greenwood, D. Phenotypic resistance to antimicrobial agents. J. Antimicrob. Chemother. 1985, 15, 653–655. [Google Scholar] [CrossRef][Green Version]
- Witkin, E.M. Inherited differences in sensitivity to radiation in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1946, 32, 59–68. [Google Scholar] [CrossRef]
- Luria, S.E.; Latarjet, R. Ultraviolet irradiation of bacteriophage during intracellular growth. J. Bacteriol. 1947, 53, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.R.M.; Hu, Y. New strategies for antibacterial drug design: Targeting non-multiplying latent bacteria. Drugs R D 2006, 7, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.R.; Halls, G.; Hu, Y. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 2011, 163, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef] [PubMed]
- Kaprelyants, A.S.; Gottschal, J.C.; Kell, D.B. Dormancy in non-sporulating bacteria. FEMS Microbiol. Rev. 1993, 10, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Kaprelyants, A.S.; Weichart, D.H.; Harwood, C.R.; Barer, M.R. Viability and activity in readily culturable bacteria: A review and discussion of the practical issues. Antonie Van Leeuwenhoek 1998, 73, 169–187. [Google Scholar] [CrossRef]
- Kell, D.; Potgieter, M.; Pretorius, E. Individuality, phenotypic differentiation, dormancy and ‘persistence’ in culturable bacterial systems: Commonalities shared by environmental, laboratory, and clinical microbiology. F1000Research 2015, 4, 179. [Google Scholar] [CrossRef]
- Bakkeren, E.; Diard, M.; Hardt, W.D. Evolutionary causes and consequences of bacterial antibiotic persistence. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef]
- Kussell, E.; Kishony, R.; Balaban, N.Q.; Leibler, S. Bacterial persistence: A model of survival in changing environments. Genetics 2005, 169, 1807–1814. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef]
- Veening, J.W.; Smits, W.K.; Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Wang, H.; Ray, J.C.J. Lineage space and the propensity of bacterial cells to undergo growth transitions. PLoS Comput. Biol. 2018, 14, e1006380. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, H.J.; Gallie, J.; Kost, C.; Ferguson, G.C.; Rainey, P.B. Experimental evolution of bet hedging. Nature 2009, 462, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.F.; Ziv, N.; Siegal, M.L. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012, 10, e1001325. [Google Scholar] [CrossRef]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006, 6, 53. [Google Scholar] [CrossRef]
- Jubair, M.; Morris, J.G., Jr.; Ali, A. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS ONE 2012, 7, e45187. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun. 2015, 6, 7983. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Kolar, M. Starvation- and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed. Pap. Med. Fac. Palacky Univ. Olomouc 2017, 161, 58–67. [Google Scholar] [CrossRef]
- Brown, D.R. Nitrogen starvation induces persister cell formation in Escherichia coli. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Koch, A.L. The adaptive responses of Escherichia coli to a feast and famine existence. Adv. Microb. Physiol. 1971, 6, 147–217. [Google Scholar] [CrossRef] [PubMed]
- Poindexter, J.S. Oligotrophy: Fast and famine existence. Adv. Microb. Ecol. 1981, 5, 63–89. [Google Scholar]
- Gelens, L.; Hill, L.; Vandervelde, A.; Danckaert, J.; Loris, R. A general model for toxin-antitoxin module dynamics can explain persister cell formation in E. coli. PLoS Comput. Biol. 2013, 9, e1003190. [Google Scholar] [CrossRef]
- Gerdes, K.; Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Holden, D.W.; Errington, J. Type II toxin-antitoxin systems and persister cells. mBio 2018, 9. [Google Scholar] [CrossRef]
- Kedzierska, B.; Hayes, F. Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef]
- Moyed, H.S.; Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef]
- Goormaghtigh, F.; Fraikin, N.; Putrins, M.; Hallaert, T.; Hauryliuk, V.; Garcia-Pino, A.; Sjodin, A.; Kasvandik, S.; Udekwu, K.; Tenson, T.; et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Fino, C.; Sorensen, M.A.; Semsey, S.; Gerdes, K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Fino, C.; Vestergaard, M.; Ingmer, H.; Pierrel, F.; Gerdes, K.; Harms, A. PasT of Escherichia coli sustains antibiotic tolerance and aerobic respiration as a bacterial homolog of mitochondrial Coq10. Microbiologyopen 2020. [Google Scholar] [CrossRef] [PubMed]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.; Michiels, J. The Persistence-inducing toxin hokb forms dynamic pores that cause ATP leakage. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wilmaerts, D.; Dewachter, L.; De Loose, P.J.; Bollen, C.; Verstraeten, N.; Michiels, J. HokB monomerization and membrane repolarization control persister awakening. Mol. Cell 2019, 75, 1031–1042 e1034. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1, 16051. [Google Scholar] [CrossRef]
- Braetz, S.; Schwerk, P.; Thompson, A.; Tedin, K.; Fulde, M. The role of ATP pools in persister cell formation in (fluoro)quinolone-susceptible and -resistant strains of Salmonella enterica ser. Typhimurium. Vet. Microbiol. 2017, 210, 116–123. [Google Scholar] [CrossRef]
- Shan, Y.; Gandt, A.B.; Rowe, S.E.; Deisinger, J.P.; Conlon, B.P.; Lewis, K. ATP-dependent persister formation in Escherichia coli. mBio 2017, 8. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Fraysse, L.; Moinier, D.; Dukan, S. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J. Bacteriol. 2008, 190, 887–893. [Google Scholar] [CrossRef]
- Coquel, A.S.; Jacob, J.P.; Primet, M.; Demarez, A.; Dimiccoli, M.; Julou, T.; Moisan, L.; Lindner, A.B.; Berry, H. Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput. Biol. 2013, 9, e1003038. [Google Scholar] [CrossRef]
- Leszczynska, D.; Matuszewska, E.; Kuczynska-Wisnik, D.; Furmanek-Blaszk, B.; Laskowska, E. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE 2013, 8, e54737. [Google Scholar] [CrossRef]
- Baig, U.I.; Bhadbhade, B.J.; Mariyam, D.; Watve, M.G. Protein aggregation in E. coli: Short term and long term effects of nutrient density. PLoS ONE 2014, 9, e107445. [Google Scholar] [CrossRef]
- Govers, S.K.; Mortier, J.; Adam, A.; Aertsen, A. Protein aggregates encode epigenetic memory of stressful encounters in individual Escherichia coli cells. PLoS Biol. 2018, 16, e2003853. [Google Scholar] [CrossRef]
- Pu, Y.; Li, Y.; Jin, X.; Tian, T.; Ma, Q.; Zhao, Z.; Lin, S.Y.; Chen, Z.; Li, B.; Yao, G.; et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 2019, 73, 143–156. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Yin, H.; Chang, Z. Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 2019, 5, 8. [Google Scholar] [CrossRef]
- Yamamoto, N.; Isshiki, R.; Kawai, Y.; Tanaka, D.; Sekiguchi, T.; Matsumoto, S.; Tsuneda, S. Stochastic expression of lactate dehydrogenase A induces Escherichia coli persister formation. J. Biosci. Bioeng. 2018, 126, 30–37. [Google Scholar] [CrossRef]
- Zalis, E.A.; Nuxoll, A.S.; Manuse, S.; Clair, G.; Radlinski, L.C.; Conlon, B.P.; Adkins, J.; Lewis, K. Stochastic Variation in Expression of the Tricarboxylic Acid Cycle Produces Persister Cells. mBio 2019, 10. [Google Scholar] [CrossRef]
- Carret, G.; Flandrois, J.P.; Lobry, J.R. Biphasic kinetics of bacterial killing by quinolones. J. Antimicrob. Chemother. 1991, 27, 319–327. [Google Scholar] [CrossRef]
- Kell, D.B.; Oliver, S.G. How drugs get into cells: Tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front. Pharmacol. 2014, 5, 231. [Google Scholar] [CrossRef]
- Lanthaler, K.; Bilsland, E.; Dobson, P.D.; Moss, H.J.; Pir, P.; Kell, D.B.; Oliver, S.G. Genome-wide assessment of the carriers involved in the cellular uptake of drugs: A model system in yeast. BMC Biol. 2011, 9, 70. [Google Scholar] [CrossRef]
- Kell, D.B.; Dobson, P.D.; Oliver, S.G. Pharmaceutical drug transport: The issues and the implications that it is essentially carrier-mediated only. Drug Discov. Today 2011, 16, 704–714. [Google Scholar] [CrossRef]
- Dobson, P.D.; Lanthaler, K.; Oliver, S.G.; Kell, D.B. Implications of the dominant role of transporters in drug uptake by cells. Curr. Top. Med. Chem. 2009, 9, 163–181. [Google Scholar] [CrossRef]
- Dobson, P.D.; Kell, D.B. Carrier-mediated cellular uptake of pharmaceutical drugs: An exception or the rule? Nat. Rev. Drug Discov. 2008, 7, 205–220. [Google Scholar] [CrossRef]
