Microbial Association with Genus Actinomyces in Primary and Secondary Endodontic Lesions, Review
Abstract
1. Introduction
2. Materials and Methods
- Population—patients with teeth with primary and secondary endodontic infections;
- Intervention—microbial associations with the genus Actinomyces;
- Control—patients with teeth that have no Actinomyces infections;
- Outcome—odds ratio of microbial genera that are found in association with the genus Actinomyces in primary and secondary endodontic infections.
2.1. Eligibility Criteria
- Studies were included if they identified both bacteria of the genus Actinomyces and bacteria of other genera in dental elements subjected to endodontic treatment or retreatment, or in the teeth subjected to apicectomy or extraction following endodontic failure;
- Studies were excluded if they did not report the prevalence data for bacteria of the genus Actinomyces in the primary and secondary lesions of the dental elements, did not consider the microbial composition of each analyzed sample, tested the presence of only a few species of bacteria, were not written in English or were published before 1980.
2.2. Research Methodology
2.3. Screening Methodology
- (1)
- Primary outcome—which genera of bacteria are found in association with the genus Actinomyces in primary and secondary endodontic infections? What is the odds ratio of microbial genera that are found in association with the genus Actinomyces in primary and secondary endodontic infections?
- (2)
- Secondary outcome—the determination of the prevalence of the species of the genus Actinomyces that has the greatest prevalence in endodontic lesions.
2.4. Statistical Analysis Protocol
3. Results
3.1. Study Characteristics and Data Extraction
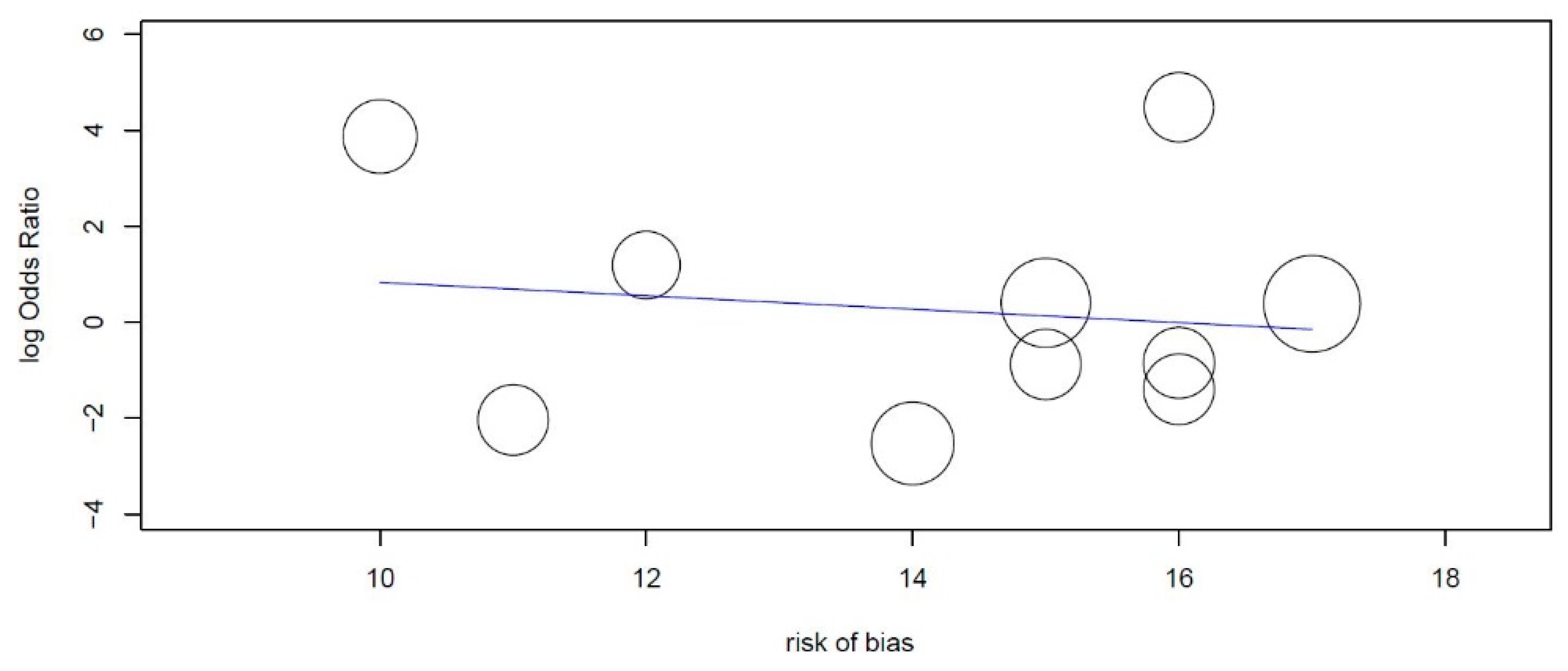
3.2. Risk of Bias
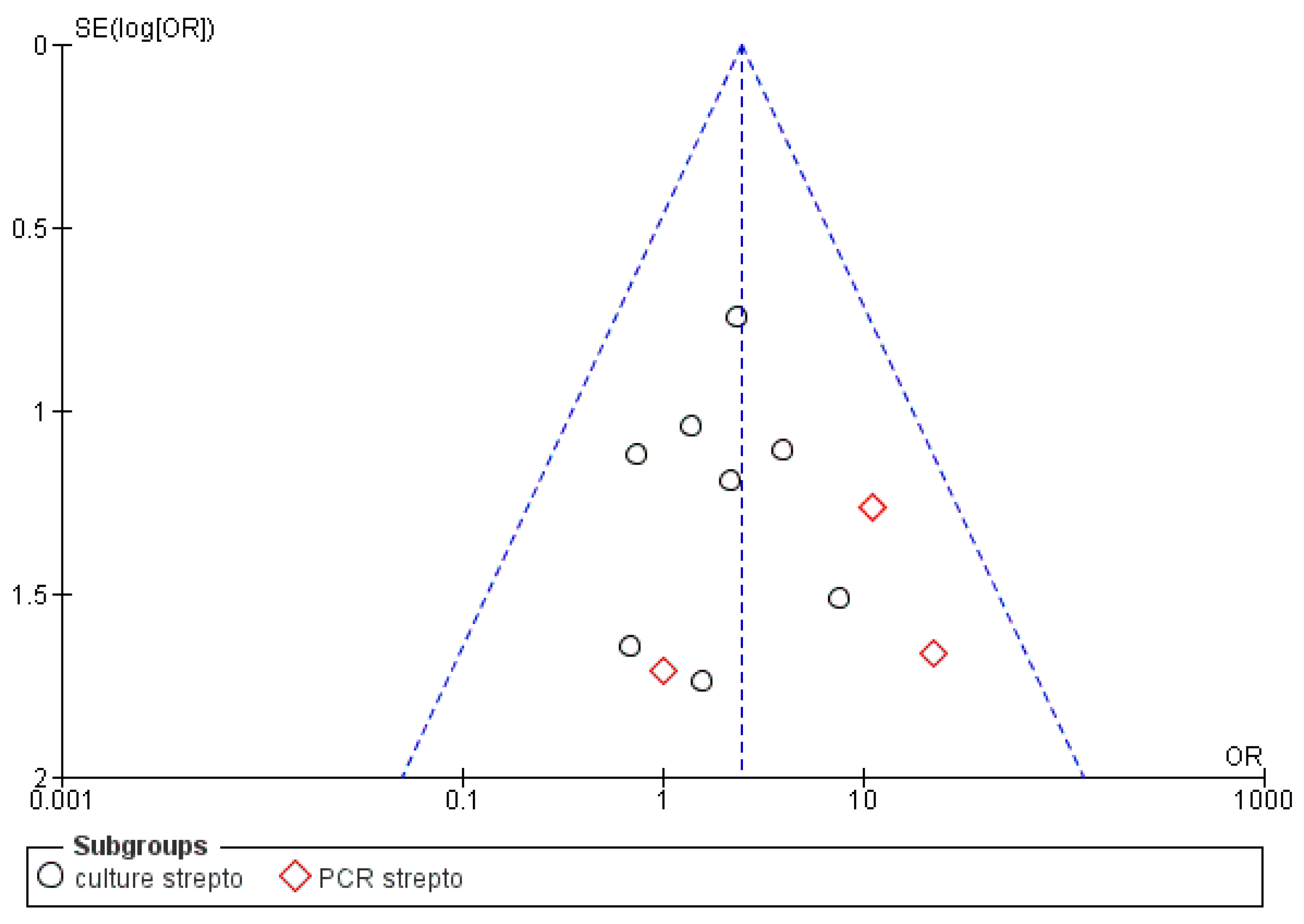
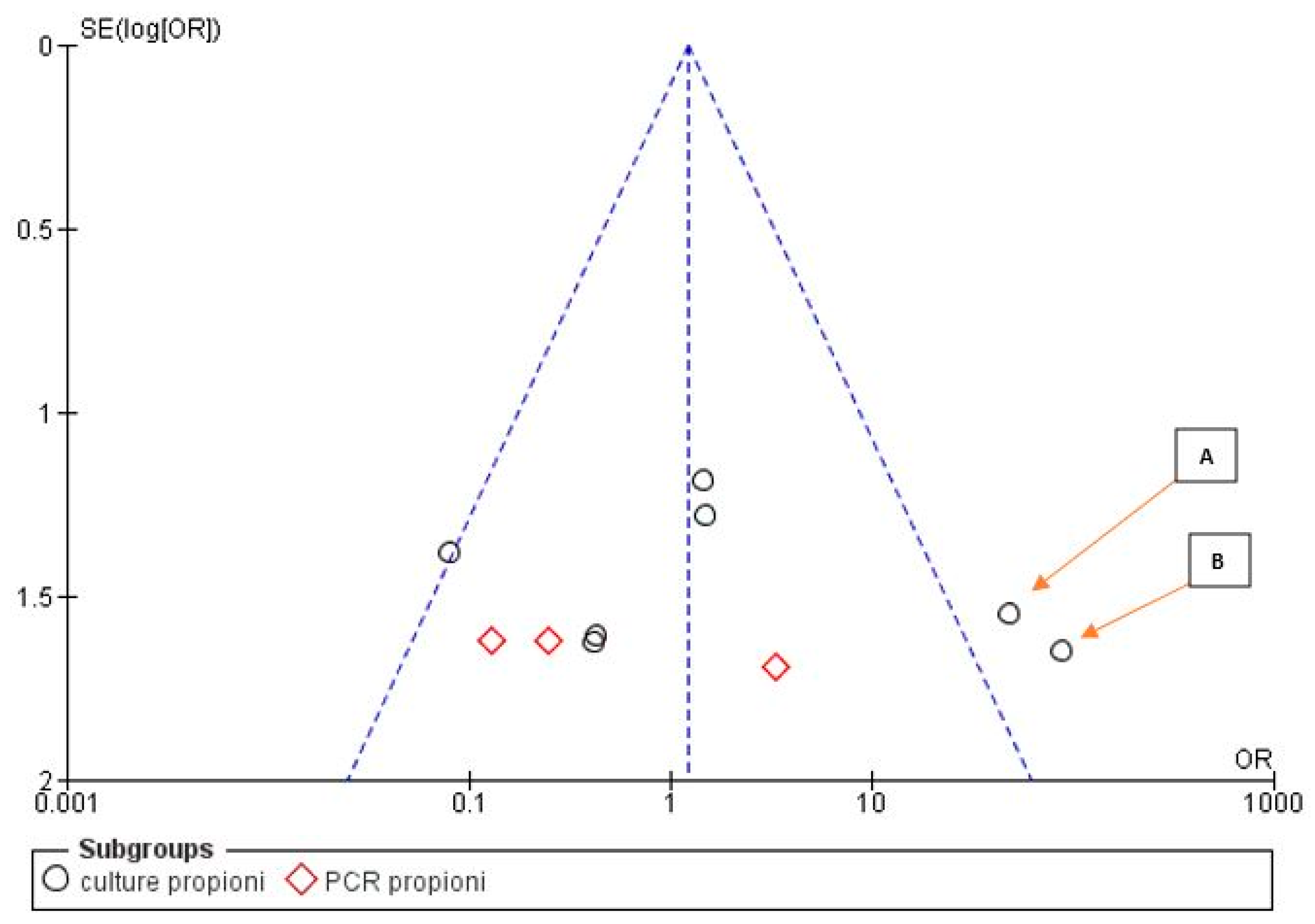
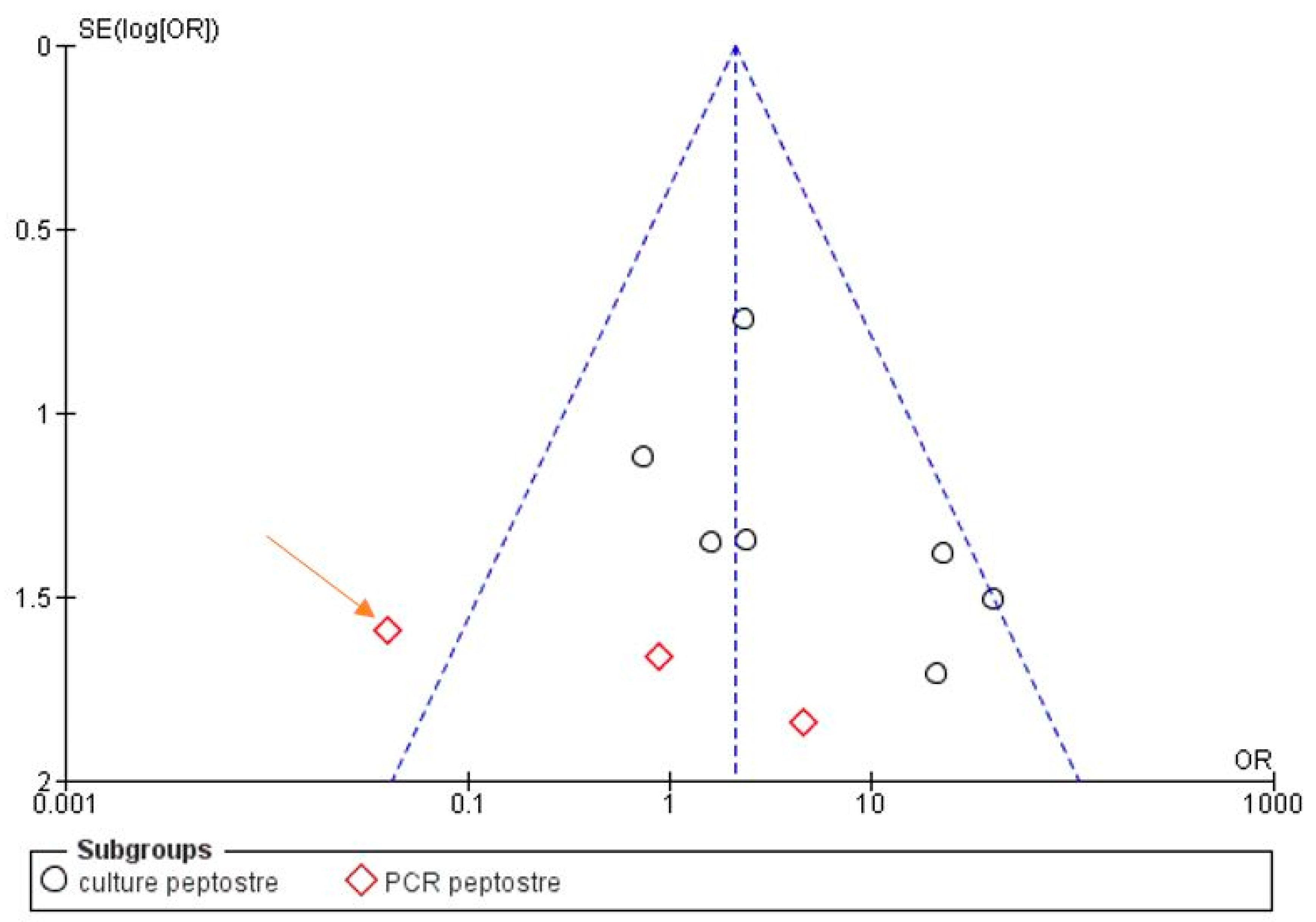
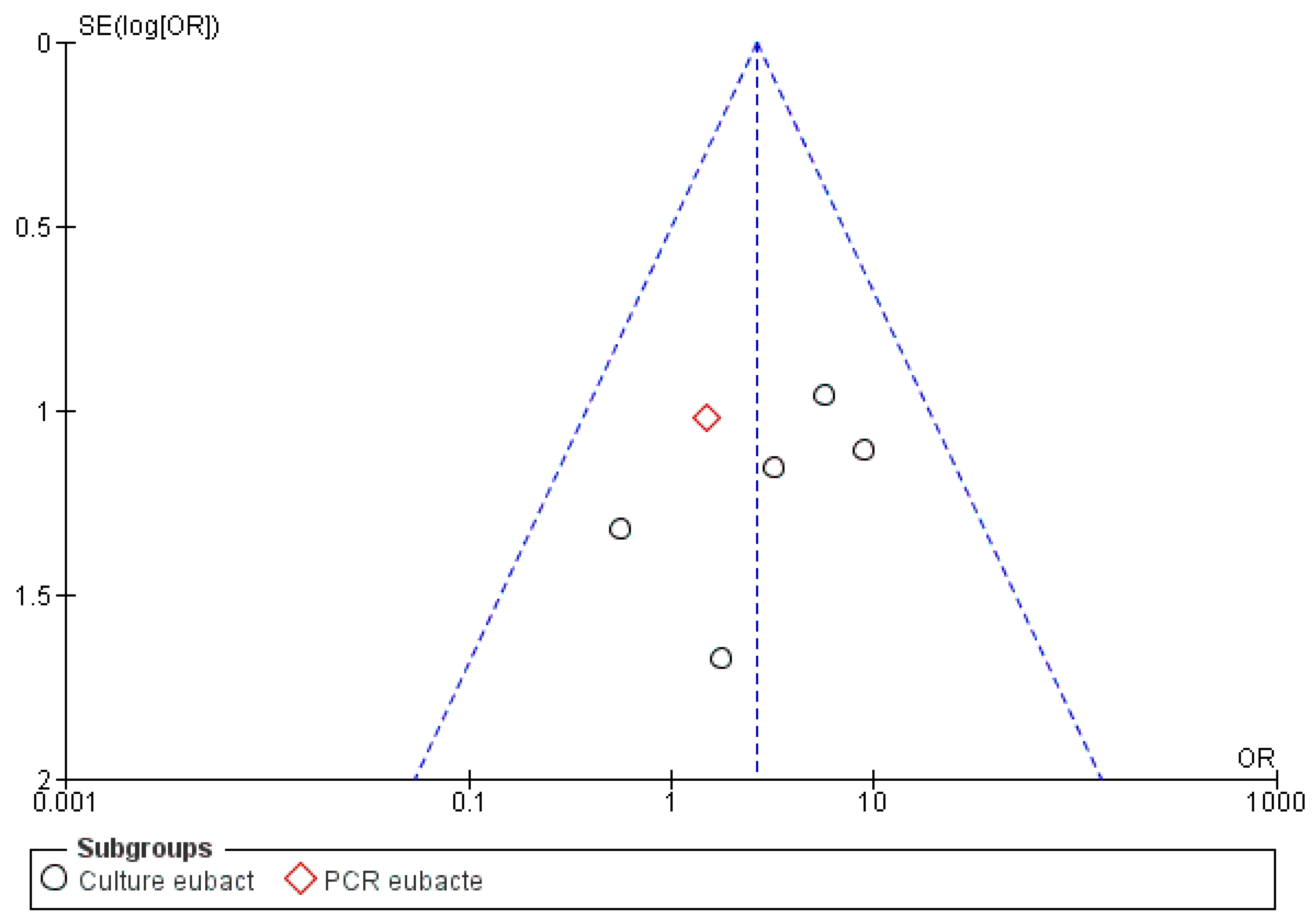
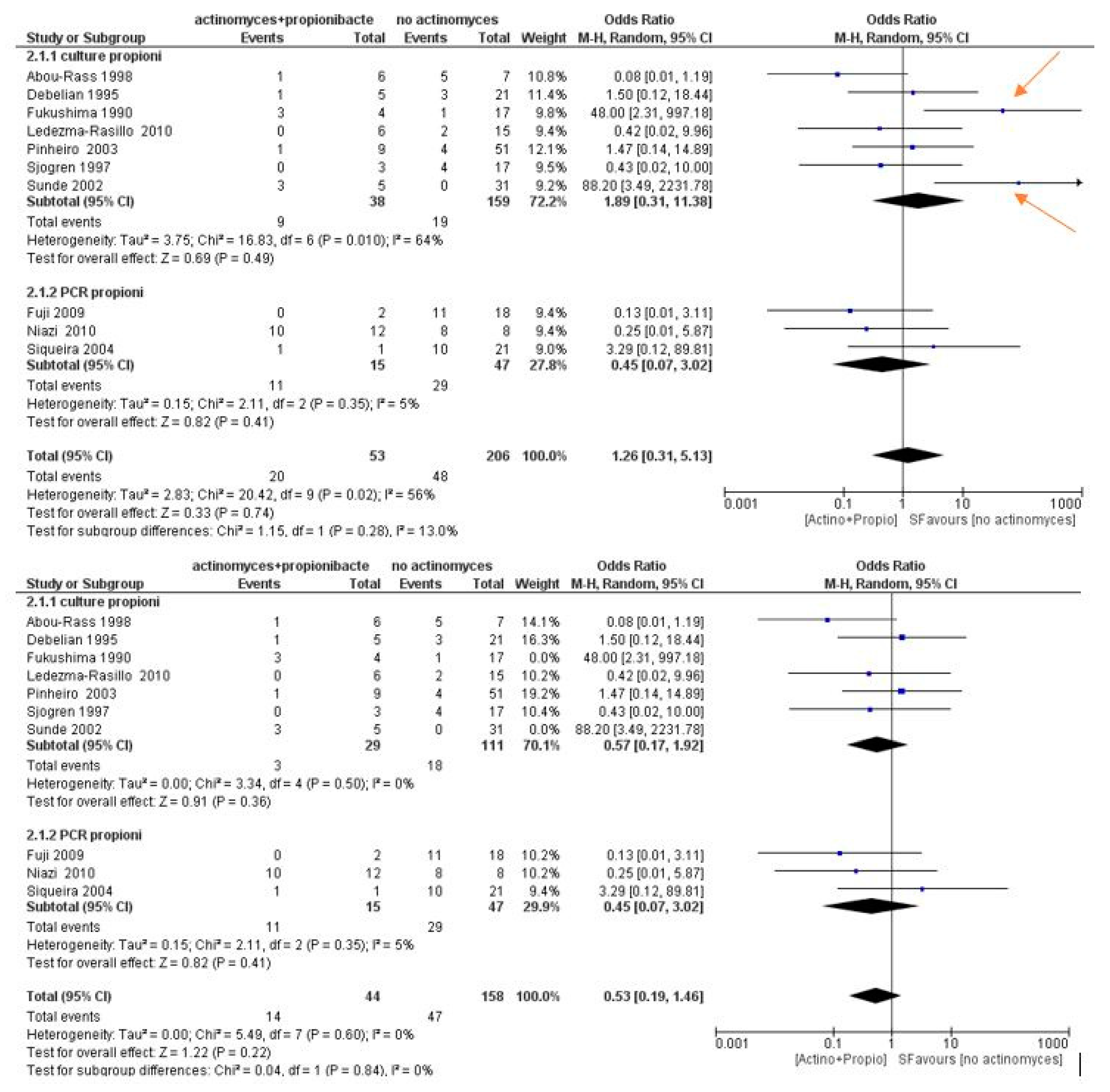
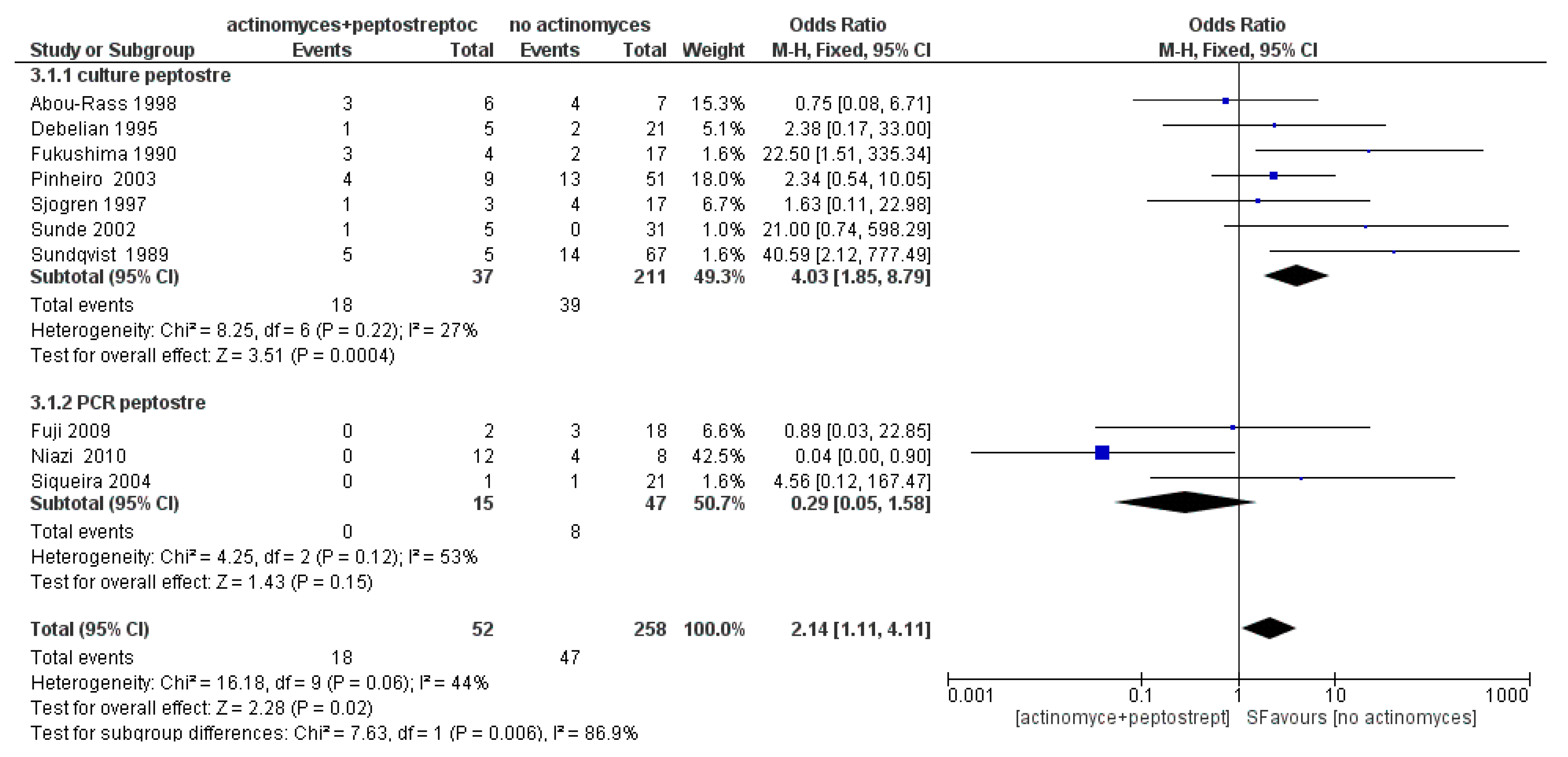
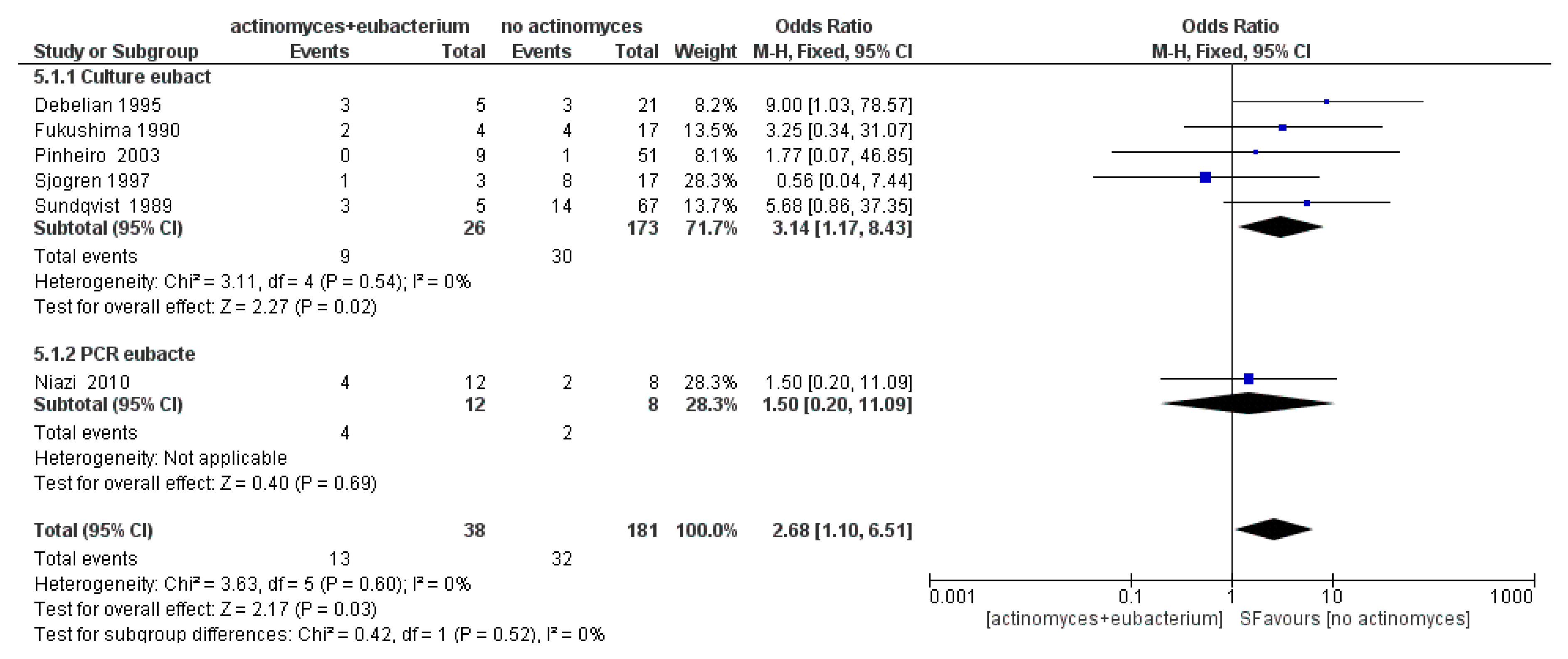
3.3. Meta-Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheung, G.S.; Ho, M.W. Microbial flora of root canal-treated teeth associated with asymptomatic periapical radiolucent lesions. Oral Microbiol. Immunol. 2001, 16, 332–337. [Google Scholar] [CrossRef]
- Sundqvist, G.; Figdor, D.; Persson, S.; Sjogren, U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1998, 85, 86–93. [Google Scholar] [CrossRef]
- Sjogren, U.; Figdor, D.; Persson, S.; Sundqvist, G. Influence of infection at the time of root filling on the outcome of endodontic treatment of teeth with apical periodontitis. Int. Endod. J. 1997, 30, 297–306. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Clinical implications and microbiology of bacterial persistence after treatment procedures. J. Endod. 2008, 34, 1291–1301.e3. [Google Scholar] [CrossRef]
- Kerekes, K.; Tronstad, L. Long-term results of endodontic treatment performed with a standardized technique. J. Endod. 1979, 5, 83–90. [Google Scholar] [CrossRef]
- Narayanan, L.L.; Vaishnavi, C. Endodontic microbiology. J. Conserv. Dent. 2010, 13, 233–239. [Google Scholar] [CrossRef]
