Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development
Abstract
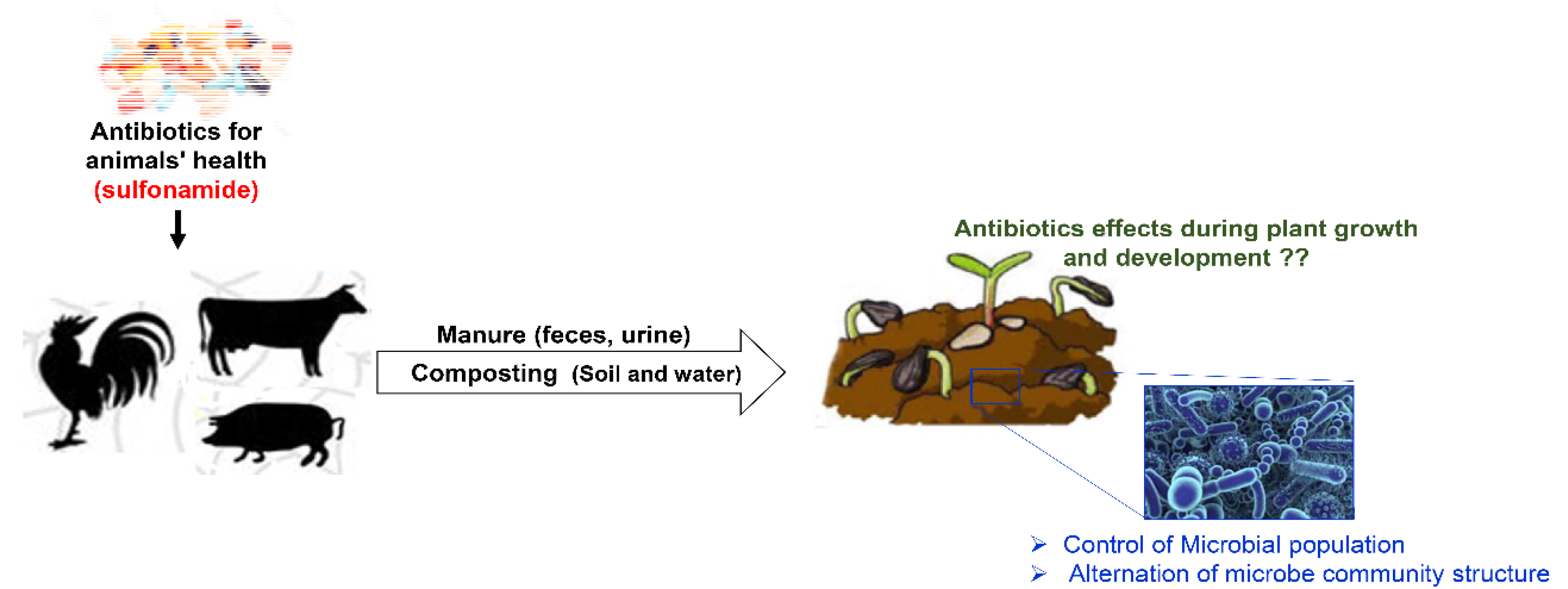
1. Introduction
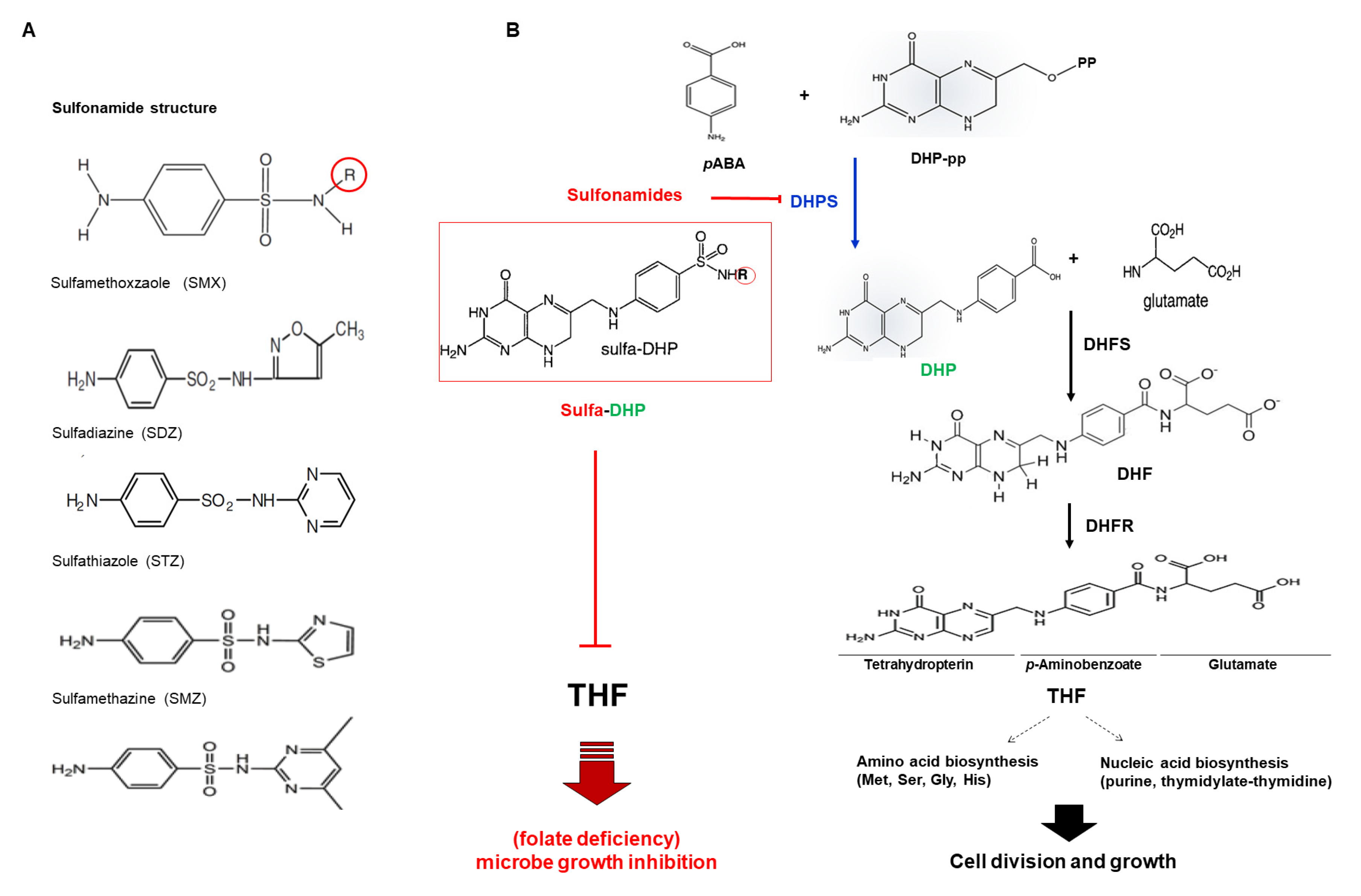
1.1. Para-Aminobenzoic Acid (pABA) as an Endogenous Analog of Sulfonamide
1.2. Folate in Plants
2. Results
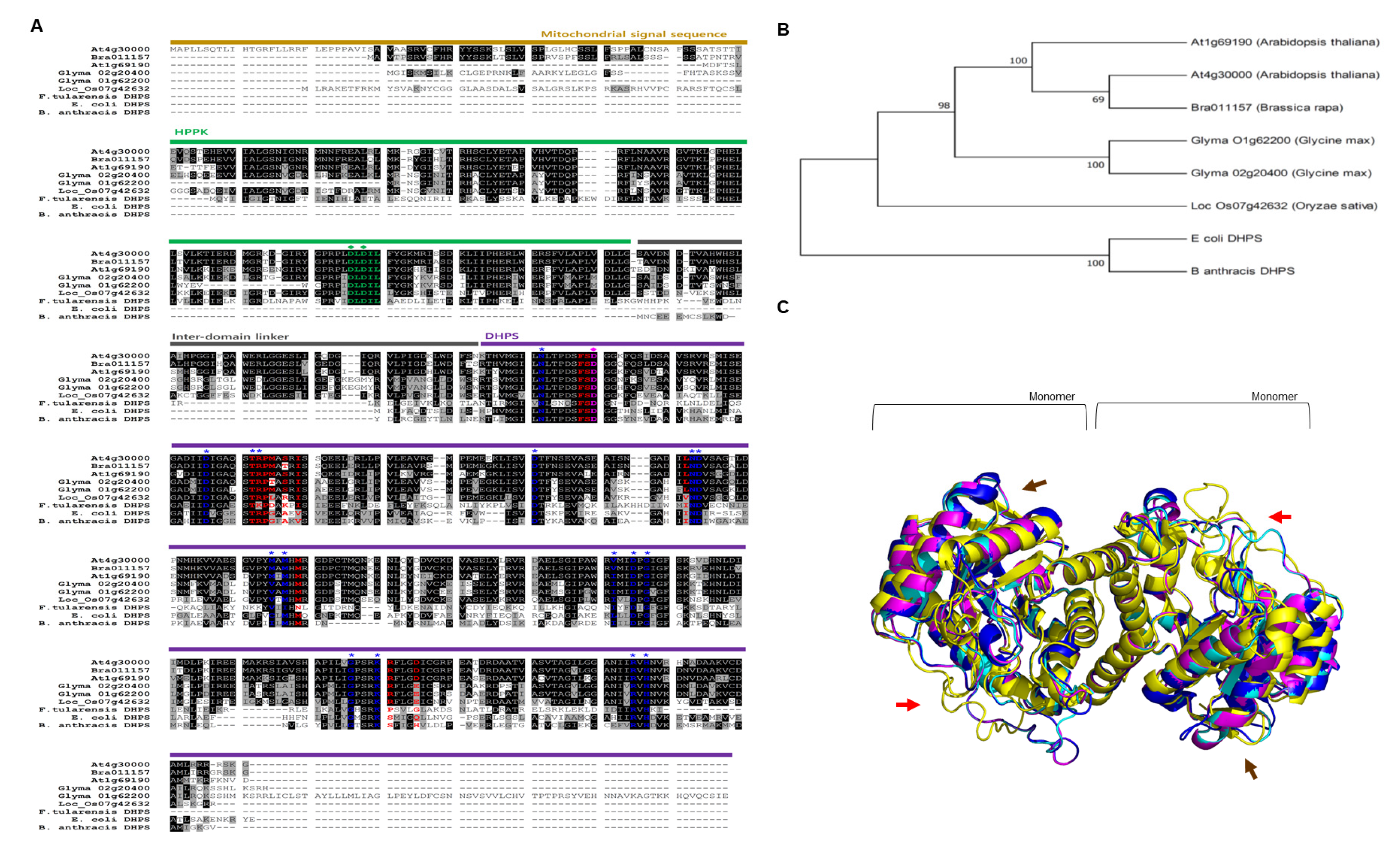
2.1. Plant Dihydropteroate Synthase (DHPS)
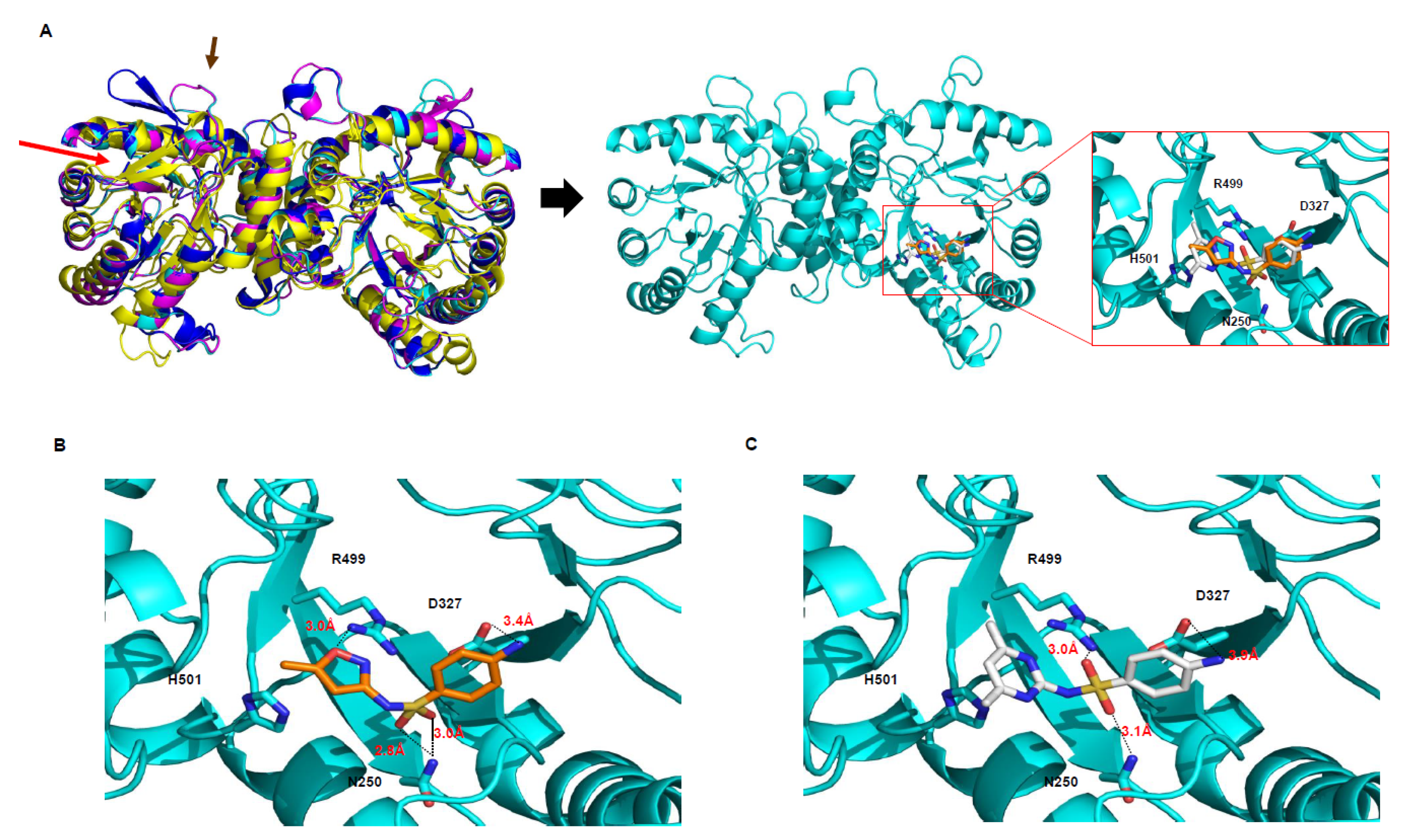
2.2. DHPS as a Sulfonamide Target
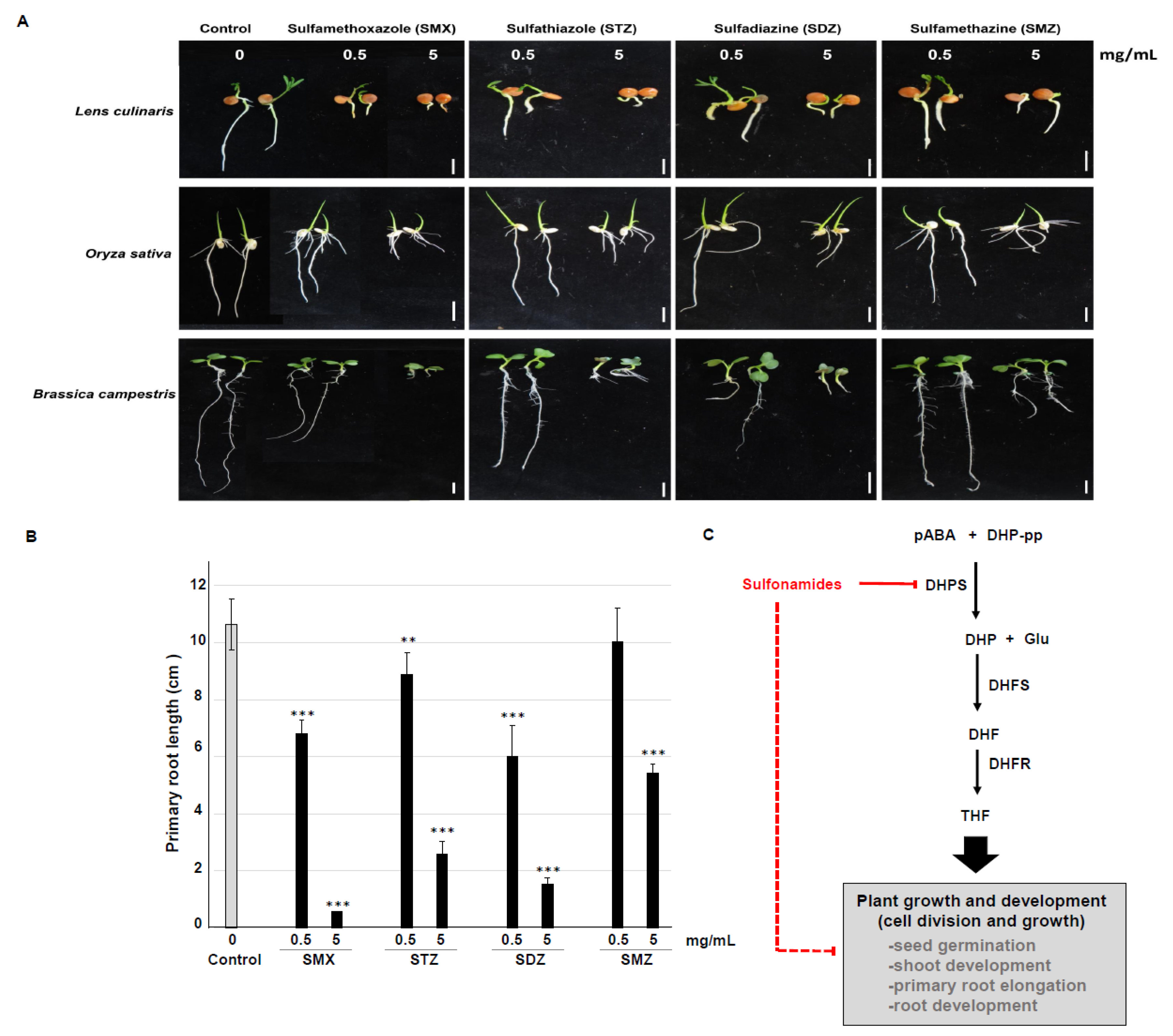
2.3. Phytotoxicity of Sulfonamides During Plant Growth and Development