- Rahal, J.J., Jr.; Simberkoff, M.S. Bactericidal and bacteriostatic action of chloramphenicol against memingeal pathogens. Antimicrob. Agents Chemother. 1979, 16, 13–18. [Google Scholar] [CrossRef]
- Preblud, S.R.; Gill, C.J.; Campos, J.M. Bactericidal activities of chloramphenicol and eleven other antibiotics against Salmonella spp. Antimicrob. Agents Chemother. 1984, 25, 327–330. [Google Scholar] [CrossRef]
- Soriano, F.; Ponte, M.; Garcia-Hierro, P. Bactericidal activity of chloramphenicol, clindamycin, metronidazole and cefoxitin against Bacteroides fragilis. J. Antimicrob. Chemother. 1980, 6, 679–680. [Google Scholar] [CrossRef]
- Mulet, X.; Macia, M.D.; Mena, A.; Juan, C.; Perez, J.L.; Oliver, A. Azithromycin in Pseudomonas aeruginosa biofilms: Bactericidal activity and selection of nfxB mutants. Antimicrob. Agents Chemother. 2009, 53, 1552–1560. [Google Scholar] [CrossRef]
- Drago, L.; De Vecchi, E.; Nicola, L.; Legnani, D.; Gismondo, M.R. Kinetic bactericidal activity of telithromycin, azithromycin and clarithromycin against respiratory pathogens. APMIS 2005, 113, 655–663. [Google Scholar] [CrossRef]
- Imamura, Y.; Higashiyama, Y.; Tomono, K.; Izumikawa, K.; Yanagihara, K.; Ohno, H.; Miyazaki, Y.; Hirakata, Y.; Mizuta, Y.; Kadota, J.; et al. Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob. Agents Chemother. 2005, 49, 1377–1380. [Google Scholar] [CrossRef]
- Gu, J.W.; Scully, B.E.; Neu, H.C. Bactericidal activity of clarithromycin and its 14-hydroxy metabolite against Haemophilus influenzae and streptococcal pathogens. J. Clin. Pharmacol. 1991, 31, 1146–1150. [Google Scholar] [CrossRef]
- Lacy, M.K.; Owens, R.C., Jr.; Xu, X.; Nicolau, D.P.; Quintiliani, R.; Nightingale, C.H. Comparison of bactericidal activity after multidose administration of clarithromycin, azithromycin, and ceruroxime axetil against Streptococcus pneumoniae. Int. J. Antimicrob. Agents 1998, 10, 279–283. [Google Scholar] [CrossRef]
- Tateda, K.; Ishii, Y.; Matsumoto, T.; Furuya, N.; Nagashima, M.; Matsunaga, T.; Ohno, A.; Miyazaki, S.; Yamaguchi, K. Direct evidence for antipseudomonal activity of macrolides: Exposure-dependent bactericidal activity and inhibition of protein synthesis by erythromycin, clarithromycin, and azithromycin. Antimicrob. Agents Chemother. 1996, 40, 2271–2275. [Google Scholar] [CrossRef]
- Piccolomini, R.; Catamo, G.; Di Bonaventura, G. Bacteriostatic and bactericidal in vitro activities of clarithromycin and erythromycin against periodontopathic Actinobacillus actinomycetemcomitans. Antimicrob. Agents Chemother. 1998, 42, 3000–3001. [Google Scholar] [CrossRef][Green Version]
- Baltch, A.L.; Smith, R.P.; Ritz, W. Inhibitory and bactericidal activities of levofloxacin, ofloxacin, erythromycin, and rifampin used singly and in combination against Legionella pneumophila. Antimicrob. Agents Chemother. 1995, 39, 1661–1666. [Google Scholar] [CrossRef][Green Version]
- Bonacorsi, S.; Bingen, E. Bactericidal activity of erythromycin associated with sulphisoxazole against the infectious agents most frequently responsible for acute infantile otitis media. J. Antimicrob. Chemother. 1994, 33, 885–886. [Google Scholar] [CrossRef]
- Bartmann, K.; Tarbuc, R. The bactericidal activity of ampicillin, amoxicillin, tetracycline, and doxycycline against H. influenzae under various conditions of culture. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1985, 259, 78–89. [Google Scholar] [CrossRef]
- Ringertz, S.; Dornbusch, K. In vitro susceptibility to tetracycline and doxycycline in clinical isolates of Haemophilus influenzae. Scand. J. Infect. Dis. Suppl. 1988, 53, 7–11. [Google Scholar]
- Hamburger, M.; Carleton, J. Bactericidal action of penicillin and tetracycline against gram-positive cocci in broth and in blood clots. with a few observations concerning “persisters”. Arch. Intern. Med. 1960, 105, 372–382. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 2013, 57, 3230–3239. [Google Scholar] [CrossRef]
- Nemeth, J.; Oesch, G.; Kuster, S.P. Bacteriostatic versus bactericidal antibiotics for patients with serious bacterial infections: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2015, 70, 382–395. [Google Scholar] [CrossRef]
- Postgate, J.R. Viable counts and viability. Meth. Microbiol. 1969, 1, 611–628. [Google Scholar]
- Postgate, J.R. Death in microbes and macrobes. In The Survival of Vegetative Microbes; Gray, T.R.G., Postgate, J.R., Eds.; Cambridge University Press: Cambridge, UK, 1976; pp. 1–19. [Google Scholar]
- Barer, M.R.; Kaprelyants, A.S.; Weichart, D.H.; Harwood, C.R.; Kell, D.B. Microbial stress and culturability: Conceptual and operational domains. Microbiol. UK 1998, 144, 2009–2010. [Google Scholar] [CrossRef]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Wald-Dickler, N.; Holtom, P.; Spellberg, B. Busting the myth of “Static vs Cidal”: A systemic literature review. Clin. Infect. Dis. 2018, 66, 1470–1474. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, P.S.; Lazar, V.; Papp, B.; Arnoldini, M.; Zur Wiesch, P.A.; Busa-Fekete, R.; Fekete, G.; Pál, C.; Ackermann, M.; Bonhoeffer, S. Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob. Agents Chemother. 2014, 58, 4573–4582. [Google Scholar] [CrossRef]
- Coetzee, J.F.; Magstadt, D.R.; Sidhu, P.K.; Follett, L.; Schuler, A.M.; Krull, A.C.; Cooper, V.L.; Engelken, T.J.; Kleinhenz, M.D.; O’Connor, A.M. Association between antimicrobial drug class for treatment and retreatment of bovine respiratory disease (BRD) and frequency of resistant BRD pathogen isolation from veterinary diagnostic laboratory samples. PLoS ONE 2019, 14, e0219104. [Google Scholar] [CrossRef]
- Brown, M.R.W.; Williams, P. Influence of substrate limitation and growth phase on sensitivity to antimicrobial agents. J. Antimicrob. Chemother. 1985, 15 (Suppl. A), 7–14. [Google Scholar] [CrossRef]
- Brown, M.R.W.; Collier, P.J.; Gilbert, P. Influence of growth rate on susceptibility to antimicrobial agents: Modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 1990, 34, 1623–1628. [Google Scholar] [CrossRef]
- Stokes, J.M.; Gutierrez, A.; Lopatkin, A.J.; Andrews, I.W.; French, S.; Matic, I.; Brown, E.D.; Collins, J.J. A multiplexable assay for screening antibiotic lethality against drug-tolerant bacteria. Nat. Methods 2019, 16, 303–306. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Collins, J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 2009, 12, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Foti, J.J.; Devadoss, B.; Winkler, J.A.; Collins, J.J.; Walker, G.C. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 2012, 336, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Brynildsen, M.P.; Winkler, J.A.; Spina, C.S.; MacDonald, I.C.; Collins, J.J. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 2013, 31, 160–165. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef]
- Zhao, X.; Drlica, K. Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 2014, 21, 1–6. [Google Scholar] [CrossRef]
- Hong, Y.; Zeng, J.; Wang, X.; Drlica, K.; Zhao, X. Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. USA 2019, 116, 10064–10071. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Camacho, D.M.; Kohanski, M.A.; Callura, J.M.; Collins, J.J. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 2012, 46, 561–572. [Google Scholar] [CrossRef]
- Dorsey-Oresto, A.; Lu, T.; Mosel, M.; Wang, X.; Salz, T.; Drlica, K.; Zhao, X. YihE kinase is a central regulator of programmed cell death in bacteria. Cell Rep. 2013, 3, 528–537. [Google Scholar] [CrossRef]
- Votyakova, T.V.; Kaprelyants, A.S.; Kell, D.B. Influence of viable cells on the resuscitation of dormant cells in Micrococcus luteus cultures held in an extended stationary phase: The population effect. Appl. Environ. Microbiol. 1994, 60, 3284–3291. [Google Scholar] [CrossRef]
- Kaprelyants, A.S.; Kell, D.B. Dormancy in stationary-phase cultures of Micrococcus luteus: Flow cytometric analysis of starvation and resuscitation. Appl. Environ. Microbiol. 1993, 59, 3187–3196. [Google Scholar] [CrossRef]
- Volzing, K.G.; Brynildsen, M.P. Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. mBio 2015, 6. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Kashevarova, N.M.; Tyuleneva, E.A.; Shumkov, M.S. Stationary-phase genes upregulated by polyamines are responsible for the formation of Escherichia coli persister cells tolerant to netilmicin. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Wang, Y.; Bojer, M.S.; George, S.E.; Wang, Z.; Jensen, P.R.; Wolz, C.; Ingmer, H. Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci. Rep. 2018, 8, 10849. [Google Scholar] [CrossRef]