- Torabinejad, M.; Ung, B.; Kettering, J.D. In vitro bacterial penetration of coronally unsealed endodontically treated teeth. J. Endod. 1990, 16, 566–569. [Google Scholar] [CrossRef]
- Tronstad, L.; Kreshtool, D.; Barnett, F. Microbiological monitoring and results of treatment of extraradicular endodontic infection. Endod. Dent. Traumatol. 1990, 6, 129–136. [Google Scholar] [CrossRef]
- Nair, P.N. On the causes of persistent apical periodontitis: A review. Int. Endod. J. 2006, 39, 249–281. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr. Aetiology of root canal treatment failure: Why well-treated teeth can fail. Int. Endod. J. 2001, 34, 1–10. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr. Endodontic infections: Concepts, paradigms, and perspectives. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2002, 94, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Molander, A.; Reit, C.; Dahlen, G.; Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 1998, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, Y.; Chen, W.; Zhu, C.; Liang, J. Bacterial flora and extraradicular biofilm associated with the apical segment of teeth with post-treatment apical periodontitis. J. Endod. 2012, 38, 954–959. [Google Scholar] [CrossRef]
- Pinheiro, E.T.; Gomes, B.P.; Ferraz, C.C.; Sousa, E.L.; Teixeira, F.B.; Souza-Filho, F.J. Microorganisms from canals of root-filled teeth with periapical lesions. Int. Endod. J. 2003, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Stuart, C.H.; Schwartz, S.A.; Beeson, T.J.; Owatz, C.B. Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J. Endod. 2006, 32, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, Q.; Zhang, C.; Cheung, G.S.; Shen, Y. Prevalence, phenotype, and genotype of Enterococcus faecalis isolated from saliva and root canals in patients with persistent apical periodontitis. J. Endod. 2010, 36, 1950–1955. [Google Scholar] [CrossRef]
- Ates, M.; Akdeniz, B.G.; Sen, B.H. The effect of calcium chelating or binding agents on Candida albicans. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2005, 100, 626–630. [Google Scholar] [CrossRef]
- Tong, Z.; Ling, J.; Lin, Z.; Li, X.; Mu, Y. The effect of MTADN on 10 Enterococcus faecalis isolates and biofilm: An in vitro study. J. Endod. 2013, 39, 674–678. [Google Scholar] [CrossRef]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef]
- Chavez de Paz, L.E. Redefining the persistent infection in root canals: Possible role of biofilm communities. J. Endod. 2007, 33, 652–662. [Google Scholar] [CrossRef]
- Gomes, B.P.; Pinheiro, E.T.; Sousa, E.L.; Jacinto, R.C.; Zaia, A.A.; Ferraz, C.C.; de Souza-Filho, F.J. Enterococcus faecalis in dental root canals detected by culture and by polymerase chain reaction analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006, 102, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Rolph, H.J.; Lennon, A.; Riggio, M.P.; Saunders, W.P.; MacKenzie, D.; Coldero, L.; Bagg, J. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 2001, 39, 3282–3289. [Google Scholar] [CrossRef] [PubMed]