3. Discussion
3.1. The Comparison Between DHPS Proteins
3.2. Folate and DHPS in Plants
3.3. Sulfonamides and Plant Growth Inhibition
4. Materials and Methods
4.1. Model Building and Refinement with the 3D Structure of DHPS
4.2. Plant Growth Conditions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Tacic, A.; Nikolic, V.; Nikolic, L.; Savic, I. Antimicrobial sulfonamide drugs. Adv. Technol. 2017, 6, 58–71. [Google Scholar] [CrossRef]
- Patel, O.G.; Mberu, E.K.; Nzila, A.M.; Macreadie, I.G. Sulfa drugs strike more than once. Trends Parasitol. 2004, 20, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ezzariai, A.; Hafidi, M.; Khadra, A.; Aemig, Q.; El Fels, L.; Barret, M.; Merlina, G.; Patureau, D.; Pinelli, E. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. J. Hazard. Mater. 2018, 359, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Nkoa, R. Agricultural benefits and environmental risks of soil fertilization with anaerobic digestates: A review. Agron. Sustain. Dev. 2014, 34, 473–492. [Google Scholar] [CrossRef]
- Sukul, P.; Spiteller, M. Sulfonamides in the environment as veterinary drugs. Rev. Environ. Contam. Toxicol. 2006, 187, 67–101. [Google Scholar] [CrossRef] [PubMed]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Thiele-Bruhn, S. Pharmaceutical antibiotic compounds in soils - A review. J. Plant Nutr. Soil Sci. 2003, 166, 145–167. [Google Scholar] [CrossRef]
- Korea Animal Health Products Association. Available online: http://www.kahpa.or.kr/ENG/ (accessed on 1 January 2020).