- Feng, J.; Li, T.; Yee, R.; Yuan, Y.; Bai, C.; Cai, M.; Shi, W.; Embers, M.; Brayton, C.; Saeki, H.; et al. Stationary phase persister/biofilm microcolony of Borrelia burgdorferi causes more severe disease in a mouse model of Lyme arthritis: Implications for understanding persistence, Post-treatment Lyme Disease Syndrome (PTLDS), and treatment failure. Discov. Med. 2019, 27, 125–138. [Google Scholar]
- Aedo, S.J.; Orman, M.A.; Brynildsen, M.P. Stationary phase persister formation in Escherichia coli can be suppressed by piperacillin and PBP3 inhibition. BMC Microbiol. 2019, 19, 140. [Google Scholar] [CrossRef]
- Morgan, J.; Smith, M.; Mc Auley, M.T.; Salcedo-Sora, J.E. Disrupting folate metabolism reduces the capacity of bacteria in exponential growth to develop persisters to antibiotics. Microbiology 2018, 164, 1432–1445. [Google Scholar] [CrossRef]
- Vilcheze, C.; Hartman, T.; Weinrick, B.; Jain, P.; Weisbrod, T.R.; Leung, L.W.; Freundlich, J.S.; Jacobs, W.R., Jr. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, 4495–4500. [Google Scholar] [CrossRef]
- Choudhary, E.; Sharma, R.; Kumar, Y.; Agarwal, N. Conditional silencing by CRISPRi reveals the role of DNA gyrase in formation of drug-tolerant persister population in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2019, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Soriano, F.; Greenwood, D. Action and interaction of penicillin and gentamicin on enterococci. J. Clin. Pathol. 1979, 32, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T. The 4-quinoline antibacterials. In The Scientific Basis of Antimicrobial Chemotherapry; Greenwood, D., O’Grady, F., Eds.; Cambridge University Press: Avon, UK, 1985; pp. 69–94. [Google Scholar]
- Eagle, H.; Musselman, A.D. The rate of bactericidal action of penicillin in vitro as a function of its concentration, and its paradoxically reduced activity at high concentrations against certain organisms. J. Exp. Med. 1948, 88, 99–131. [Google Scholar] [CrossRef] [PubMed]
- Liebens, V.; Defraine, V.; Knapen, W.; Swings, T.; Beullens, S.; Corbau, R.; Marchand, A.; Chaltin, P.; Fauvart, M.; Michiels, J. Identification of 1-((2,4-Dichlorophenethyl)Amino)-3-Phenoxypropan-2-ol, a novel antibacterial compound active against persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Meylan, S.; Andrews, I.W.; Collins, J.J. Targeting antibiotic tolerance, pathogen by pathogen. Cell 2018, 172, 1228–1238. [Google Scholar] [CrossRef]
- Salcedo-Sora, J.E.; Jindal, S.; O’Hagan, S.; Kell, D.B. A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: Surrogate ligands for bacterial membrane transporters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Steve, O.; Swainston, N.; Handl, J.; Kell, D.B. A ‘rule of 0.5’ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics 2015, 11, 323–339. [Google Scholar] [CrossRef]
- Whittle, M.; Gillet, V.J.; Willett, P.; Alex, A.; Loesel, J. Enhancing the effectiveness of virtual screening by fusing nearest neighbor lists: A comparison of similarity coefficients. J. Chem. Inf. Comput. Sci. 2004, 44, 1840–1848. [Google Scholar] [CrossRef]
- Willett, P. Similarity searching using 2D structural fingerprints. Methods Mol. Biol. 2011, 672, 133–158. [Google Scholar] [CrossRef]
- “persister, n.”. OED Online. June 2020. Oxford University Press. Available online: https://www.oed.com/view/Entry/141469?redirectedFrom=persister (accessed on 12 August 2020).
- Van den Bergh, B.; Michiels, J.E.; Wenseleers, T.; Windels, E.M.; Boer, P.V.; Kestemont, D.; De Meester, L.; Verstrepen, K.J.; Verstraeten, N.; Fauvart, M.; et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 2016, 1, 16020. [Google Scholar] [CrossRef] [PubMed]
- Swaminath, S.; Paul, A.; Pradhan, A.; Sebastian, J.; Nair, R.R.; Ajitkumar, P. Mycobacterium smegmatis moxifloxacin persister cells produce high levels of hydroxyl radical, generating genetic resisters selectable not only with moxifloxacin, but also with ethambutol and isoniazid. Microbiology 2020, 166, 180–198. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Gefen, O.; Balaban, N.Q. The importance of being persistent: Heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 2009, 33, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Kussell, E.; Leibler, S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005, 309, 2075–2078. [Google Scholar] [CrossRef] [PubMed]
- Radzikowski, J.L.; Vedelaar, S.; Siegel, D.; Ortega, A.D.; Schmidt, A.; Heinemann, M. Bacterial persistence is an active sigmaS stress response to metabolic flux limitation. Mol. Syst. Biol. 2016, 12, 882. [Google Scholar] [CrossRef] [PubMed]
- Dam, S.; Pages, J.M.; Masi, M. Stress responses, outer membrane permeability control and antimicrobial resistance in Enterobacteriaceae. Microbiology 2018, 164, 260–267. [Google Scholar] [CrossRef]
- Kim, J.S.; Chowdhury, N.; Yamasaki, R.; Wood, T.K. Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 2018, 20, 2038–2048. [Google Scholar] [CrossRef]
- Kubistova, L.; Dvoracek, L.; Tkadlec, J.; Melter, O.; Licha, I. Environmental stress affects the formation of Staphylococcus aureus persisters tolerant to antibiotics. Microb. Drug Resist. 2018, 24, 547–555. [Google Scholar] [CrossRef]
- Amato, S.M.; Orman, M.A.; Brynildsen, M.P. Metabolic control of persister formation in Escherichia coli. Mol. Cell 2013, 50, 475–487. [Google Scholar] [CrossRef]
- Fisher, J.F.; Mobashery, S. Constructing and deconstructing the bacterial cell wall. Protein Sci. 2020, 29, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Antimicrobial resistance in gram-positive bacteria. Am. J. Med. 2006, 119, S11–S19, discussion S62–S70. [Google Scholar] [CrossRef] [PubMed]
- Karas, J.A.; Chen, F.; Schneider-Futschik, E.K.; Kang, Z.; Hussein, M.; Swarbrick, J.; Hoyer, D.; Giltrap, A.M.; Payne, R.J.; Li, J.; et al. Synthesis and structure-activity relationships of teixobactin. Ann. N. Y. Acad. Sci. 2020, 1459, 86–105. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Maria, J.P.S., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.E.; Peschel, A.; et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef]
- Samara, E.; Moriarty, T.F.; Decosterd, L.A.; Richards, R.G.; Gautier, E.; Wahl, P. Antibiotic stability over six weeks in aqueous solution at body temperature with and without heat treatment that mimics the curing of bone cement. Bone Joint Res. 2017, 6, 296–306. [Google Scholar] [CrossRef]
- Tang, H.J.; Chang, M.C.; Ko, W.C.; Huang, K.Y.; Lee, C.L.; Chuang, Y.C. In vitro and in vivo activities of newer fluoroquinolones against Vibrio vulnificus. Antimicrob. Agents Chemother. 2002, 46, 3580–3584. [Google Scholar] [CrossRef]
- MacKenzie, F.M.; Milne, K.E.; Gould, I.M. The post-exposure response of Enterobacteriaceae to ceftibuten. J. Antimicrob. Chemother. 1999, 43, 71–77. [Google Scholar] [CrossRef][Green Version]
- Bergan, T.; Carlsen, I.B. Effect of antibiotics eliminated by first order kinetics. J. Antimicrob. Chemother. 1985, 15 (Suppl. A), 147–152. [Google Scholar] [CrossRef]
- Gunnison, J.B.; Fraher, M.A.; Jawetz, E. Persistence of Staphylococcus aureus in Penicillin in Vitro. J. Gen. Microbiol. 1964, 35, 335–349. [Google Scholar] [CrossRef]
- Prasetyoputri, A.; Jarrad, A.M.; Cooper, M.A.; Blaskovich, M.A.T. The eagle effect and antibiotic-induced persistence: Two sides of the same coin? Trends Microbiol. 2019, 27, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Jarrad, A.M.; Blaskovich, M.A.T.; Prasetyoputri, A.; Karoli, T.; Hansford, K.A.; Cooper, M.A. Detection and investigation of eagle effect resistance to vancomycin in Clostridium difficile with an ATP-bioluminescence assay. Front. Microbiol. 2018, 9, 1420. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; Tan, J.; Dick, T. Eagle Effect in nonreplicating persister mycobacteria. Antimicrob. Agents Chemother. 2015, 59, 7786–7789. [Google Scholar] [CrossRef][Green Version]