- Downes, J.; Munson, M.A.; Spratt, D.A.; Kononen, E.; Tarkka, E.; Jousimies-Somer, H.; Wade, W.G. Characterisation of Eubacterium-like strains isolated from oral infections. J. Med. Microbiol. 2001, 50, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Relman, D.A. Emerging infections and newly-recognised pathogens. Neth. J. Med. 1997, 50, 216–220. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Alovisi, M.; Crincoli, V.; Aiuto, R.; Malagnino, G.; Quarta, C.; Laneve, E.; Sovereto, D.; Lo Russo, L.; Troiano, G.; et al. Prevalence of the Genus Propionibacterium in Primary and Persistent Endodontic Lesions: A Systematic Review. J. Clin. Med. 2020, 9, 739. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Crincoli, V.; Laino, L.; Alovisi, M.; Sovereto, D.; Lo Muzio, L.; Troiano, G. Prevalence of Bacteria of Genus Actinomyces in Persistent Extraradicular Lesions-Systematic Review. J. Clin. Med. 2020, 9, 457. [Google Scholar] [CrossRef]
- Dioguardi, M.; Crincoli, V.; Laino, L.; Alovisi, M.; Sovereto, D.; Mastrangelo, F.; Russo, L.L.; Muzio, L.L. The Role of Periodontitis and Periodontal Bacteria in the Onset and Progression of Alzheimer’s Disease: A Systematic Review. J. Clin. Med. 2020, 9, 495. [Google Scholar] [CrossRef]
- Skerman, V.B.D.; McGowan, V.; Sneath, P.H.A. Approved lists of bacterial names. Med. J. Aust. 1980, 2, 3–4. [Google Scholar] [CrossRef]
- Lo, C.K.; Mertz, D.; Loeb, M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med. Res. Methodol. 2014, 14, 45. [Google Scholar] [CrossRef]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Sunde, P.T.; Olsen, I.; Debelian, G.J.; Tronstad, L. Microbiota of periapical lesions refractory to endodontic therapy. J. Endod. 2002, 28, 304–310. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2004, 97, 85–94. [Google Scholar] [CrossRef]
- Ledezma-Rasillo, G.; Flores-Reyes, H.; Gonzalez-Amaro, A.M.; Garrocho-Rangel, A.; Ruiz-Rodriguez Mdel, S.; Pozos-Guillen, A.J. Identification of cultivable microorganisms from primary teeth with necrotic pulps. J. Clin. Pediatr. Dent. 2010, 34, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, G.; Johansson, E.; Sjogren, U. Prevalence of black-pigmented bacteroides species in root canal infections. J. Endod. 1989, 15, 13–19. [Google Scholar] [CrossRef]
- Abou-Rass, M.; Bogen, G. Microorganisms in closed periapical lesions. Int. Endod. J. 1998, 31, 39–47. [Google Scholar] [CrossRef]
- Niazi, S.A.; Clarke, D.; Do, T.; Gilbert, S.C.; Mannocci, F.; Beighton, D. Propionibacterium acnes and Staphylococcus epidermidis isolated from refractory endodontic lesions are opportunistic pathogens. J. Clin. Microbiol. 2010, 48, 3859–3869. [Google Scholar] [CrossRef]
- Fujii, R.; Saito, Y.; Tokura, Y.; Nakagawa, K.I.; Okuda, K.; Ishihara, K. Characterization of bacterial flora in persistent apical periodontitis lesions. Oral Microbiol. Immunol. 2009, 24, 502–505. [Google Scholar] [CrossRef]
- Fukushima, H.; Yamamoto, K.; Hirohata, K.; Sagawa, H.; Leung, K.P.; Walker, C.B. Localization and identification of root canal bacteria in clinically asymptomatic periapical pathosis. J. Endod. 1990, 16, 534–538. [Google Scholar] [CrossRef]