- Kim, K.R.; Owens, G.; Kwon, S.I.; So, K.H.; Lee, D.B.; Ok, Y.S. Occurrence and environmental fate of veterinary antibiotics in the terrestrial environment. Water. Air. Soil Pollut. 2011, 214, 163–174. [Google Scholar] [CrossRef]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Botelho, R.G.; Monteiro, S.H.; Tornisielo, V.L. Veterinary Antibiotics in the Environment. Emerg. Pollut. Environ. Curr. Furth. Implic. 2015. [Google Scholar] [CrossRef]
- Tong, X.N.; Wang, X.Z.; He, X.J.; Wang, Z.; Li, W.X. Effects of antibiotics on microbial community structure and microbial functions in constructed wetlands treated with artificial root exudates. Environ. Sci. Process. Impacts 2020, 22, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Pessarakli, M.; Haghighi, M.; Sheibanirad, A. Plant Responses under Environmental Stress Conditions. Adv. Plants Agric. Res. 2015, 2, 276–286. [Google Scholar] [CrossRef]
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants. Abiotic Biot. Stress Plants 2019, 1–6. [Google Scholar] [CrossRef]
- Klotz, I.M. The Mode of Action of Sulfonamides. J. Am. Chem. Soc. 1944, 66, 459–464. [Google Scholar] [CrossRef]
- Hanson, A.D.; Gregory, J.F. Folate Biosynthesis, Turnover, and Transport in Plants. Annu. Rev. Plant Biol. 2011, 62, 105–125. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Derrick, J.P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 2002, 24, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Bourne, C.R. Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics 2014, 3, 1. [Google Scholar] [CrossRef]
- Kirillova, L.L.; Nazarova, G.N.; Ivanova, E.P. Para-aminobenzoic acid stimulates seed germination plant growth, development, photosynthesis and nitrogen assimilation in the amaranth (Amaranthus L.). Sel’skokhozyaistvennaya Biol. 2016, 51, 688–695. [Google Scholar] [CrossRef]
- Bhargava, S.; Tyagi, S.C. Nutriepigenetic regulation by folate-homocysteine-methionine axis: A review. Mol. Cell. Biochem. 2014, 387, 55–61. [Google Scholar] [CrossRef]
- Patel, O.; Satchell, J.; Baell, J.; Fernley, R.; Coloe, P.; Macreadie, I. Inhibition studies of sulfonamide-containing folate analogs in yeast. Microb. Drug Resist. 2003, 9, 139–146. [Google Scholar] [CrossRef]
- Ravanel, S.; Douce, R.; Rébeillé, F. Metabolism of folates in plants. Adv. Bot. Res. 2011, 59, 67–106. [Google Scholar] [CrossRef]
- Rébeillé, F.; Ravanel, S.; Jabrin, S.; Douce, R.; Storozhenko, S.; Van Der Straeten, D. Folates in plants: Biosynthesis, distribution, and enhancement. Physiol. Plant. 2006, 126, 330–342. [Google Scholar] [CrossRef]
- Fischer, M.; Thöny, B.; Leimkühler, S. The biosynthesis of folate and pterins and their enzymology. Compr. Nat. Prod. II Chem. Biol. 2010, 7, 599–648. [Google Scholar] [CrossRef]
- Sadaka, C.; Ellsworth, E.; Hansen, P.R.; Ewin, R.; Damborg, P.; Watts, J.L. Review on abyssomicins: Inhibitors of the chorismate pathway and folate biosynthesis. Molecules 2018, 23, 1371. [Google Scholar] [CrossRef] [PubMed]
- Rébeillé, F.; Macherel, D.; Mouillon, J.M.; Garin, J.; Douce, R. Folate biosynthesis in higher plants: Purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 1997, 16, 947–957. [Google Scholar] [CrossRef]
- Mouillon, J.M.; Ravanel, S.; Douce, R.; Rébeillé, F. Folate synthesis in higher-plant mitochondria: Coupling between the dihydropterin pyrophosphokinase and the dihydropteroate synthase activities. Biochem. J. 2002, 363, 313. [Google Scholar] [CrossRef]
- Babaoglu, K.; Qi, J.; Lee, R.E.; White, S.W. Crystal structure of 7,8-dihydropteroate synthase from Bacillus anthracis: Mechanism and novel inhibitor design. Struct. 2004, 12, 1705–1717. [Google Scholar] [CrossRef]
- Achari, A.; Somers, D.O.; Champness, J.N.; Bryant, P.K.; Rosemond, J.; Stammers, D.K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997, 4, 490–497. [Google Scholar] [CrossRef]
- Shaw, G.X.; Li, Y.; Shi, G.; Wu, Y.; Cherry, S.; Needle, D.; Zhang, D.; Tropea, J.E.; Waugh, D.S.; Yan, H.; et al. Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis. FEBS J. 2014, 281, 4123–4137. [Google Scholar] [CrossRef]
- Zessel, K.; Mohring, S.; Hamscher, G.; Kietzmann, M.; Stahl, J. Biocompatibility and antibacterial activity of photolytic products of sulfonamides. Chemosphere 2014, 100, 167–174. [Google Scholar] [CrossRef]
- Brown, G.M. The biosynthesis of folic acid. II. Inhibition by sulfonamides. J. Biol. Chem. 1962, 237. [Google Scholar]