- Grandiere-Perez, L.; Jacqueline, C.; Lemabecque, V.; Patey, O.; Potel, G.; Caillon, J. Eagle effect in Corynebacterium diphtheriae. J. Infect. Dis. 2005, 191, 2118–2120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ericson, J.E.; Hornik, C.P.; Greenberg, R.G.; Clark, R.H.; Tremoulet, A.H.; Le, J.; Cohen-Wolkowiez, M.; Smith, P.B.; Benjamin, D.K., Jr.; Best Pharmaceuticals for Children Act—Pediatric Trials Network. Paradoxical antibiotic effect of ampicillin: Use of a population pharmacokinetic model to evaluate a clinical correlate of the eagle effect in infants with bacteremia. Pediatr. Infect. Dis. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General mechanisms leading to persister formation and awakening. Trends Genet. 2019, 35, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.D.; Schroeder, K.; Jonas, K. Protein aggregation in bacteria. FEMS Microbiol. Rev. 2020, 44, 54–72. [Google Scholar] [CrossRef]
- Yamamotoya, T.; Dose, H.; Tian, Z.; Faure, A.; Toya, Y.; Honma, M.; Igarashi, K.; Nakahigashi, K.; Soga, T.; Mori, H.; et al. Glycogen is the primary source of glucose during the lag phase of E. coli proliferation. Biochim. Biophys. Acta 2012, 1824, 1442–1448. [Google Scholar] [CrossRef]
- Boehm, A.; Arnoldini, M.; Bergmiller, T.; Roosli, T.; Bigosch, C.; Ackermann, M. Genetic manipulation of glycogen allocation affects replicative lifespan in E. coli. PLoS Genet. 2016, 12, e1005974. [Google Scholar] [CrossRef]
- Simsek, E.; Kim, M. Power-law tail in lag time distribution underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 2019, 116, 17635–17640. [Google Scholar] [CrossRef]
- Koch, A.L.; Verwer, R.W.; Nanninga, N. Incorporation of diaminopimelic acid into the old poles of Escherichia coli. J. Gen. Microbiol. 1982, 128, 2893–2898. [Google Scholar] [CrossRef]
- Pradel, N.; Santini, C.L.; Bernadac, A.; Shih, Y.L.; Goldberg, M.B.; Wu, L.F. Polar positional information in Escherichia coli spherical cells. Biochem. Biophys. Res. Commun. 2007, 353, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.W.; Yie, A.M.; Eder, E.K.; Dennis, R.G.; Basting, P.J.; Martinez, K.A., II; Jones, B.D.; Slonczewski, J.L. Periplasmic acid stress increases cell division asymmetry (polar aging) of Escherichia coli. PLoS ONE 2015, 10, e0144650. [Google Scholar] [CrossRef] [PubMed]
- Rang, C.U.; Peng, A.Y.; Chao, L. Temporal dynamics of bacterial aging and rejuvenation. Curr. Biol. 2011, 21, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Yu, Y.; Lee, H.; Jeon, J.H.; Wanner, B.L.; Ritchie, K. Asymmetric polar localization dynamics of the serine chemoreceptor protein Tsr in Escherichia coli. PLoS ONE 2018, 13, e0195887. [Google Scholar] [CrossRef]
- Ping, L.; Weiner, B.; Kleckner, N. Tsr-GFP accumulates linearly with time at cell poles, and can be used to differentiate ‘old’ versus ‘new’ poles, in Escherichia coli. Mol. Microbiol. 2008, 69, 1427–1438. [Google Scholar] [CrossRef]
- Rang, C.U.; Proenca, A.; Buetz, C.; Shi, C.; Chao, L. Minicells as a damage disposal mechanism in Escherichia coli. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Marchou, B.; Bellido, F.; Charnas, R.; Lucain, C.; Pechere, J.C. Contribution of beta-lactamase hydrolysis and outer membrane permeability to ceftriaxone resistance in Enterobacter cloacae. Antimicrob. Agents Chemother. 1987, 31, 1589–1595. [Google Scholar] [CrossRef]
- Pagani, L.; Debiaggi, M.; Tenni, R.; Cereda, P.M.; Landini, P.; Romero, E. Beta-lactam resistant Pseudomonas aeruginosa strains emerging during therapy: Synergistic resistance mechanisms. Microbiologica 1988, 11, 47–53. [Google Scholar]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.B.; et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell 2016, 62, 284–294. [Google Scholar] [CrossRef]
- Vilhena, C.; Kaganovitch, E.; Shin, J.Y.; Grunberger, A.; Behr, S.; Kristoficova, I.; Brameyer, S.; Kohlheyer, D.; Jung, K. A single-cell view of the BtsSR/YpdAB pyruvate sensing network in Escherichia coli and its biological relevance. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 2007, 51, 2092–2099. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, Y. PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2010, 65, 1237–1242. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, B.; Guo, J.; Cong, H.; He, S.; Zhou, H.; Zhu, F.; Wang, Q.; Zhang, L. L-Tryptophan represses persister formation via inhibiting bacterial motility and promoting antibiotics absorption. Future Microbiol. 2019, 14, 757–771. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Yan, B.; Lyu, L.; Takiff, H.E.; Gao, Q. The potassium transporter KdpA affects persister formation by regulating ATP levels in Mycobacterium marinum. Emerg. Microbes. Infect. 2020, 9, 129–139. [Google Scholar] [CrossRef]
- Kim, W.; Zou, G.; Hari, T.P.A.; Wilt, I.K.; Zhu, W.; Galle, N.; Faizi, H.A.; Hendricks, G.L.; Tori, K.; Pan, W.; et al. A selective membrane-targeting repurposed antibiotic with activity against persistent methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 16529–16534. [Google Scholar] [CrossRef]
- Kim, W.; Steele, A.D.; Zhu, W.; Csatary, E.E.; Fricke, N.; Dekarske, M.M.; Jayamani, E.; Pan, W.; Kwon, B.; Sinitsa, I.F.; et al. Discovery and optimization of nTZDpa as an Antibiotic effective against bacterial persisters. ACS Infect. Dis. 2018, 4, 1540–1545. [Google Scholar] [CrossRef]
- Lewis, K. New approaches to antimicrobial discovery. Biochem. Pharmacol 2017, 134, 87–98. [Google Scholar] [CrossRef]
- Yaakov, G.; Lerner, D.; Bentele, K.; Steinberger, J.; Barkai, N. Coupling phenotypic persistence to DNA damage increases genetic diversity in severe stress. Nat. Ecol. Evol. 2017, 1, 16. [Google Scholar] [CrossRef]
- Megaw, J.; Gilmore, B.F. Archaeal persisters: Persister cell formation as a stress response in Haloferax volcanii. Front. Microbiol. 2017, 8, 1589. [Google Scholar] [CrossRef]
- Greenwood, D. The activity of polymyxins against dense populations of Escherichia coli. J. Gen. Microbiol. 1975, 91, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Moyed, H.S.; Broderick, S.H. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1986, 166, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, J.S.; Hooper, D.C.; McHugh, G.L.; Bozza, M.A.; Swartz, M.N. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 1990, 34, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- John, J.F., Jr.; Atkins, L.T.; Maple, P.A.; Bratoeva, M. Activities of newer fluoroquinolones against Shigella sonnei. Antimicrob. Agents Chemother. 1992, 36, 2346–2348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meng, X.; Nightingale, C.H.; Sweeney, K.R.; Quintiliani, R. Loss of bactericidal activities of quinolones during the post-antibiotic effect induced by rifampicin. J. Antimicrob. Chemother. 1994, 33, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, J.R.; Hershberger, E.; Rybak, M.J. Analysis of vancomycin population susceptibility profiles, killing activity, and postantibiotic effect against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 1914–1918. [Google Scholar] [CrossRef]
- Debbia, E.A.; Roveta, S.; Schito, A.M.; Gualco, L.; Marchese, A. Antibiotic persistence: The role of spontaneous DNA repair response. Microb. Drug Resist. 2001, 7, 335–342. [Google Scholar] [CrossRef]
- Spoering, A.L.; Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef]
- Bedenic, B.; Vranes, J. Post-exposure effects of cefepime and cefpirome on isogenic Escherichia coli hosts producing SHV-extended-spectrum beta-lactamases. J. Chemother. 2003, 15, 342–349. [Google Scholar] [CrossRef]
- Cha, R.; Akins, R.L.; Rybak, M.J. Linezolid, levofloxacin, and vancomycin against vancomycin-tolerant and fluoroquinolone-resistant Streptococcus pneumoniae in an in vitro pharmacodynamic model. Pharmacotherapy 2003, 23, 1531–1537. [Google Scholar] [CrossRef]
- Hu, Y.; Coates, A.R.; Mitchison, D.A. Sterilizing activities of fluoroquinolones against rifampin-tolerant populations of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Sieradzki, K.; Leski, T.; Dick, J.; Borio, L.; Tomasz, A. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: Multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 2003, 41, 1687–1693. [Google Scholar] [CrossRef] [PubMed]
- Spoering, A.L.; Vulic, M.; Lewis, K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 2006, 188, 5136–5144. [Google Scholar] [CrossRef] [PubMed]
- Gefen, O.; Gabay, C.; Mumcuoglu, M.; Engel, G.; Balaban, N.Q. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 6145–6149. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Lewis, K.; Vulic, M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 2008, 52, 2718–2726. [Google Scholar] [CrossRef]