- Debelian, G.J.; Olsen, I.; Tronstad, L. Bacteremia in conjunction with endodontic therapy. Endod. Dent. Traumatol. 1995, 11, 142–149. [Google Scholar] [CrossRef]
- Molven, O.; Halse, A. Success rates for gutta-percha and Kloroperka N-0 root fillings made by undergraduate students: Radiographic findings after 10–17 years. Int. Endod. J. 1988, 21, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Sjogren, U.; Happonen, R.P.; Kahnberg, K.E.; Sundqvist, G. Survival of Arachnia propionica in periapical tissue. Int. Endod. J. 1988, 21, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Koppang, H.S.; Koppang, R.; Solheim, T.; Aarnes, H.; Stolen, S.O. Cellulose fibers from endodontic paper points as an etiological factor in postendodontic periapical granulomas and cysts. J. Endod. 1989, 15, 369–372. [Google Scholar] [CrossRef]
- Yusuf, H. The significance of the presence of foreign material periapically as a cause of failure of root treatment. Oral Surg. Oral Med. Oral Pathol. 1982, 54, 566–574. [Google Scholar] [CrossRef]
- Sedgley, C.M.; Messer, H. Long-term retention of a paper point in the periapical tissues: A case report. Endod. Dent. Traumatol. 1993, 9, 120–123. [Google Scholar] [CrossRef]
- Nair, P.N.; Sjogren, U.; Schumacher, E.; Sundqvist, G. Radicular cyst affecting a root-filled human tooth: A long-term post-treatment follow-up. Int. Endod. J. 1993, 26, 225–233. [Google Scholar] [CrossRef]
- Nair, P.N. New perspectives on radicular cysts: Do they heal? Int. Endod. J. 1998, 31, 155–160. [Google Scholar] [CrossRef]
- Su, L.; Gao, Y.; Yu, C.; Wang, H.; Yu, Q. Surgical endodontic treatment of refractory periapical periodontitis with extraradicular biofilm. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010, 110, e40–e44. [Google Scholar] [CrossRef]
- Ricucci, D.; Siqueira, J.F., Jr. Apical actinomycosis as a continuum of intraradicular and extraradicular infection: Case report and critical review on its involvement with treatment failure. J. Endod. 2008, 34, 1124–1129. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Polymerase chain reaction detection of Propionibacterium propionicus and Actinomyces radicidentis in primary and persistent endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2003, 96, 215–222. [Google Scholar] [CrossRef]
- Zakaria, M.N.; Takeshita, T.; Shibata, Y.; Maeda, H.; Wada, N.; Akamine, A.; Yamashita, Y. Microbial community in persistent apical periodontitis: A 16S rRNA gene clone library analysis. Int. Endod. J. 2015, 48, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J.; Frolander, F.; Sundquist, G. Oxygen tolerance of anaerobic bacteria isolated from necrotic dental pulps. Acta Odontol. Scand. 1977, 35, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Liet, S.; Sorin, S.M. Evaluation of clinical results based upon culturing root canals. J. Br. Endod. Soc. 1969, 3, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Heling, B.; Shapira, J. Roentgenologic and clinical evaluation of endodontically treated teeth, with or without negative culture. Quintessence Int. Dent. Dig. 1978, 9, 79–84. [Google Scholar]