- Gorelova, V.; Bastien, O.; De Clerck, O.; Lespinats, S.; Rébeillé, F.; Van Der Straeten, D. Evolution of folate biosynthesis and metabolism across algae and land plant lineages. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Chu, L.M. Phytotoxicity of veterinary antibiotics to seed germination and root elongation of crops. Ecotoxicol. Environ. Saf. 2016, 126, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ying, G.G.; Tao, R.; Zhao, J.L.; Yang, J.F.; Zhao, L.F. Effects of six selected antibiotics on plant growth and soil microbial and enzymatic activities. Environ. Pollut. 2009, 157, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Brain, R.A.; Johnson, D.J.; Richards, S.M.; Hanson, M.L.; Sanderson, H.; Lam, M.W.; Young, C.; Mabury, S.A.; Sibley, P.K.; Solomon, K.R. Microcosm evaluation of the effects of an eight pharmaceutical mixture to the aquatic macrophytes Lemna gibba and Myriophyllum sibiricum. Aquat. Toxicol. 2004, 70, 23–40. [Google Scholar] [CrossRef]
- Brain, R.A.; Ramirez, A.J.; Fulton, B.A.; Chambliss, C.K.; Brooks, B.W. Herbicidal effects of sulfamethoxazole in Lemna gibba: Using p-aminobenzoic acid as a biomarker of effect. Environ. Sci. Technol. 2008, 42, 8965–8970. [Google Scholar] [CrossRef]
- Grenni, P.; Patrolecco, L.; Rauseo, J.; Spataro, F.; Di Lenola, M.; Aimola, G.; Zacchini, M.; Pietrini, F.; Di Baccio, D.; Stanton, I.C.; et al. Sulfamethoxazole persistence in a river water ecosystem and its effects on the natural microbial community and Lemna minor plant. Microchem. J. 2019, 149, 103999. [Google Scholar] [CrossRef]
- Hillis, D.G.; Fletcher, J.; Solomon, K.R.; Sibley, P.K. Effects of ten antibiotics on seed germination and root elongation in three plant species. Arch. Environ. Contam. Toxicol. 2011, 60, 220–232. [Google Scholar] [CrossRef]
- Teixeira, J.; Ferraz, P.; Gouveia, C.; Azevedo, F.; Neves, S.; Fidalgo, F.; Silva, A.M.T. Targeting key metabolic points for an enhanced phytoremediation of wastewaters pre-treated by the photo-Fenton process using Solanum nigrum L. Ecotoxicol. Environ. Saf. 2015, 120, 124–129. [Google Scholar] [CrossRef]
- Dudley, S.; Sun, C.; Jiang, J.; Gan, J. Metabolism of sulfamethoxazole in Arabidopsis thaliana cells and cucumber seedlings. Environ. Pollut. 2018, 242, 1748–1757. [Google Scholar] [CrossRef]
- Chen, H.R.; Rairat, T.; Loh, S.H.; Wu, Y.C.; Vickroy, T.W.; Chou, C.C. Assessment of veterinary drugs in plants using pharmacokinetic approaches: The absorption, distribution and elimination of tetracycline and sulfamethoxazole in ephemeral vegetables. PLoS ONE 2017, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Caban, J.R.; Kuppusamy, S.; Kim, J.H.; Yoon, Y.E.; Kim, S.Y.; Lee, Y.B. Hairy Vetch Incorporated as Green Manure Inhibits Sulfathiazole Uptake by Lettuce in Soil. Water. Air. Soil Pollut. 2018, 229, 104. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, W.; Ma, Q.; Zhou, H.; Jiang, C. Toxicity of sulfadiazine and copper and their interaction to wheat (Triticum aestivum L.) seedlings. Ecotoxicol. Environ. Saf. 2017, 142, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Chen, Q.; Sun, R.; Zhou, Q.; Liu, J. Eco-toxic effects of sulfadiazine sodium, sulfamonomethoxine sodium and enrofloxacin on wheat, Chinese cabbage and tomato. Ecotoxicology 2009, 18, 878–885. [Google Scholar] [CrossRef]
- Pufal, G.; Memmert, J.; Leonhardt, S.D.; Minden, V. Negative bottom-up effects of sulfadiazine, but not penicillin and tetracycline, in soil substitute on plants and higher trophic levels. Environ. Pollut. 2019, 245, 531–544. [Google Scholar] [CrossRef]
- Minden, V.; Deloy, A.; Volkert, A.M.; Leonhardt, S.D.; Pufal, G. Antibiotics impact plant traits, even at small concentrations. AoB Plants 2017, 9. [Google Scholar] [CrossRef]
- Michelini, L.; Meggio, F.; Reichel, R.; Thiele-Bruhn, S.; Pitacco, A.; Scattolin, L.; Montecchio, L.; Alberghini, S.; Squartini, A.; Ghisi, R. Sulfadiazine uptake and effects in common hazel (Corylus avellana L.). Environ. Sci. Pollut. Res. 2015, 22, 13362–13371. [Google Scholar] [CrossRef]
- Sharma, N.; Arrigoni, G.; Ebinezer, L.B.; Trentin, A.R.; Franchin, C.; Giaretta, S.; Carletti, P.; Thiele-Bruhn, S.; Ghisi, R.; Masi, A. A proteomic and biochemical investigation on the effects of sulfadiazine in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2019, 178, 146–158. [Google Scholar] [CrossRef]
- Michelini, L.; Reichel, R.; Werner, W.; Ghisi, R.; Thiele-Bruhn, S. Sulfadiazine uptake and effects on salix fragilis l. and zea mays l. plants. Water. Air. Soil Pollut. 2012, 223, 5243–5257. [Google Scholar] [CrossRef]
- Susarla, S.; Medina, V.F.; McCutcheon, S.C. Phytoremediation: An ecological solution to organic chemical contamination. Ecol. Eng. 2002, 18, 647–658. [Google Scholar] [CrossRef]
- Piotrowicz-Cieślak, A.I.; Adomas, B.; Nałaȩcz-Jawecki, G.; Michalczyk, D.J. Phytotoxicity of sulfamethazine soil pollutant to six legume plant species. J. Toxicol. Environ. Heal. Part A Curr. Issues 2010, 73, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, Y.H.; Liu, C.X.; Wang, Z.; Dong, J.; Zhu, G.F.; Huang, X. Potential effect and accumulation of veterinary antibiotics in Phragmites australis under hydroponic conditions. Ecol. Eng. 2013, 53, 138–143. [Google Scholar] [CrossRef]