- Dorr, T.; Lewis, K.; Vulic, M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009, 5, e1000760. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef]
- Dorr, T.; Vulic, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef]
- Joers, A.; Kaldalu, N.; Tenson, T. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J. Bacteriol. 2010, 192, 3379–3384. [Google Scholar] [CrossRef]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010, 391, 209–213. [Google Scholar] [CrossRef]
- Ma, C.; Sim, S.; Shi, W.; Du, L.; Xing, D.; Zhang, Y. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 2010, 303, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Manuel, J.; Zhanel, G.G.; de Kievit, T. Cadaverine suppresses persistence to carboxypenicillins in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2010, 54, 5173–5179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moker, N.; Dean, C.R.; Tao, J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 2010, 192, 1946–1955. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2011, 2, e00100–e00111. [Google Scholar] [CrossRef]
- Kim, J.S.; Heo, P.; Yang, T.J.; Lee, K.S.; Jin, Y.S.; Kim, S.K.; Shin, D.; Kweon, D.H. Bacterial persisters tolerate antibiotics by not producing hydroxyl radicals. Biochem. Biophys. Res. Commun. 2011, 413, 105–110. [Google Scholar] [CrossRef]
- Luidalepp, H.; Joers, A.; Kaldalu, N.; Tenson, T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 2011, 193, 3598–3605. [Google Scholar] [CrossRef]
- Shapiro, J.A.; Nguyen, V.L.; Chamberlain, N.R. Evidence for persisters in Staphylococcus epidermidis RP62a planktonic cultures and biofilms. J. Med. Microbiol. 2011, 60, 950–960. [Google Scholar] [CrossRef]
- Hong, S.H.; Wang, X.; O’Connor, H.F.; Benedik, M.J.; Wood, T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012, 5, 509–522. [Google Scholar] [CrossRef]
- Lechner, S.; Lewis, K.; Bertram, R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 2012, 22, 235–244. [Google Scholar] [CrossRef]
- Lechner, S.; Patra, P.; Klumpp, S.; Bertram, R. Interplay between population dynamics and drug tolerance of Staphylococcus aureus persister cells. J. Mol. Microbiol. Biotechnol. 2012, 22, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Leung, V.; Levesque, C.M. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 2012, 194, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Niepa, T.H.; Gilbert, J.L.; Ren, D. Controlling Pseudomonas aeruginosa persister cells by weak electrochemical currents and synergistic effects with tobramycin. Biomaterials 2012, 33, 7356–7365. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Kawata, K.; Taniuchi, A.; Kakinuma, K.; May, T.; Okabe, S. RelE-mediated dormancy is enhanced at high cell density in Escherichia coli. J. Bacteriol. 2012, 194, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Vulic, M.; Keren, I.; Lewis, K. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 2012, 56, 4922–4926. [Google Scholar] [CrossRef] [PubMed]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef]
- Knudsen, G.M.; Ng, Y.; Gram, L. Survival of bactericidal antibiotic treatment by a persister subpopulation of Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 7390–7397. [Google Scholar] [CrossRef]
- Li, J.; Ji, L.; Shi, W.; Xie, J.; Zhang, Y. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2477–2481. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 2013, 57, 4398–4409. [Google Scholar] [CrossRef]
- Que, Y.A.; Hazan, R.; Strobel, B.; Maura, D.; He, J.; Kesarwani, M.; Panopoulos, P.; Tsurumi, A.; Giddey, M.; Wilhelmy, J.; et al. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS ONE 2013, 8, e80140. [Google Scholar] [CrossRef]
- Feng, J.; Wang, T.; Shi, W.; Zhang, S.; Sullivan, D.; Auwaerter, P.G.; Zhang, Y. Identification of novel activity against Borrelia burgdorferi persisters using an FDA approved drug library. Emerg. Microbes Infect. 2014, 3, e49. [Google Scholar] [CrossRef] [PubMed]
- Fleck, L.E.; North, E.J.; Lee, R.E.; Mulcahy, L.R.; Casadei, G.; Lewis, K. A screen for and validation of prodrug antimicrobials. Antimicrob. Agents Chemother. 2014, 58, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- Goneau, L.W.; Yeoh, N.S.; MacDonald, K.W.; Cadieux, P.A.; Burton, J.P.; Razvi, H.; Reid, G. Selective target inactivation rather than global metabolic dormancy causes antibiotic tolerance in uropathogens. Antimicrob. Agents Chemother. 2014, 58, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Chauhan, A.; Letoffe, S.; Fischer, F.; de Reuse, H.; Beloin, C.; Ghigo, J.M. pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J. Infect. Dis. 2014, 210, 1357–1366. [Google Scholar] [CrossRef]
- Marques, C.N.; Morozov, A.; Planzos, P.; Zelaya, H.M. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ. Microbiol. 2014, 80, 6976–6991. [Google Scholar] [CrossRef]
- Mordukhova, E.A.; Pan, J.G. Stabilization of homoserine-O-succinyltransferase (MetA) decreases the frequency of persisters in Escherichia coli under stressful conditions. PLoS ONE 2014, 9, e110504. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Deshayes, S.; Hawker, S.; Blacker, A.; Kasko, A.M.; Wong, G.C. Engineering persister-specific antibiotics with synergistic antimicrobial functions. ACS Nano 2014, 8, 8786–8793. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Kashevarova, N.M.; Karavaeva, E.A.; Shumkov, M.S. Putrescine controls the formation of Escherichia coli persister cells tolerant to aminoglycoside netilmicin. FEMS Microbiol. Lett. 2014, 361, 25–33. [Google Scholar] [CrossRef][Green Version]
- Tripathi, A.; Dewan, P.C.; Siddique, S.A.; Varadarajan, R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014, 289, 4191–4205. [Google Scholar] [CrossRef]
- Willenborg, J.; Willms, D.; Bertram, R.; Goethe, R.; Valentin-Weigand, P. Characterization of multi-drug tolerant persister cells in Streptococcus suis. BMC Microbiol. 2014, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.M.; Brynildsen, M.P. Persister heterogeneity arising from a single metabolic stress. Curr. Biol. 2015, 25, 2090–2098. [Google Scholar] [CrossRef] [PubMed]
- Caskey, J.R.; Embers, M.E. Persister development by Borrelia burgdorferi populations in vitro. Antimicrob. Agents Chemother. 2015, 59, 6288–6295. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, G.S.; Yao, X.; Wang, J.; Peng, B.; Bader, R.A.; Ren, D. Human granulocyte macrophage colony-stimulating factor enhances antibiotic susceptibility of Pseudomonas aeruginosa Persister Cells. Sci. Rep. 2015, 5, 17315. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Auwaerter, P.G.; Zhang, Y. Drug combinations against Borrelia burgdorferi persisters in vitro: Eradication achieved by using daptomycin, cefoperazone and doxycycline. PLoS ONE 2015, 10, e0117207. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Kallifidas, D.; Bai, F.; Ukhanova, M.; Mai, V.; Jin, S.; Luesch, H.; Huigens, R.W., III. Halogenated phenazines that potently eradicate biofilms, MRSA persister cells in non-biofilm cultures, and Mycobacterium tuberculosis. Angew. Chem. Int. Ed. Engl. 2015, 54, 14819–14823. [Google Scholar] [CrossRef]
- Germain, E.; Roghanian, M.; Gerdes, K.; Maisonneuve, E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. USA 2015, 112, 5171–5176. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, A.; Ortega-Muro, F.; Alameda-Martin, L.; Mitchison, D.; Coates, A. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front. Microbiol. 2015, 6, 641. [Google Scholar] [CrossRef]
- Kim, W.; Conery, A.L.; Rajamuthiah, R.; Fuchs, B.B.; Ausubel, F.M.; Mylonakis, E. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS ONE 2015, 10, e0127640. [Google Scholar] [CrossRef]
- Kuczynska-Wisnik, D.; Stojowska, K.; Matuszewska, E.; Leszczynska, D.; Algara, M.M.; Augustynowicz, M.; Laskowska, E. Lack of intracellular trehalose affects formation of Escherichia coli persister cells. Microbiology 2015, 161, 786–796. [Google Scholar] [CrossRef]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef]