- Hancock, H.H., 3rd; Sigurdsson, A.; Trope, M.; Moiseiwitsch, J. Bacteria isolated after unsuccessful endodontic treatment in a North American population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 91, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Bowden, G.H.; Hamilton, I.R. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 1998, 9, 54–85. [Google Scholar] [CrossRef]
- Hall, V.; Collins, M.D.; Hutson, R.A.; Inganas, E.; Falsen, E.; Duerden, B.I. Actinomyces oricola sp. nov., from a human dental abscess. Int. J. Syst. Evol. Microbiol. 2003, 53, 1515–1518. [Google Scholar] [CrossRef]
- Yeguez, J.F.; Martinez, S.A.; Sands, L.R.; Hellinger, M.D. Pelvic actinomycosis presenting as malignant large bowel obstruction: A case report and a review of the literature. Am. Surg. 2000, 66, 85–90. [Google Scholar]
- Acevedo, F.; Baudrand, R.; Letelier, L.M.; Gaete, P. Actinomycosis: A great pretender. Case reports of unusual presentations and a review of the literature. Int. J. Infect. Dis. 2008, 12, 358–362. [Google Scholar] [CrossRef]
- Valour, F.; Senechal, A.; Dupieux, C.; Karsenty, J.; Lustig, S.; Breton, P.; Gleizal, A.; Boussel, L.; Laurent, F.; Braun, E.; et al. Actinomycosis: Etiology, clinical features, diagnosis, treatment, and management. Infect. Drug Resist. 2014, 7, 183–197. [Google Scholar] [CrossRef]
- Sundqvist, G.; Reuterving, C.O. Isolation of Actinomyces israelii from periapical lesion. J. Endod. 1980, 6, 602–606. [Google Scholar] [CrossRef]
- Moghimi, M.; Salentijn, E.; Debets-Ossenkop, Y.; Karagozoglu, K.H.; Forouzanfar, T. Treatment of cervicofacial actinomycosis: A report of 19 cases and review of literature. Med. Oral Patol. Oral Cir. Bucal. 2013, 18, e627–e632. [Google Scholar] [CrossRef] [PubMed]
- Hirshberg, A.; Tsesis, I.; Metzger, Z.; Kaplan, I. Periapical actinomycosis: A clinicopathologic study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2003, 95, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.M.; Krost, B.S. Actinomycosis presenting as a midline palatal defect. J. Oral Maxillofac. Surg. 1995, 53, 701–703. [Google Scholar] [CrossRef]
- Belmont, M.J.; Behar, P.M.; Wax, M.K. Atypical presentations of actinomycosis. Head. Neck. 1999, 21, 264–268. [Google Scholar] [CrossRef]




| Provider Database | Keywords | Search Details | No. of Records | Articles after Removal of Overlapping Articles | Number of Records after Restriction by Year of Publication (Last 40 Years) | Numbers of Articles That Have Investigated the Role of Bacteria in Endodontic Infections | Number of Studies That Consider the Microbial Composition of Each Analyzed Sample |
|---|---|---|---|---|---|---|---|
| PubMed | “persistent intraradicular infection” OR “primary endodontic infection” | “persistent intraradicular infection” [All Fields] OR “primary endodontic infection” [All Fields] | 37 | ||||
| PubMed | “endodontic failure” OR “endodontic microbiologic” | “endodontic failure” [All Fields] OR (endodontic [All Fields] AND microbiologic [All Fields]) | 203 | ||||
| PubMed | “Actinomyces” AND “endodontic” OR “apical parodontitis” | “Actinomyces” [All Fields] AND “endodontic” [All Fields] OR “apical parodontitis” [All Fields] | 117 | ||||