- Michelini, L.; La Rocca, N.; Rascio, N.; Ghisi, R. Structural and functional alterations induced by two sulfonamide antibiotics on barley plants. Plant Physiol. Biochem. 2013, 67, 55–62. [Google Scholar] [CrossRef]
- Migliore, L.; Rotini, A.; Cerioli, N.L.; Cozzolino, S.; Fiori, M. Phytotoxic antibiotic sulfadimethoxine elicits a complex hormetic response in the weed Lythrum salicaria L. Dose-Response 2010, 8, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Migliore, L.; Civitareale, C.; Brambilla, G.; Cozzolino, S.; Casoria, P.; Gaudio, L. Effects of sulphadimethoxine on cosmopolitan weeds (Amaranthus retroflexus L., Plantago major L. and Rumex acetosella L.). Agric. Ecosyst. Environ. 1997, 65, 163–168. [Google Scholar] [CrossRef]
- Ahmed, M.B.M.; Rajapaksha, A.U.; Lim, J.E.; Vu, N.T.; Kim, I.S.; Kang, H.M.; Lee, S.S.; Ok, Y.S. Distribution and accumulative pattern of tetracyclines and sulfonamides in edible vegetables of cucumber, tomato, and lettuce. J. Agric. Food Chem. 2015, 63, 398–405. [Google Scholar] [CrossRef]
- Migliore, L.; Brambilla, G.; Cozzolino, S.; Gaudio, L. Effect on plants of sulphadimethoxine used in intensive farming (Panicum miliaceum, Pisum sativum and Zea mays). Agric. Ecosyst. Environ. 1995. [Google Scholar] [CrossRef]
- Michelini, L.; Gallina, G.; Capolongo, F.; Ghisi, R. Accumulation and Response of Willow Plants Exposed to Environmental Relevant Sulfonamide Concentrations. Int. J. Phytoremediation 2014, 16, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.; Reinhold, D. Metabolism of Sulfamethoxazole by the Model Plant Arabidopsis thaliana. Environ. Sci. Technol. 2019, 53, 4901–4911. [Google Scholar] [CrossRef]
- Pemble, C.W.; Mehta, P.K.; Mehra, S.; Li, Z.; Nourse, A.; Lee, R.E.; White, S.W. Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase•dihydropteroate synthase bifunctional enzyme from Francisella tularensis. PLoS One 2010, 5, e14165. [Google Scholar] [CrossRef]
- Hevener, K.E.; Yun, M.K.; Qi, J.; Kerr, I.D.; Babaoglu, K.; Hurdle, J.G.; Balakrishna, K.; White, S.W.; Lee, R.E. Structural studies of pterin-based inhibitors of dihydropteroate synthase. J. Med. Chem. 2010, 53, 166–177. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef] [PubMed]
- Albani, D.; Giorgetti, L.; Pitto, L.; Luo, M.; Cantoni, R.M.; Erra Pujada, M.; Rotino, G.L.; Cella, R. Proliferation-dependent pattern of expression of a dihydrofolate reductase-thymidylate synthase gene from Daucus carota. Eur. J. Histochem. 2005. [Google Scholar] [CrossRef]
- Jabrin, S.; Ravanel, S.; Gambonnet, B.; Douce, R.; Rébeillé, F. One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol. 2003, 131, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Vaniushin, B.F. DNA methylation and epigenetics. Genetika 2006, 42, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lv, S.; Meng, Y. Epigenetic performers in plants‡. Dev. Growth Differ. 2010, 52, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, X.; Miki, D.; Cutler, S.; La, H.; Hou, Y.J.; Oh, J.E.; Zhu, J.K. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef]
- Meng, H.; Jiang, L.; Xu, B.; Guo, W.; Li, J.; Zhu, X.; Qi, X.; Duan, L.; Meng, X.; Fan, Y.; et al. Arabidopsis plastidial folylpolyglutamate synthetase is required for seed reserve accumulation and seedling establishment in darkness. PLoS One 2014, 9, e101905. [Google Scholar] [CrossRef]
- Reyes-Hernández, B.J.; Srivastava, A.C.; Ugartechea-Chirino, Y.; Shishkova, S.; Ramos-Parra, P.A.; Lira-Ruan, V.; Díaz de la Garza, R.I.; Dong, G.; Moon, J.C.; Blancaflor, E.B.; et al. The root indeterminacy-to-determinacy developmental switch is operated through a folate-dependent pathway in Arabidopsis thaliana. New Phytol. 2014, 202, 1223–1236. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, Y.; Sun, H.; Han, Y.; Li, J.; Li, C.; Guo, W.; Meng, H.; Li, S.; Fan, Y.; et al. The mitochondrial folylpolyglutamate synthetase gene is required for nitrogen utilization during early seedling development in arabidopsis. Plant Physiol. 2013, 161, 971–989. [Google Scholar] [CrossRef]
- Bernal, M.P.; Alburquerque, J.A.; Moral, R. Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour. Technol. 2009, 100, 5444–5453. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Scozzafava, A. Carbonic anhydrase inhibitors: Aromatic sulfonamides and disulfonamides act as efficient tumor growth inhibitors. J. Enzyme Inhib. 2000, 15, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashida, M.; Hussain, S.; Hamayoun, M.; Altaf, A.; Iqbal, J. Sulfa drugs as inhibitors of carbonic anhydrase: New targets for the old drugs. Biomed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Boxall, A.B.A.; Johnson, P.; Smith, E.J.; Sinclair, C.J.; Stutt, E.; Levy, L.S. Uptake of veterinary medicines from soils into plants. J. Agric. Food Chem. 2006, 54, 2288–2297. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998. [Google Scholar] [CrossRef]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef]