- Leung, V.; Ajdic, D.; Koyanagi, S.; Levesque, C.M. The formation of Streptococcus mutans persisters induced by the quorum-sensing peptide pheromone is affected by the LexA regulator. J. Bacteriol. 2015, 197, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Mok, W.W.; Orman, M.A.; Brynildsen, M.P. Impacts of global transcriptional regulators on persister metabolism. Antimicrob. Agents Chemother. 2015, 59, 2713–2719. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Yu, Y.; Losada, L. The in vitro antibiotic tolerant persister population in Burkholderia pseudomallei is altered by environmental factors. Front. Microbiol. 2015, 6, 1338. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; He, X.; Zou, X.; Wang, G.; Li, S.; Wu, Y. Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 3267–3272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sandvik, E.L.; Fazen, C.H.; Henry, T.C.; Mok, W.W.; Brynildsen, M.P. Non-monotonic survival of Staphylococcus aureus with respect to ciprofloxacin concentration arises from prophage-dependent killing of persisters. Pharmaceuticals 2015, 8, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Lazinski, D.; Rowe, S.; Camilli, A.; Lewis, K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 2015, 6. [Google Scholar] [CrossRef]
- Sharma, B.; Brown, A.V.; Matluck, N.E.; Hu, L.T.; Lewis, K. Borrelia burgdorferi, the causative agent of lyme disease, forms drug-tolerant persister cells. Antimicrob. Agents Chemother. 2015, 59, 4616–4624. [Google Scholar] [CrossRef]
- Silva-Herzog, E.; McDonald, E.M.; Crooks, A.L.; Detweiler, C.S. Physiologic stresses reveal a salmonella persister state and TA family toxins modulate tolerance to these stresses. PLoS ONE 2015, 10, e0141343. [Google Scholar] [CrossRef]
- Verstraeten, N.; Knapen, W.J.; Kint, C.I.; Liebens, V.; Van den Bergh, B.; Dewachter, L.; Michiels, J.E.; Fu, Q.; David, C.C.; Fierro, A.C.; et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 2015, 59, 9–21. [Google Scholar] [CrossRef]
- Wang, W.; Chen, J.; Chen, G.; Du, X.; Cui, P.; Wu, J.; Zhao, J.; Wu, N.; Zhang, W.; Li, M.; et al. Transposon mutagenesis identifies novel genes associated with Staphylococcus aureus persister formation. Front. Microbiol. 2015, 6, 1437. [Google Scholar] [CrossRef] [PubMed]
- Yee, R.; Cui, P.; Shi, W.; Feng, J.; Zhang, Y. Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics 2015, 4, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.; Halliday, N.; Williams, P.; Atkins, H.S.; Bancroft, G.J.; Titball, R.W. Burkholderia pseudomallei kynB plays a role in AQ production, biofilm formation, bacterial swarming and persistence. Res. Microbiol. 2016, 167, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 2016, 63, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Kwan, B.W.; Wood, T.K. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 2016, 6, 20519. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Wood, T.L.; Martinez-Vazquez, M.; Garcia-Contreras, R.; Wood, T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 2016, 113, 1984–1992. [Google Scholar] [CrossRef]
- Curtis, T.D.; Gram, L.; Knudsen, G.M. The small colony variant of Listeria monocytogenes is more tolerant to antibiotics and has altered survival in RAW 264.7 murine macrophages. Front. Microbiol. 2016, 7, 1056. [Google Scholar] [CrossRef]
- Duan, X.; Huang, X.; Wang, X.; Yan, S.; Guo, S.; Abdalla, A.E.; Huang, C.; Xie, J. l-Serine potentiates fluoroquinolone activity against Escherichia coli by enhancing endogenous reactive oxygen species production. J. Antimicrob. Chemother. 2016, 71, 2192–2199. [Google Scholar] [CrossRef]
- Duan, X.; Li, Y.; Du, Q.; Huang, Q.; Guo, S.; Xu, M.; Lin, Y.; Liu, Z.; Xie, J. Mycobacterium Lysine epsilon-aminotransferase is a novel alarmone metabolism related persister gene via dysregulating the intracellular amino acid level. Sci. Rep. 2016, 6, 19695. [Google Scholar] [CrossRef]
- Goncalves, F.D.; de Carvalho, C.C. Phenotypic modifications in Staphylococcus aureus cells exposed to high concentrations of vancomycin and teicoplanin. Front. Microbiol. 2016, 7, 13. [Google Scholar] [CrossRef]
- Jaiswal, S.; Paul, P.; Padhi, C.; Ray, S.; Ryan, D.; Dash, S.; Suar, M. The Hha-TomB toxin-antitoxin system shows conditional toxicity and promotes persister cell formation by inhibiting apoptosis-like death in S. Typhimurium. Sci. Rep. 2016, 6, 38204. [Google Scholar] [CrossRef]
- Kim, J.S.; Cho, D.H.; Heo, P.; Jung, S.C.; Park, M.; Oh, E.J.; Sung, J.; Kim, P.J.; Lee, S.C.; Lee, D.H.; et al. Fumarate-mediated persistence of Escherichia coli against antibiotics. Antimicrob. Agents Chemother. 2016, 60, 2232–2240. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.G.; Gwon, G.; Wood, T.K.; Lee, J. Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 2016, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Liebens, V.; Frangipani, E.; Van der Leyden, A.; Fauvart, M.; Visca, P.; Michiels, J. Membrane localization and topology of the DnpA protein control fluoroquinolone tolerance in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Mina, E.G.; Marques, C.N. Interaction of Staphylococcus aureus persister cells with the host when in a persister state and following awakening. Sci. Rep. 2016, 6, 31342. [Google Scholar] [CrossRef] [PubMed]
- Molina-Quiroz, R.C.; Lazinski, D.W.; Camilli, A.; Levy, S.B. Transposon-sequencing analysis unveils novel genes involved in the generation of persister cells in uropathogenic Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 6907–6910. [Google Scholar] [CrossRef]
- Mukherjee, D.; Zou, H.; Liu, S.; Beuerman, R.; Dick, T. Membrane-targeting AM-0016 kills mycobacterial persisters and shows low propensity for resistance development. Future Microbiol. 2016, 11, 643–650. [Google Scholar] [CrossRef]
- Niepa, T.H.; Snepenger, L.M.; Wang, H.; Sivan, S.; Gilbert, J.L.; Jones, M.B.; Ren, D. Sensitizing Pseudomonas aeruginosa to antibiotics by electrochemical disruption of membrane functions. Biomaterials 2016, 74, 267–279. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Persister formation in Escherichia coli can be inhibited by treatment with nitric oxide. Free Radic. Biol. Med. 2016, 93, 145–154. [Google Scholar] [CrossRef]
- Prax, M.; Mechler, L.; Weidenmaier, C.; Bertram, R. Glucose augments killing efficiency of daptomycin challenged Staphylococcus aureus persisters. PLoS ONE 2016, 11, e0150907. [Google Scholar] [CrossRef]
- Springer, M.T.; Singh, V.K.; Cheung, A.L.; Donegan, N.P.; Chamberlain, N.R. Effect of clpP and clpC deletion on persister cell number in Staphylococcus aureus. J. Med. Microbiol. 2016, 65, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Torrey, H.L.; Keren, I.; Via, L.E.; Lee, J.S.; Lewis, K. High persister mutants in Mycobacterium tuberculosis. PLoS ONE 2016, 11, e0155127. [Google Scholar] [CrossRef] [PubMed]
- VandenBerg, K.E.; Ahn, S.; Visick, J.E. (p)ppGpp-dependent persisters increase the fitness of Escherichia coli bacteria deficient in isoaspartyl protein repair. Appl. Environ. Microbiol. 2016, 82, 5444–5454. [Google Scholar] [CrossRef] [PubMed]
- Walawalkar, Y.D.; Vaidya, Y.; Nayak, V. Response of Salmonella Typhi to bile-generated oxidative stress: Implication of quorum sensing and persister cell populations. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef]
- Xu, T.; Han, J.; Zhang, J.; Chen, J.; Wu, N.; Zhang, W.; Zhang, Y. Absence of protoheme IX farnesyltransferase CtaB causes virulence attenuation but enhances pigment production and persister survival in MRSA. Front. Microbiol. 2016, 7, 1625. [Google Scholar] [CrossRef]
- Abouelhassan, Y.; Basak, A.; Yousaf, H.; Huigens, R.W., III. Identification of N-arylated NH125 analogues as rapid eradicating agents against MRSA persister cells and potent biofilm killers of gram-positive pathogens. Chembiochem 2017, 18, 352–357. [Google Scholar] [CrossRef]
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front. Microbiol. 2017, 8, 924. [Google Scholar] [CrossRef]
- Bamford, R.A.; Smith, A.; Metz, J.; Glover, G.; Titball, R.W.; Pagliara, S. Investigating the physiology of viable but non-culturable bacteria by microfluidics and time-lapse microscopy. BMC Biol. 2017, 15, 121. [Google Scholar] [CrossRef]
- Chung, E.S.; Wi, Y.M.; Ko, K.S. Variation in formation of persister cells against colistin in Acinetobacter baumannii isolates and its relationship with treatment failure. J. Antimicrob. Chemother. 2017, 72, 2133–2135. [Google Scholar] [CrossRef]
- Curtis, T.D.; Takeuchi, I.; Gram, L.; Knudsen, G.M. The influence of the toxin/antitoxin mazEF on growth and survival of Listeria monocytogenes under stress. Toxins 2017, 9. [Google Scholar] [CrossRef]