| Scopus | “persistent intraradicular infection” | TITLE-ABS-KEY (persistent AND interradicular AND infection) | 23 | ||||
| Scopus | “persistent extraradicular infection” | TITLE-ABS-KEY (persistent AND extravascular AND infection) | 18 | ||||
| Scopus | “Actinomyces” AND “endodontic” | TITLE-ABS-KEY (“Actinomyces” AND “endodontic”) | 145 | ||||
| EBSCO | persistent extraradicular infection | 7 | |||||
| EBSCO | persistent intraradicular infection | 14 | |||||
| EBSCO | “Actinomyces” AND “endodontic” | 113 | |||||
| Web of science | persistent extraradicular infection | 19 | |||||
| Web of science | persistent intraradicular infection | 19 | |||||
| Web of science | “Actinomyces” AND “endodontic” | 117 | |||||
| Articles included in the references of the identified full-text publications | 51 | ||||||
| Total records | 883 | 475 | 462 | 81 | 11 |
| First Author, Date, Journal | Type of Endodontic Lesion | Total Number of Samples | Number of Samples with Actinomyces | Prevalence of Microbial Genera in Association with Genus Actinomyces | Prevalence of Microbial Genera in the Total Samples Analyzed | Identification Method |
|---|---|---|---|---|---|---|
| Sunde, 2002, Journal of Endodontics | Refractory apical periodontitis | 36 | 5 | Clostridium: 2/5 | Clostridium: 2/36 | Culture |
| Propionibacterium: 3/5 | Propionibacterium: 3/36 | |||||
| Gemella: 1/5 | Gemella: 2/36 | |||||
| Peptostreptococcus: 1/5 | Peptostreptococcus: 1/36 | |||||
| Vibrio: 1/5 | Vibrio: 1/36 | |||||
| Leptotrichia: 1/5 | Leptotrichia: 1/36 | |||||
| Staphylococcus: 2/5 | Staphylococcus: 3/36 | |||||
| Streptococcus: 1/5 | Streptococcus: 2/36 | |||||
| Siqueira, 2004, Oral surgery, oral medicine, oral pathology, oral radiology and endodontics | Root-filled teeth with persistent periradicular lesions | 22 | 1 | Propionibacterium, Pseudoramibacter, Enterococcus | Propionibacterium: 11/22 Pseudoramibacter: 12/22 Enterococcus: 17/22 | PCR |
| Ledezma-Rasillo, 2010, The Journal of clinical pediatric dentistry | Primary teeth with necrotic pulps | 21 | 6 | Bifidobacterium: 5/6 | Bifidobacterium: 17/21 | Culture |
| Veillonella: 1/6 | Veillonella: 2/21 | |||||
| Clostridium: 3/6 | Clostridium: 7/21 | |||||
| Streptococcus: 2/6 | Streptococcus: 6/21 | |||||
| Gemella: 1/6 | Gemella: 1/21 | |||||
| Sundqvist, 1989, Journal of endodontics | Teeth with apical periodontitis | 72 | 5 | Peptostreptococcus: 5/5 | Peptostreptococcus: 19/72 | Culture |
| Lactobacillus: 2/5 | Lactobacillus: 12/72 | |||||
| Bacteroides: 5/5 | Bacteroides: 22/72 | |||||
| Wolinella: 1/5 | Wolinella: 6/72 | |||||
| Streptococcus: 1/5 | Streptococcus: 8/72 | |||||
| Eubacterium: 3/5 | Eubacterium: 17/72 | |||||
| Fusobacterium: 3/5 | Fusobacterium: 16/72 | |||||
| Abou-Rass, 1998, International endodontic journal | Closed periapical lesions associated with refractory endodontic therapy | 13 | 6 | Streptococcus: 3/6 | Streptococcus:7/13 | Culture |
| Staphylococcus: 1/6 | Staphylococcus: 4/13 | |||||
| Peptostreptococcus: 1/6 | Peptostreptococcus: 1/13 | |||||
| Gram-negative enteric rods: 1/6 | Gram-negative enteric rods: 1/13 | |||||
| Propionibacterium: 1/6 | Propionibacterium: 6/13 | |||||