- Schuttelkopf, A.W.; van Aalten, D.M. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef]
- Cheong, M.S.; Yoon, Y.-E.; Kim, J.W.; Hong, Y.K.; Kim, S.C.; Lee, Y.B. Chlortetracycline inhibits seed germination and seedling growth in Brassica campestris by disrupting H2O2 signaling. Appl. Boil. Chem. 2020, 63, 1–8. [Google Scholar] [CrossRef]

| Sulfonamide | Plant Species | Physiological Phenotype of Plants | Reference |
|---|---|---|---|
| Sulfamethoxazole (SMX) | Cichaorium endivia, Cucumnus sativus | seed germination | [34] |
| Oryza sativa | seed germination and plant growth | [35] | |
| Myriophyllum sibiricum, Lemma gibba | plant growth and development | [36,37,38] | |
| Daucus carota, Lactuca sativa | root and shoot development, seed germination, and plant growth | [34,39,40] | |
| Cucumis sativus, Arabidopsis thaliana, Ipomoea aquatica, Brassica rapa | seed germination and growth inhibition | [35,41,42] | |
| Medicago sativa | root and shoot development | [39] | |
| Lemna minor, Lemma gibba | reduced plant growth | [36,37,38] | |
| Lens culinaris, Oryza sativa, Brassica campestris | seedling growth inhibition, primary root growth inhibition, and lateral root exposing | in this study | |
| Sulfathiazole (STZ) | Lactuca sativa | plant growth | [43] |
| Lens culinaris, Oryza sativa, Brassica campestris | seedling growth inhibition, primary root growth inhibition, and lateral root exposing | in this study | |
| Sulfadiazine (SDZ) | Triticum aestivum Cyphomandra betacea | root and shoot elongation | [44,45] |
| Triticum aestivum, Apera spica-venti, Brassica napus | plant growth and chlorophyll content | [46,47] | |
| Salix fragilis, Zea mays, Corylus avellana, Arabidopsis thaliana | plant growth and root alternation | [44,48,49,50] | |
| Lens culinaris, Oryza sativa, Brassica campestris | seedling growth inhibition, primary root growth inhibition, and lateral root exposing | in this study | |
| Sulfamethazine (SMZ) | Cichaorium endivia, Oryza sativa | seed germination | [35] |
| Cucumnus sativus | seed germination and plant growth | [34,35] | |
| Phragmites autralis, Daucus carota, Lactuca sativa, Medicago sativa | root growth and photosynthesis activity (hormetic response) | [39,40,51] | |
| Sulfamethazine (SMZ) | Medicago sativa | root and shoot development | [39] |
| Lupinus luteus, Pisum sativum, Lens culinaris, Glycine max, Vigna angularis, Medicago sativa | root decay and necrosis | [47,52] | |
| Phragmites australis | root development and leaf chlorophyll content | [53] | |
| Hordeum vulgare | root development | [54] | |
| Lemma minor | plant growth | [38] | |
| Lens culinaris, Oryza sativa, Brassica campestris | seedling growth inhibition, primary root growth inhibition, and lateral root exposing | in this study | |
| Sulfadimethoxine | Lythrum salicaria | root growth and shoot development (hormetic response) | [55] |
| Amaranthus retroflexus, Plantago major, Remex acetosella | root growth and shoot development | [56] | |
| Cucumis sativus, Solanum ktcioersicum | seedlings growth and development | [57] | |
| Pamicum milliaceum, Pisum sativum, Zea mays | root and stem growth inhibition, leave development, and biomass reduction | [47,58] | |
| Hordeum vulgare | root hair and root growth, root development, and photosynthetic pigment | [47,59] | |
| Salix fragilis | root morphology | [41,60] | |
| Lactuca sativa, Medicago sativa | root growth | [39] | |
| Sulfamethoxine | Amaranthus retroflexus | plant growth and development, post-germination | [40] |
| Cucumis sativus | seed germination and growth inhibition | [45] | |
| Panicum miliaceum, Brassica rapa, Ipomoea aquatica | plant growth and development | [42,58] | |
| Panicum miliaceum, Plantago major, Zea mays, Hordeum disthicum, Rumex acetosella, Pisum sativum | plant growth and development | [40,47,55] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheong, M.S.; Seo, K.H.; Chohra, H.; Yoon, Y.E.; Choe, H.; Kantharaj, V.; Lee, Y.B. Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development. Antibiotics 2020, 9, 456. https://doi.org/10.3390/antibiotics9080456
Cheong MS, Seo KH, Chohra H, Yoon YE, Choe H, Kantharaj V, Lee YB. Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development. Antibiotics. 2020; 9(8):456. https://doi.org/10.3390/antibiotics9080456
Chicago/Turabian StyleCheong, Mi Sun, Kyung Hye Seo, Hadjer Chohra, Young Eun Yoon, Hyeonji Choe, Vimalraj Kantharaj, and Yong Bok Lee. 2020. "Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development" Antibiotics 9, no. 8: 456. https://doi.org/10.3390/antibiotics9080456
APA StyleCheong, M. S., Seo, K. H., Chohra, H., Yoon, Y. E., Choe, H., Kantharaj, V., & Lee, Y. B. (2020). Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development. Antibiotics, 9(8), 456. https://doi.org/10.3390/antibiotics9080456