- Dahl, J.U.; Gray, M.J.; Bazopoulou, D.; Beaufay, F.; Lempart, J.; Koenigsknecht, M.J.; Wang, Y.; Baker, J.R.; Hasler, W.L.; Young, V.B.; et al. The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat. Microbiol. 2017, 2, 16267. [Google Scholar] [CrossRef]
- Gallo, S.W.; Donamore, B.K.; Pagnussatti, V.E.; Ferreira, C.A.; de Oliveira, S.D. Effects of meropenem exposure in persister cells of Acinetobacter calcoaceticus-baumannii. Future Microbiol. 2017, 12, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, X.; Li, B.; Yao, J.; Wood, T.K.; Wang, X. Tail-anchored inner membrane protein ElaB increases resistance to stress while reducing persistence in Escherichia coli. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed]
- Koeva, M.; Gutu, A.D.; Hebert, W.; Wager, J.D.; Yonker, L.M.; O’Toole, G.A.; Ausubel, F.M.; Moskowitz, S.M.; Joseph-McCarthy, D. An Antipersister strategy for treatment of chronic Pseudomonas aeruginosa infections. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Kudrin, P.; Varik, V.; Oliveira, S.R.; Beljantseva, J.; Del Peso Santos, T.; Dzhygyr, I.; Rejman, D.; Cava, F.; Tenson, T.; Hauryliuk, V. Subinhibitory concentrations of bacteriostatic antibiotics induce relA-dependent and relA-independent tolerance to beta-lactams. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, N.; Zhang, S.; Yuan, Y.; Zhang, W.; Zhang, Y. Variable persister gene interactions with (p)ppGpp for persister formation in Escherichia coli. Front. Microbiol. 2017, 8, 1795. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.W.; Feng, Z.; Luo, Y.; Veening, J.W.; Zhang, J.R. Transcriptional repressor PtvR regulates phenotypic tolerance to vancomycin in Streptococcus pneumoniae. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Mantilla-Calderon, D.; Hong, P.Y. Fate and Persistence of a pathogenic NDM-1-Positive Escherichia coli strain in anaerobic and aerobic sludge microcosms. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Sahukhal, G.S.; Pandey, S.; Elasri, M.O. msaABCR operon is involved in persister cell formation in Staphylococcus aureus. BMC Microbiol. 2017, 17, 218. [Google Scholar] [CrossRef]
- Shen, Q.; Zhou, W.; Hu, L.; Qi, Y.; Ning, H.; Chen, J.; Mo, H. Bactericidal activity of alpha-bromocinnamaldehyde against persisters in Escherichia coli. PLoS ONE 2017, 12, e0182122. [Google Scholar] [CrossRef]
- Silva-Valenzuela, C.A.; Lazinski, D.W.; Kahne, S.C.; Nguyen, Y.; Molina-Quiroz, R.C.; Camilli, A. Growth arrest and a persister state enable resistance to osmotic shock and facilitate dissemination of Vibrio cholerae. ISME J. 2017, 11, 2718–2728. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Guo, Y.; Tang, K.; Wen, Z.; Li, B.; Zeng, Z.; Wang, X. MqsR/MqsA toxin/antitoxin system regulates persistence and biofilm formation in Pseudomonas putida KT2440. Front. Microbiol. 2017, 8, 840. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; El Meouche, I.; Dunlop, M.J. Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 2017, 7, 43839. [Google Scholar] [CrossRef]
- Xu, T.; Wang, X.Y.; Cui, P.; Zhang, Y.M.; Zhang, W.H.; Zhang, Y. The agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front. Microbiol. 2017, 8, 2189. [Google Scholar] [CrossRef]
- Alkasir, R.; Ma, Y.; Liu, F.; Li, J.; Lv, N.; Xue, Y.; Hu, Y.; Zhu, B. Characterization and Transcriptome analysis of Acinetobacter baumannii persister cells. Microb. Drug Resist. 2018, 24, 1466–1474. [Google Scholar] [CrossRef]
- Alvarez, R.; Inostroza, O.; Garavaglia, M.; Minton, N.P.; Paredes-Sabja, D.; Gil, F. Effect of antibiotic treatment on the formation of non-spore Clostridium difficile persister-like cells. J. Antimicrob. Chemother. 2018, 73, 2396–2399. [Google Scholar] [CrossRef]
- Bojer, M.S.; Lindemose, S.; Vestergaard, M.; Ingmer, H. Quorum sensing-regulated phenol-soluble modulins limit persister cell populations in Staphylococcus aureus. Front. Microbiol. 2018, 9, 255. [Google Scholar] [CrossRef]
- Cameron, D.R.; Shan, Y.; Zalis, E.A.; Isabella, V.; Lewis, K. A Genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Cui, P.; Niu, H.; Shi, W.; Zhang, S.; Zhang, W.; Zhang, Y. Identification of genes involved in bacteriostatic antibiotic-induced persister formation. Front. Microbiol. 2018, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Donamore, B.K.; Gallo, S.W.; Ferreira, P.M.A.; Ferreira, C.A.S.; de Oliveira, S.D. Levels of persisters influenced by aeration in Acinetobacter calcoaceticus-baumannii. Future Microbiol. 2018, 13, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Baltekin, O.; Waneskog, M.; Elkhalifa, D.; Hammarlof, D.L.; Elf, J.; Koskiniemi, S. Contact-dependent growth inhibition induces high levels of antibiotic-tolerant persister cells in clonal bacterial populations. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xu, R.; Zhao, Y.; Liu, D.; Liu, Z.; Wang, X.; Chen, H.; Kong, M.G. Gas plasma pre-treatment increases antibiotic sensitivity and persister eradication in methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2018, 9, 537. [Google Scholar] [CrossRef]
- Kalita, S.; Kandimalla, R.; Bhowal, A.C.; Kotoky, J.; Kundu, S. Functionalization of beta-lactam antibiotic on lysozyme capped gold nanoclusters retrogress MRSA and its persisters following awakening. Sci. Rep. 2018, 8, 5778. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Sharma, P.; Capalash, N. Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci. Rep. 2018, 8, 11029. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Matsumoto, S.; Ling, Y.; Okuda, S.; Tsuneda, S. AldB controls persister formation in Escherichia coli depending on environmental stress. Microbiol. Immunol. 2018, 62, 299–309. [Google Scholar] [CrossRef]
- Kim, J.S.; Yamasaki, R.; Song, S.; Zhang, W.; Wood, T.K. Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 2018, 20, 2085–2098. [Google Scholar] [CrossRef]
- Kim, W.; Zhu, W.; Hendricks, G.L.; Van Tyne, D.; Steele, A.D.; Keohane, C.E.; Fricke, N.; Conery, A.L.; Shen, S.; Pan, W.; et al. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 2018, 556, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Bae, S. Molecular viability testing of viable but non-culturable bacteria induced by antibiotic exposure. Microb Biotechnol. 2018, 11, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Kawai, Y.; Miyagawa, S.; Iwamoto, Y.; Okuda, S.; Sanchez-Gorostiaga, A.; Vicente, M.; Tsuneda, S. Unique transcriptional profile of native persisters in Escherichia coli. J. Biosci. Bioeng. 2018, 125, 15–22. [Google Scholar] [CrossRef]
- Miyaue, S.; Suzuki, E.; Komiyama, Y.; Kondo, Y.; Morikawa, M.; Maeda, S. Bacterial memory of persisters: Bacterial persister cells can retain their phenotype for days or weeks after withdrawal from colony-biofilm culture. Front. Microbiol. 2018, 9, 1396. [Google Scholar] [CrossRef]
- Molina-Quiroz, R.C.; Silva-Valenzuela, C.; Brewster, J.; Castro-Nallar, E.; Levy, S.B.; Camilli, A. Cyclic AMP regulates bacterial persistence through repression of the oxidative stress response and SOS-dependent DNA repair in uropathogenic Escherichia coli. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswamy, V.P.; Keagy, L.L.; Duris, K.; Wiesmann, W.; Loughran, A.J.; Townsend, S.M.; Baker, S. Novel glycopolymer eradicates antibiotic- and CCCP-induced persister cells in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 1724. [Google Scholar] [CrossRef] [PubMed]
- Oppezzo, O.J.; Giacobone, A.F.F. Lethal effect of photodynamic treatment on persister bacteria. Photochem. Photobiol. 2018, 94, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Rycroft, J.A.; Gollan, B.; Grabe, G.J.; Hall, A.; Cheverton, A.M.; Larrouy-Maumus, G.; Hare, S.A.; Helaine, S. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat. Commun. 2018, 9, 1993. [Google Scholar] [CrossRef]
- Van Laar, T.A.; Esani, S.; Birges, T.J.; Hazen, B.; Thomas, J.M.; Rawat, M. Pseudomonas aeruginosa gshA mutant is defective in biofilm formation, swarming, and pyocyanin production. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Vulin, C.; Leimer, N.; Huemer, M.; Ackermann, M.; Zinkernagel, A.S. Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat. Commun. 2018, 9, 4074. [Google Scholar] [CrossRef]
- Wang, M.; Fang, K.; Hong, S.M.C.; Kim, I.; Jang, I.S.; Hong, S.H. Medium chain unsaturated fatty acid ethyl esters inhibit persister formation of Escherichia coli via antitoxin HipB. Appl. Microbiol. Biotechnol. 2018, 102, 8511–8524. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, Y.; Lv, B.; Sun, F.; Yao, W.; Wang, Y.; Fu, X. Hypoionic shock facilitates aminoglycoside killing of both nutrient shift- and starvation-induced bacterial persister cells by rapidly enhancing aminoglycoside uptake. Front. Microbiol. 2019, 10, 2028. [Google Scholar] [CrossRef]