| Porphyromonas: 1/6 | Porphyromonas: 1/13 | |||||
| Fusobacterium: 1/6 | Fusobacterium: 1/13 | |||||
| Niazi, 2010, Journal of clinical microbiology | Refractory endodontic lesions (9 with abscesses and 11 without abscesses) | 20 | 12 | Gemella: 3/12 | Gemella: 5/20 | PCR |
| Propionibacterium: 10/12 | Propionibacterium: 18/20 | |||||
| Staphylococcus: 9/12 | Staphylococcus:15/20 | |||||
| Streptococcus: 11/12 | Streptococcus: 15/20 | |||||
| Clostridium: 1/12 | Clostridium: 2/20 | |||||
| Capnocytophaga: 3/12 | Capnocytophaga: 3/20 | |||||
| Prevotella: 4/12 | Prevotella: 7/20 | |||||
| Selenomonas: 3/12 | Selenomonas: 3/20 | |||||
| Olsenella: 4/12 | Olsenella: 5/20 | |||||
| Bifidobacterium: 1/12 | Bifidobacterium: 2/20 | |||||
| Lactobacillus: 1/12 | Lactobacillus: 1/20 | |||||
| Abiotrophia: 1/12 | Abiotrophia: 1/20 | |||||
| Granulicatella: 2/12 | Granulicatella: 2/20 | |||||
| Kocuria: 1/12 | Kocuria: 1/20 | |||||
| Micrococcus: 1/12 | Micrococcus: 2/20 | |||||
| Rothia: 2/12 | Rothia: 2/20 | |||||
| Eubacterium: 4/12 | Eubacterium: 6/20 | |||||
| Parvimonas: 2/12 | Parvimonas: 2/20 | |||||
| Solobacterium: 2/12 | Solobacterium: 3/20 | |||||
| Veillonella: 3/12 | Veillonella: 4/20 | |||||
| Enterococcus: 1/12 | Enterococcus: 3/20 | |||||
| Bacteroides: 1/12 | Bacteroides: 1/20 | |||||
| Desulfovibrio: 1/12 | Desulfovibrio: 1/20 | |||||
| Lautropia: 1/12 | Lautropia: 1/20 | |||||
| Phascolarctobacterium: 1/12 | Phascolarctobacterium: 1/20 | |||||
| Catonella: 1/12 | Catonella: 1/20 | |||||
| Oribacterium: 1/12 | Oribacterium: 1/20 | |||||
| Slackia: 2/12 | Slackia: 4/20 | |||||
| Pseudoramibacter: 3/12 | Pseudoramibacter: 4/20 | |||||
| Mogibacterium: 3/12 | Mogibacterium: 6/20 | |||||
| Atopobium: 2/12 | Atopobium: 2/20 | |||||
| Dialister: 3/12 | Dialister: 5/20 | |||||
| Porphyromonas: 2/12 | Porphyromonas:2/20 | |||||
| Tanerella: 1/12 | Tanerella: 4/20 | |||||
| Campylobacter: 1/12 | Campylobacter: 2/20 | |||||
| Fujii, 2009, Oral microbiology and immunology | Apical periodontitis lesions of obturated teeth | 20 | 2 | Fusobacterium: 1/2 | Fusobacterium: 5/20 | PCR |
| Slackia: 1/2 | Slackia: 1/20 | |||||
| Staphylococcus: 1/2 | Staphylococcus: 8/20 | |||||
| Streptococcus: 2/2 | Streptococcus: 5/20 | |||||
| Stenotrophomonas: 1/2 | Stenotrophomonas: 1/20 | |||||
| Prevotella: 1/2 | Prevotella: 4/20 | |||||
| Pinheiro, 2003, International endodontic journal | Root-filled teeth with apical periodontitis | 60 | 9 | Streptococcus: 4/9 | Streptococcus: 17/60 | Culture |
| Enterococcus: 2/9 | Enterococcus: 28/60 | |||||
| Prevotella: 2/9 | Prevotella: 6/60 | |||||
| Peptostreptococcus: 2/9 | Peptostreptococcus: 9/60 | |||||
| Bifidobacterium: 1/9 | Bifidobacterium: 1/60 | |||||
| Veillonella: 3/9 | Veillonella: 4/60 | |||||
| Candida: 1/9 | Candida: 2/60 | |||||
| Propionibacterium: 1/9 | Propionibacterium: 5/60 | |||||
| Fusobacterium: 1/9 | Fusobacterium: 3/60 | |||||
| Gemella: 3/9 | Gemella: 4/60 | |||||
| Haemophilus: 1/9 | Haemophilus: 1/60 | |||||
| Staphylococcus: 1/9 | Staphylococcus: 3/60 | |||||
| Sjogren, 1997, International endodontic journal | Apical periodontitis | 20 | 3 | Prevotella: 1/3 | Prevotella: 3/20 | Culture |