- Chung, E.S.; Ko, K.S. Eradication of persister cells of Acinetobacter baumannii through combination of colistin and amikacin antibiotics. J. Antimicrob. Chemother. 2019, 74, 1277–1283. [Google Scholar] [CrossRef]
- Drescher, S.P.M.; Gallo, S.W.; Ferreira, P.M.A.; Ferreira, C.A.S.; Oliveira, S.D. Salmonella enterica persister cells form unstable small colony variants after in vitro exposure to ciprofloxacin. Sci. Rep. 2019, 9, 7232. [Google Scholar] [CrossRef]
- Edelmann, D.; Berghoff, B.A. Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci. Rep. 2019, 9, 14256. [Google Scholar] [CrossRef] [PubMed]
- Goormaghtigh, F.; Van Melderen, L. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci. Adv. 2019, 5, eaav9462. [Google Scholar] [CrossRef]
- Lee, J.S.; Choi, J.Y.; Chung, E.S.; Peck, K.R.; Ko, K.S. Variation in the formation of persister cells against meropenem in Klebsiella pneumoniae bacteremia and analysis of its clinical features. Diagn. Microbiol. Infect. Dis. 2019, 95, 114853. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Fu, W.; Li, S.; Ren, H.; Li, M.; Liu, C.; Zhang, B.; Xia, Y.; Fan, Z.; Xu, C.; et al. Identification of novel genes that promote persister formation by repressing transcription and cell division in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2019, 74, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.; Mlouka, M.A.B.; Berthe, T.; Di Martino, P.; Jouenne, T.; Brunel, J.M.; De, E. Anti-persister activity of squalamine against Acinetobacter baumannii. Int. J. Antimicrob. Agents 2019, 53, 337–342. [Google Scholar] [CrossRef]
- Soares, A.; Alexandre, K.; Lamoureux, F.; Lemee, L.; Caron, F.; Pestel-Caron, M.; Etienne, M. Efficacy of a ciprofloxacin/amikacin combination against planktonic and biofilm cultures of susceptible and low-level resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2019, 74, 3252–3259. [Google Scholar] [CrossRef]
- Spanka, D.T.; Konzer, A.; Edelmann, D.; Berghoff, B.A. High-throughput proteomics identifies proteins with importance to postantibiotic recovery in depolarized persister cells. Front. Microbiol. 2019, 10, 378. [Google Scholar] [CrossRef]
- Svenningsen, M.S.; Veress, A.; Harms, A.; Mitarai, N.; Semsey, S. Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci. Rep. 2019, 9, 6056. [Google Scholar] [CrossRef]
- Thao, S.; Brandl, M.T.; Carter, M.Q. Enhanced formation of shiga toxin-producing Escherichia coli persister variants in environments relevant to leafy greens production. Food Microbiol. 2019, 84, 103241. [Google Scholar] [CrossRef]
- van Tatenhove-Pel, R.J.; Zwering, E.; Solopova, A.; Kuipers, O.P.; Bachmann, H. Ampicillin-treated Lactococcus lactis MG1363 populations contain persisters as well as viable but non-culturable cells. Sci. Rep. 2019, 9, 9867. [Google Scholar] [CrossRef]
- Amraei, F.; Narimisa, N.; Kalani, B.S.; Lohrasbi, V.; Jazi, F.M. Persister cells formation and expression of type II Toxin-Antitoxin system genes in Brucella melitensis (16M) and Brucella abortus (B19). Iran. J. Pathol. 2020, 15, 127–133. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Consensus rank orderings of molecular fingerprints illustrate the ‘most genuine’ similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. ADMET DMPK 2017, 5, 85–125. [Google Scholar]
- Clevenland, W.S.; Grosse, E.; Shyu, W.M. Local regression models. In Statistical Models in S.; Chambers, J.M., Hastie, T.J., Eds.; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1992. [Google Scholar]
- Hu, Y.; Shamaei-Tousi, A.; Liu, Y.; Coates, A. A new approach for the discovery of antibiotics by targeting non-multiplying bacteria: A novel topical antibiotic for staphylococcal infections. PLoS ONE 2010, 5, e11818. [Google Scholar] [CrossRef] [PubMed]
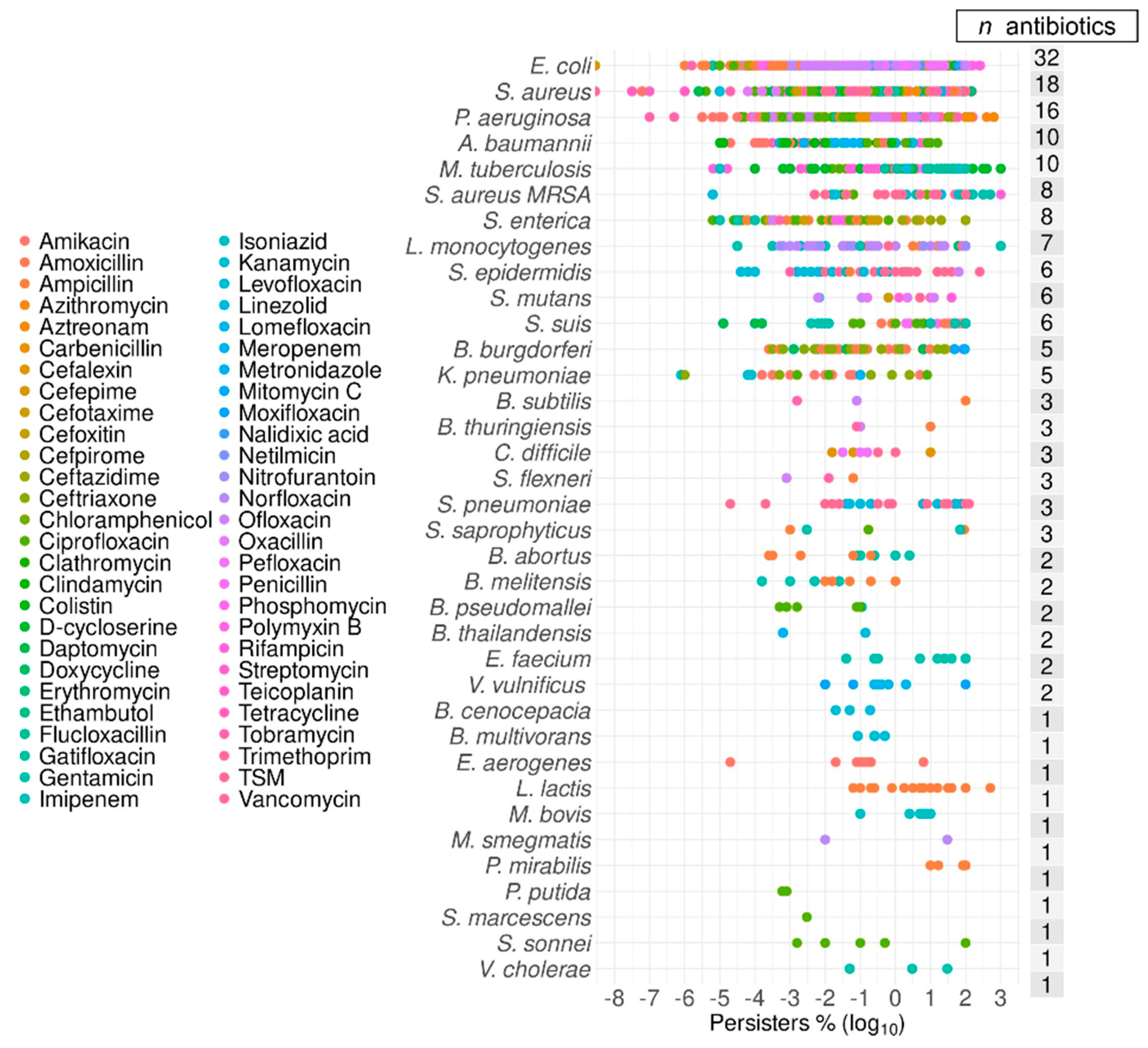
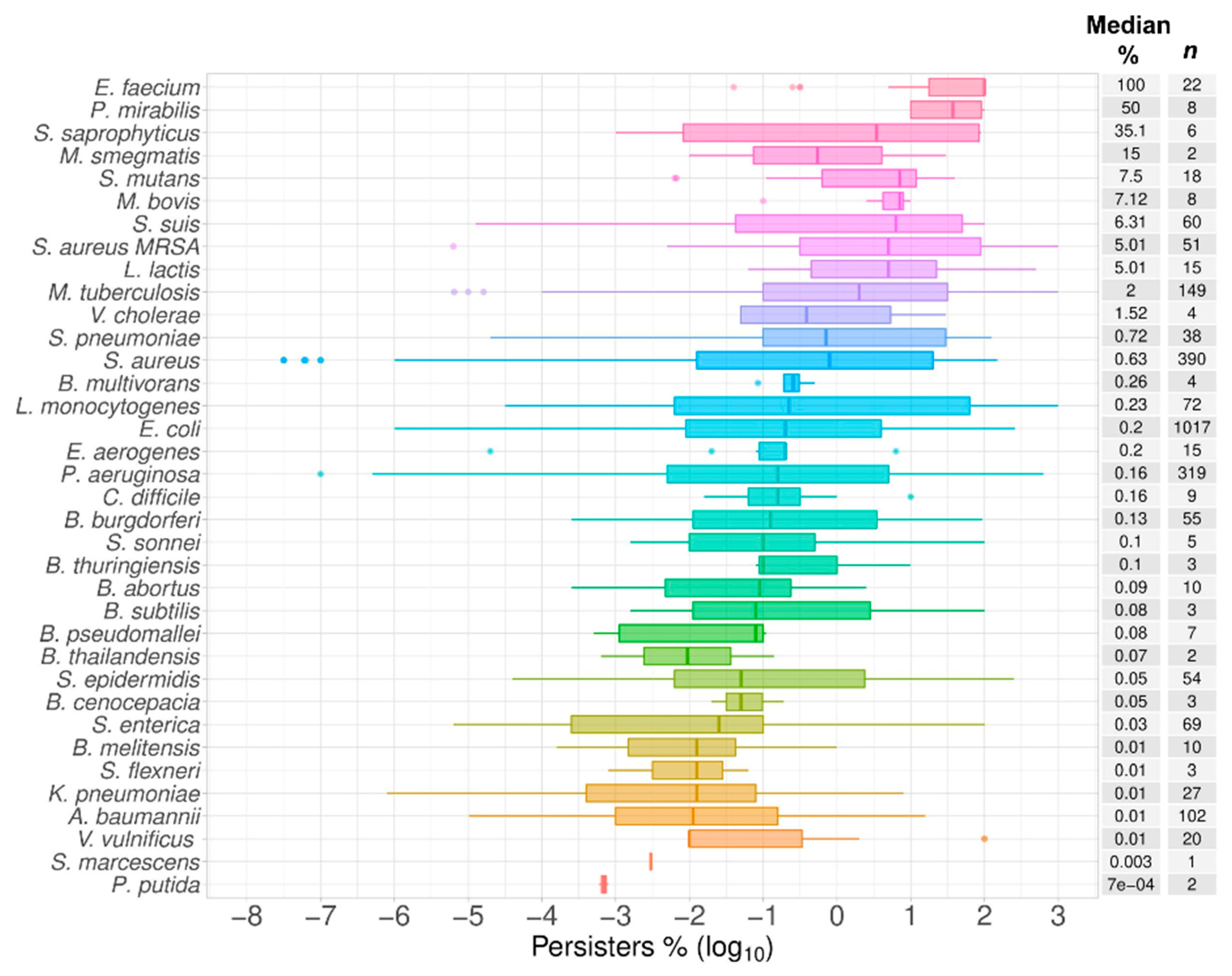
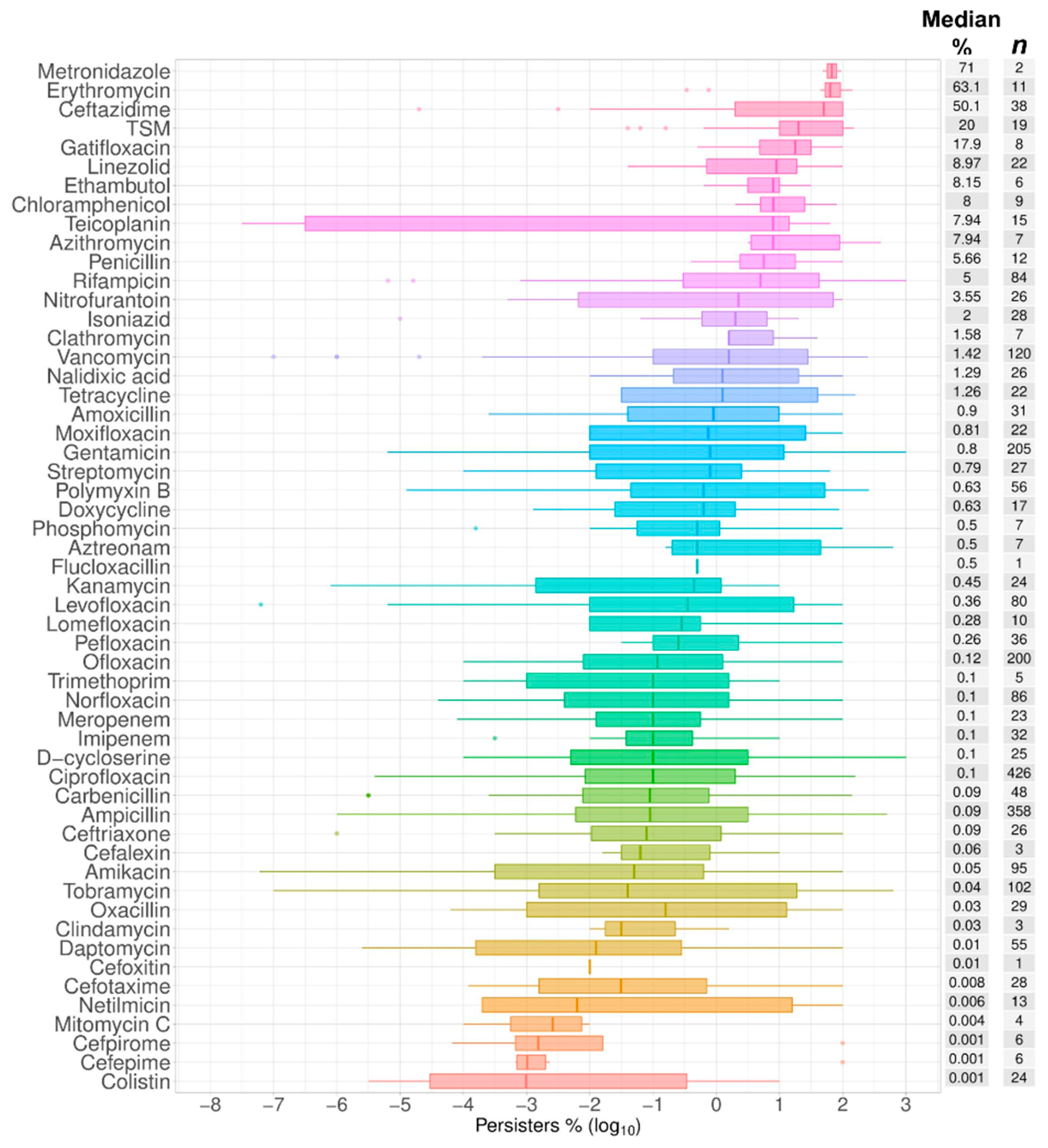
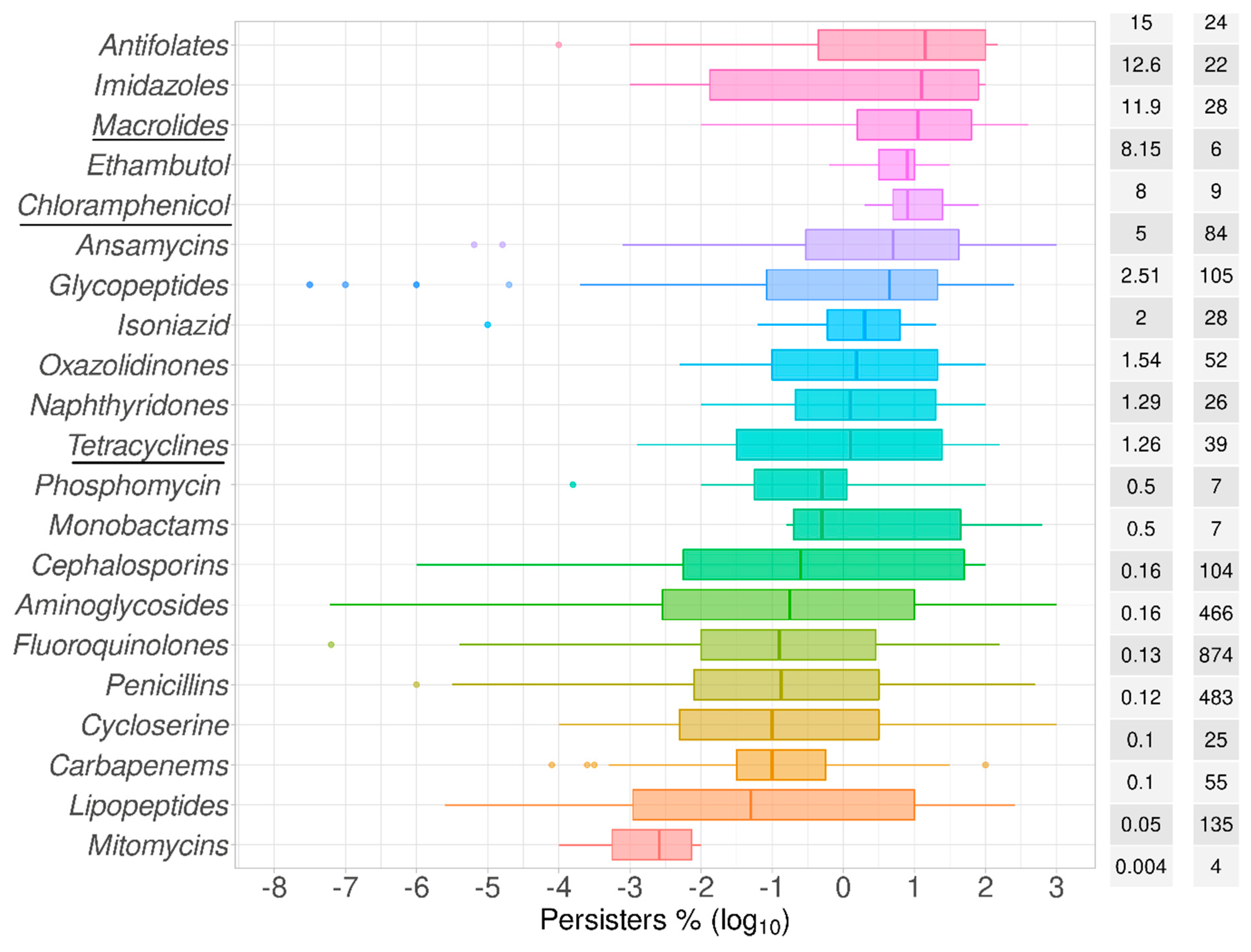
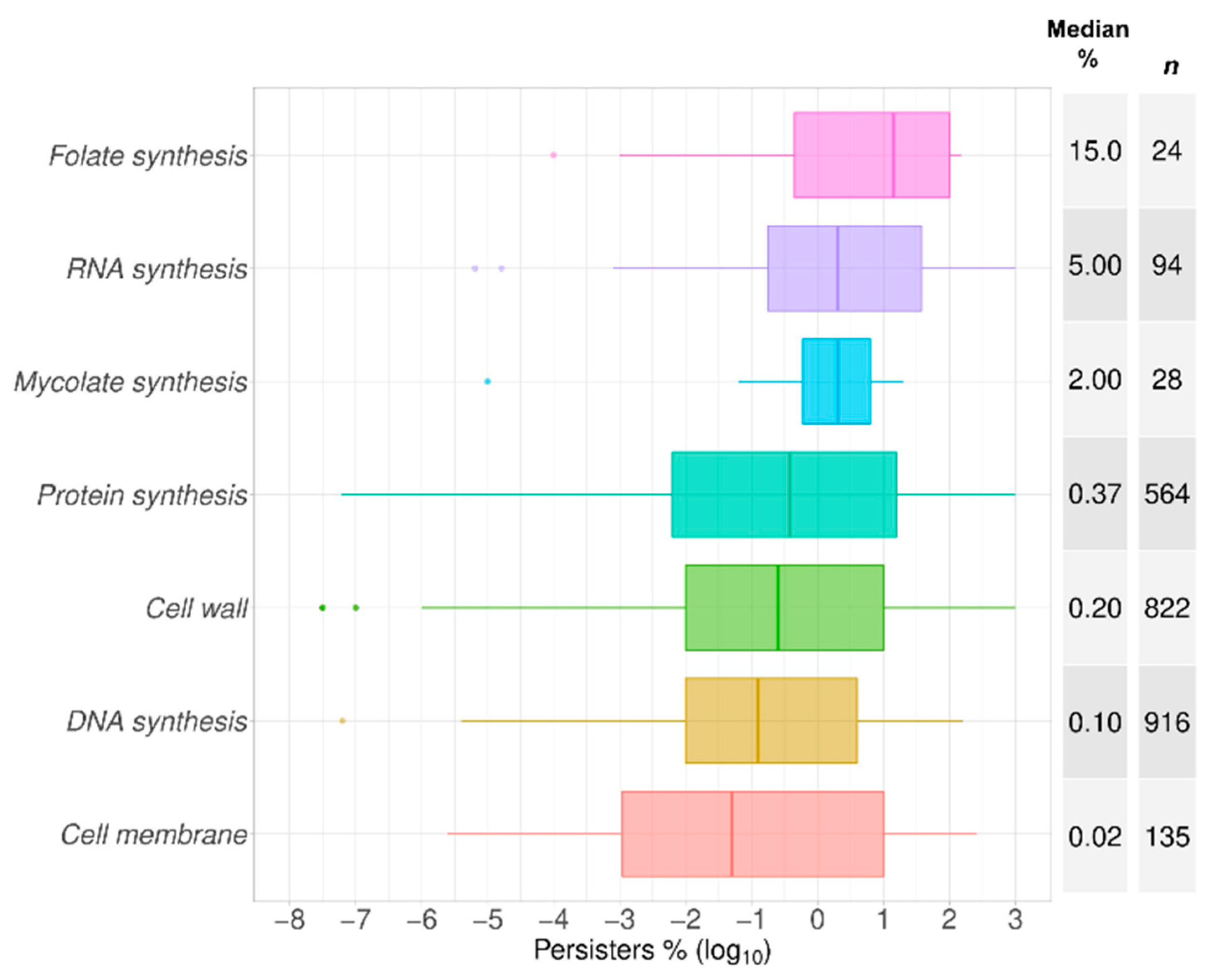
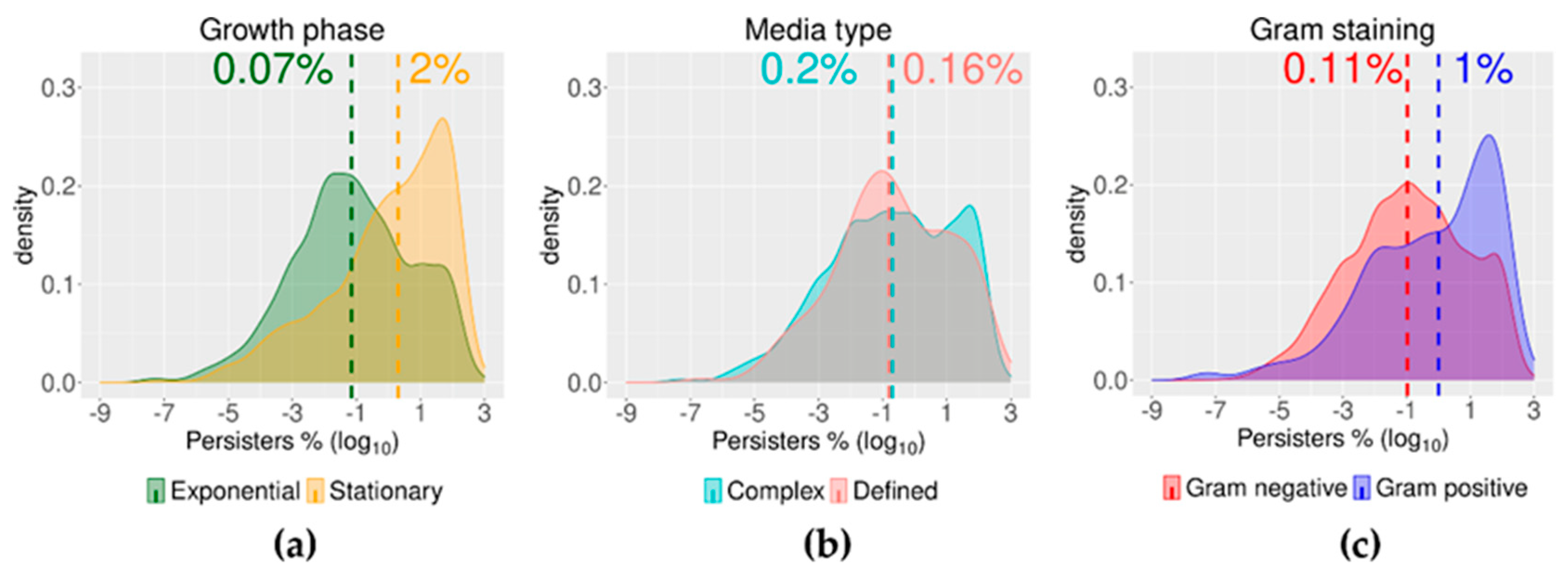
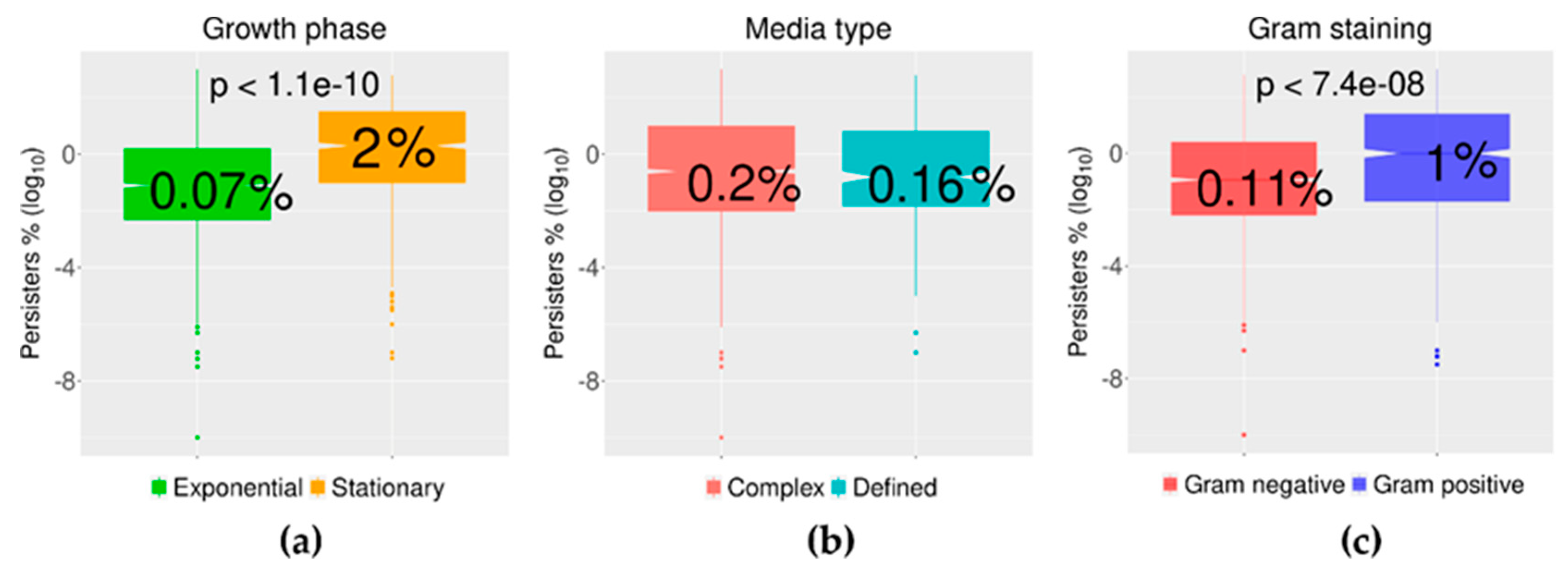
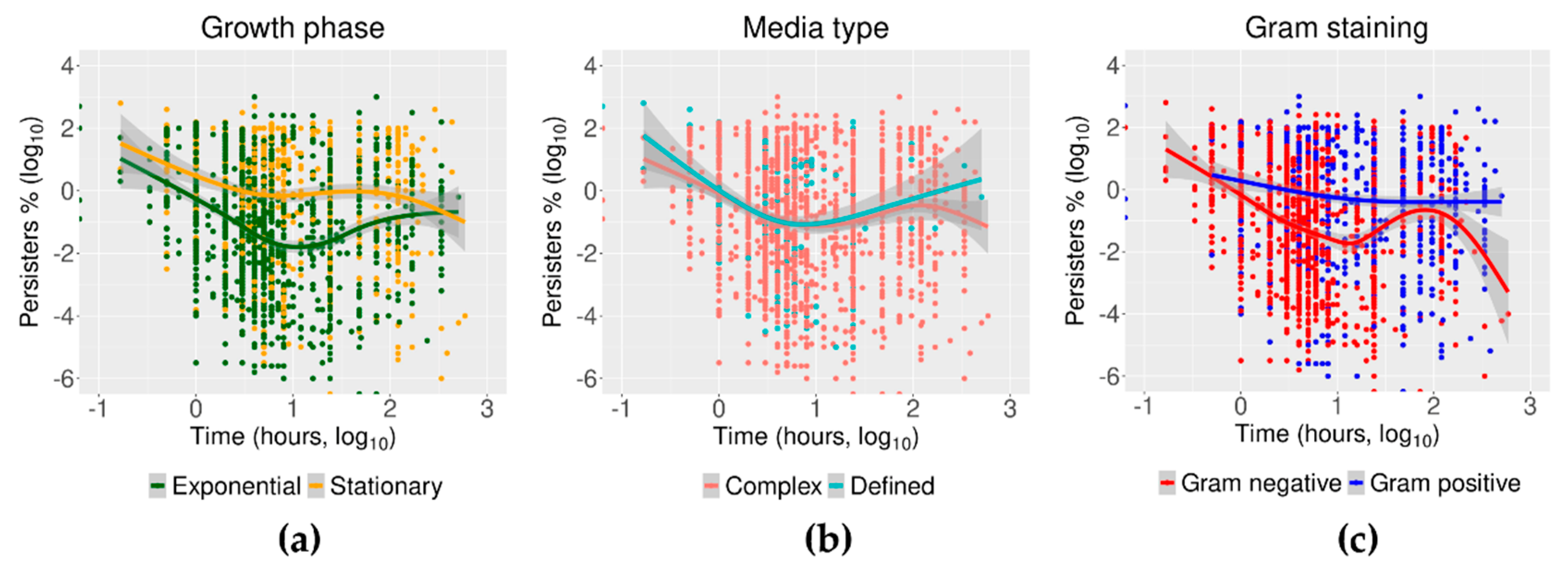
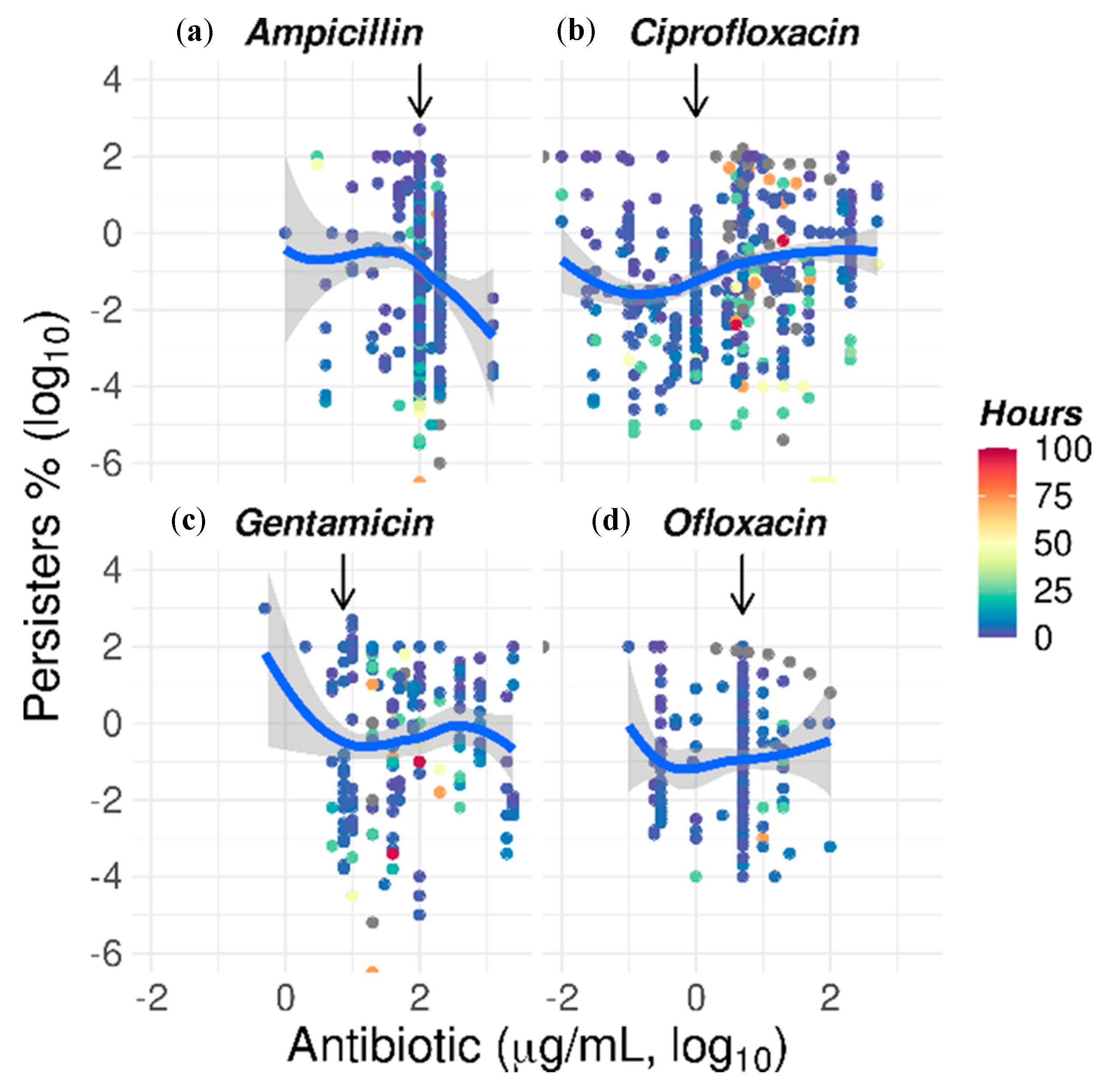
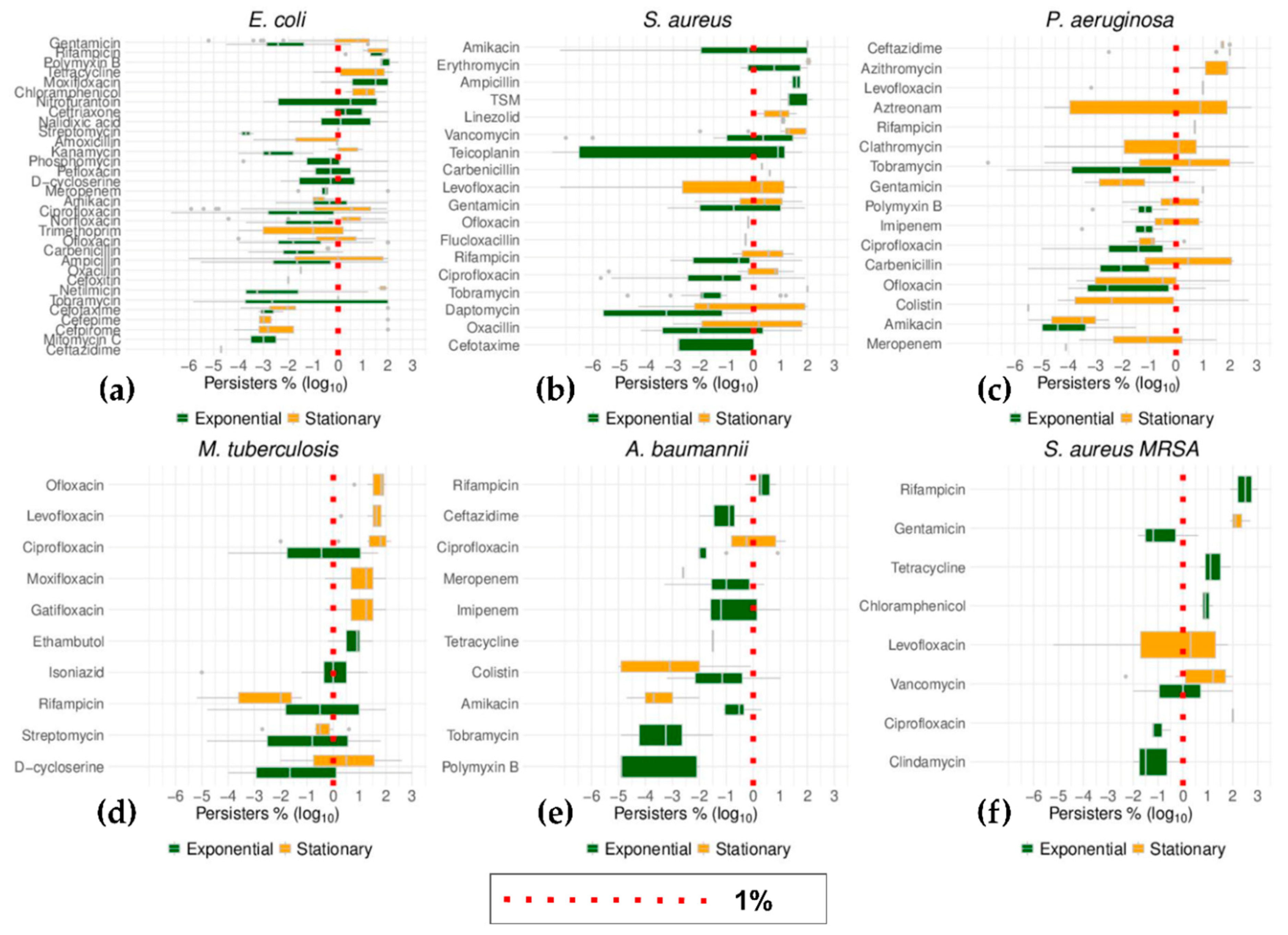
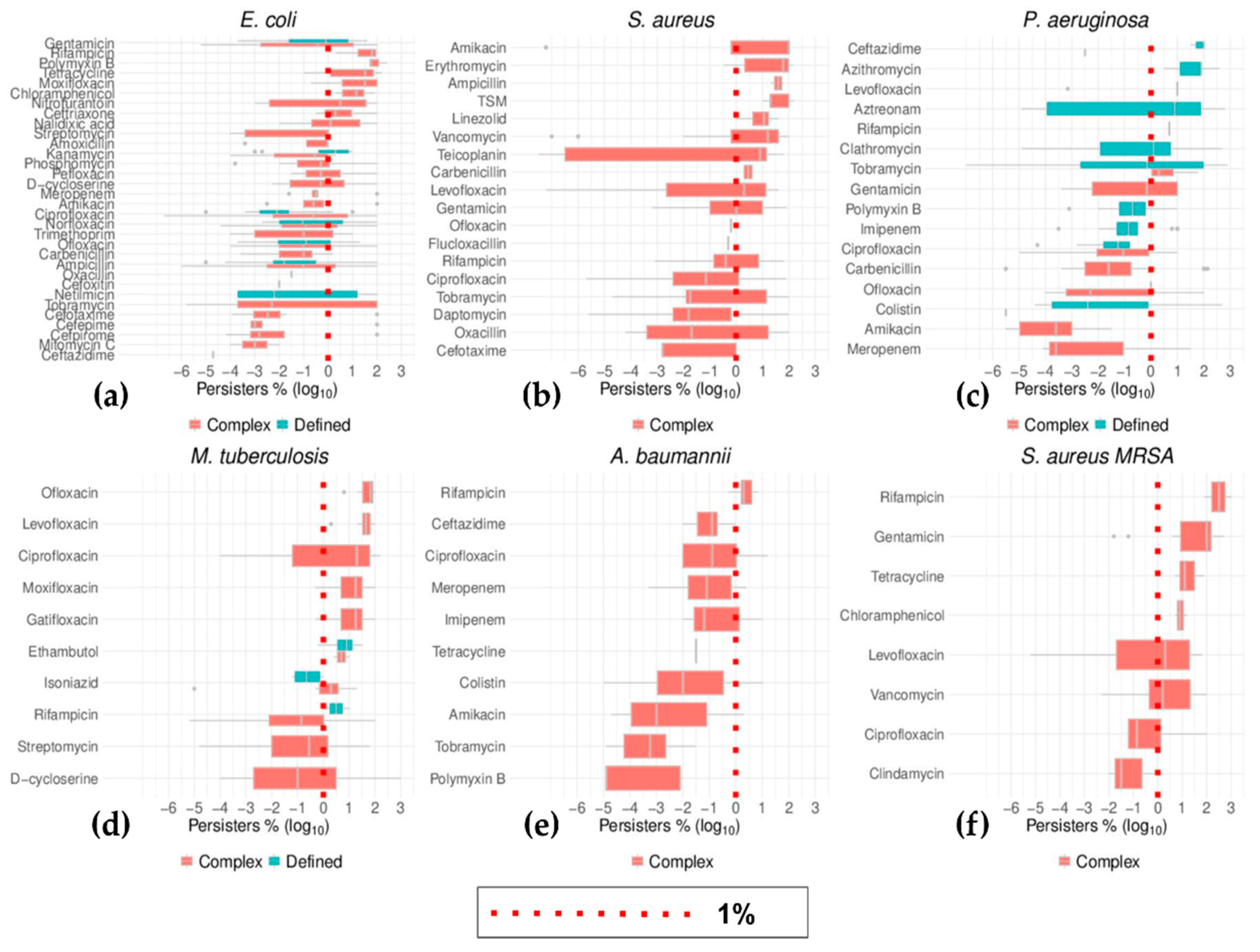
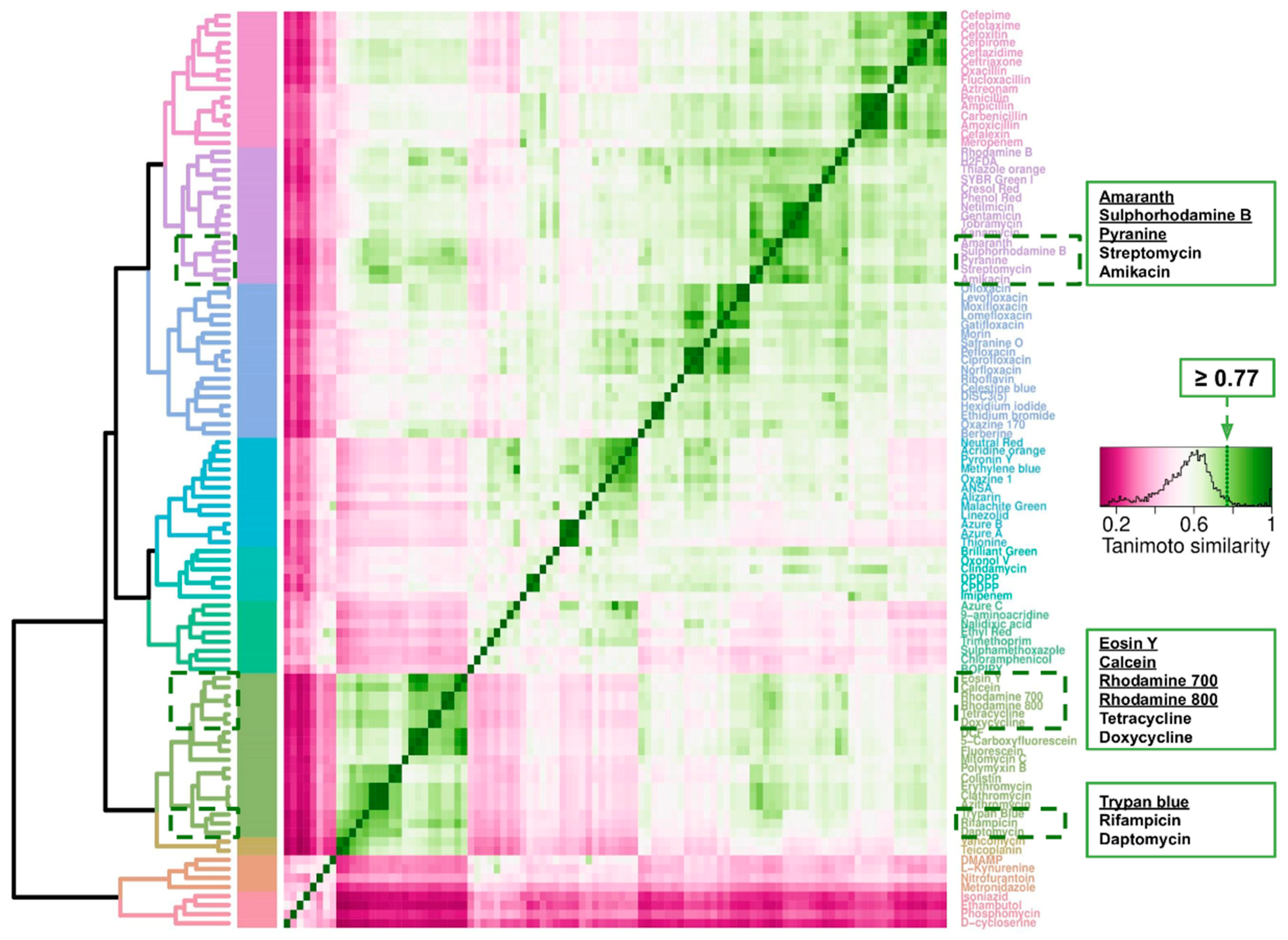
| Gram Staining | |
|---|---|
| Negative | Positive |
| Acinetobacter baumannii | Bacillus subtillis (*) |
| Borrelia burgdorferi | Bacillus thuringiensis (*) |
| Brucella abortus | Clostridium difficile (*) |
| Brucella melitensis | Enterococcus faecium |
| Burkholderia cenocepacia | Lactobacillus lactis |
| Burkholderia multivorans | Listeria monocytogenes |
| Burkholderia pseudomallei | Mycobacterium bovis |
| Burkholderia thailandensis | Mycobacterium smegmatis |
| Enterobacter aerogenes | Mycobacterium tuberculosis |
| Escherichia coli | Staphylococcus aureus |
| Klebsiella pneumoniae | Staphylococcus aureus MRSA |
| Proteus mirabilis | Staphylococcus epidermidis |
| Pseudomonas aeruginosa | Staphylococcus saprophyticus |
| Pseudomonas putida | Streptococcus mutants |
| Salmonella enterica | Streptococcus pneumoniae |
| Serratia marcescens | Streptococcus suis |
| Shigella flexneri | |
| Shigella sonnei | |
| Vibrio cholera | |
| Vibrio vulnificus | |
| Mechanism of Action | Class | Antibiotic |
|---|---|---|
| Cell Wall Synthesis | Carbapenems | Imipenem |
| Meropenem | ||
| Cephalosporins | Cefalexin | |
| Cefepime | ||
| Cefotaxime | ||
| Cefoxitin | ||
| Cefpirome | ||
| Ceftazidime | ||
| Ceftriaxone | ||
| Cycloserine | D-cycloserine | |
| Ethambutol | Ethambutol | |
| Glycopeptides | Teicoplanin | |
| Vancomycin | ||
| Monobactams | Aztreonam | |
| Penicillins | Amoxicillin | |
| Ampicillin | ||
| Carbenicillin | ||
| Flucloxacillin | ||
| Oxacillin | ||
| Penicillin | ||
| Phosphomycins | Phosphomycin | |
| Protein synthesis | Aminoglycosides | Amikacin |
| Gentamicin | ||
| Kanamycin | ||
| Netilmicin | ||
| Streptomycin | ||
| Tobramycin | ||
| Chloramphenicol (*) | Chloramphenicol | |
| Macrolides (*) | Azithromycin | |
| Clathromycin | ||
| Clindamycin | ||
| Erythromycin | ||
| Oxazolidinones | Linezolid | |
| Tetracyclines (*) | Doxycycline | |
| Tetracycline | ||
| DNA synthesis | Fluoroquinolones | Ciprofloxacin |
| Gatifloxacin | ||
| Levofloxacin | ||
| Lomefloxacin | ||
| Moxifloxacin | ||
| Norfloxacin | ||
| Ofloxacin | ||
| Pefloxacin | ||
| Imidizoles | Metronidazole | |
| Nitrofurantoin | ||
| Mitomycins | Mitomycin C | |
| Naphthyridones | Nalidixic acid | |
| Cell membrane | Lipopeptides | Colistin |
| Daptomycin | ||
| Polymyxin B | ||
| Folate synthesis | Antifolates | Trimethoprim |
| TSM | ||
| Mycolic acid synthesis | Isoniazid | Isoniazid |
| RNA synthesis | Ansamycins | Rifampicin |
| Growth Phase | Growth Media Type | Gram Staining | ||||
|---|---|---|---|---|---|---|
| Exponential | Stationary | Complex | Defined | Negative | Positive | |
| Median (%) | 0.07 | 2.00 | 0.20 | 0.16 | 0.11 | 1.00 |
| Mean (%) | 12.6 | 26.8 | 17.2 | 19.9 | 12.7 | 27.1 |
| CI95 (%) | 10.0–15.1 | 23.3–30.3 | 15.1–19.4 | 12.8–27.0 | 10.9–14.5 | 22.2–32.0 |
| n | 1675 | 908 | 2248 | 335 | 1707 | 876 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salcedo-Sora, J.E.; Kell, D.B. A Quantitative Survey of Bacterial Persistence in the Presence of Antibiotics: Towards Antipersister Antimicrobial Discovery. Antibiotics 2020, 9, 508. https://doi.org/10.3390/antibiotics9080508
Salcedo-Sora JE, Kell DB. A Quantitative Survey of Bacterial Persistence in the Presence of Antibiotics: Towards Antipersister Antimicrobial Discovery. Antibiotics. 2020; 9(8):508. https://doi.org/10.3390/antibiotics9080508
Chicago/Turabian StyleSalcedo-Sora, Jesus Enrique, and Douglas B. Kell. 2020. "A Quantitative Survey of Bacterial Persistence in the Presence of Antibiotics: Towards Antipersister Antimicrobial Discovery" Antibiotics 9, no. 8: 508. https://doi.org/10.3390/antibiotics9080508
APA StyleSalcedo-Sora, J. E., & Kell, D. B. (2020). A Quantitative Survey of Bacterial Persistence in the Presence of Antibiotics: Towards Antipersister Antimicrobial Discovery. Antibiotics, 9(8), 508. https://doi.org/10.3390/antibiotics9080508