| Eubacterium: 1/3 | Eubacterium: 9/20 | |||||
| Campylobacter: 1/3 | Campylobacter: 4/20 | |||||
| Peptostreptococcus: 1/3 | Peptostreptococcus: 5/20 | |||||
| Fukushima, 1990, Journal of endodontics | Untreated cases | 21 | 4 | Propionibacterium: 3/4 | Propionibacterium: 4/21 | Culture |
| Lactobacillus: 3/4 | Lactobacillus: 5/21 | |||||
| Eubacterium: 2/4 | Eubacterium: 6/21 | |||||
| Peptostreptococcus: 3/4 | Peptostreptococcus: 5/21 | |||||
| Peptococcus: 1/4 | Peptococcus: 2/21 | |||||
| Debelian et al., 1995, Endodontics & Dental Traumatology | Teeth with asymptomatic apical periodontitis | 26 | 5 | Propionibacterium: 1/5 | Propionibacterium: 4/26 | Culture |
| Prevotella: 2/5 | Prevotella: 5/26 | |||||
| Eubacterium: 3/5 | Eubacterium: 6/26 | |||||
| Campylobacter: 1/5 | Campylobacter: 1/26 | |||||
| Veillonella: 1/5 | Veillonella: 2/26 | |||||
| Lactobacillus: 1/5 | Lactobacillus: 1/26 | |||||
| Streptococcus: 2/5 | Streptococcus: 5/26 | |||||
| Porphyromonas: 1/5 | Porphyromonas: 2/26 | |||||
| Fusobacterium: 1/5 | Fusobacterium: 4/26 | |||||
| Clostridium: 1/5 | Clostridium: 1/26 | |||||
| Peptostreptococcus: 1/5 | Peptostreptococcus: 3/26 | |||||
| Saccharomyces: 1/5 | Saccharomyces: 1/26 |
| First Author, Date, Journal | Type of Endodontic Lesion | Total Number of Samples | Number of Samples with Actinomyces | Prevalence of Individual Species of the Genus Actinomyces, Given the Total Number of Analyzed Samples | Identification Method |
|---|---|---|---|---|---|
| Sunde, 2002, Journal of endodontics | refractory apical periodontitis | 36 | 5 | Actinomyces israelii: 3/36 Actinomyces viscosus: 2/36 Actinomyces meyeri: 1/36 Actinomyces naeslundii: 1/36 | Culture |
| Siqueira, 2004, Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics | Root-filled teeth with persistent periradicular lesions | 22 | 1 | Actinomyces radicidentis: 1/22 | PCR |
| Ledezma-Rasillo, 2010, The Journal of clinical pediatric dentistry | Primary teeth with necrotic pulps | 21 | 6 | Actinomyces israelii: 4/21 Actinomyces naeslundii: 2/21 | Culture |
| Sundqvist, 1989, Journal of endodontics | Teeth with apical periodontitis | 72 | 5 | Actinomyces species: 5/72 | Culture |
| Abou-Rass, 1998, International endodontic journal | Closed periapical lesions associated with refractory endodontic therapy | 13 | 6 | Actinomyces sp. I: 1/13 Actinomyces sp. II: 1/13 Actinomyces sp.: 5/13 | Culture |
| Niazi, 2010, Journal of clinical microbiology | Refractory endodontic lesions (9 with abscesses and 11 without abscesses) | 20 | 12 | Actinomyces gerencseriae oral taxon 618: 1/20 Actinomyces sp. oral clone CT047: 1/20 Actinomyces massiliensis: 1/20 Actinomyces meyeri: 1/36 Actinomyces radicidentis: 1/36 Actinomyces sp. oral taxon 169 clone AG004: 3/36 Actinomyces israelii: 1/36 Actinomyces sp. oral clone JA063: 1/36 Actinomyces sp. oral taxon 181 strain Hal1065: 1/36 Actinomyces strain B27SC: 2/36 Actinomyces strain C29KA: 1/36 | PCR |
| Fujii, 2009, Oral microbiology and immunology | Apical periodontitis lesions of obturated teeth | 20 | 2 | Actinomyces naeslundii: 2/20 | PCR |
| Pinheiro, 2003, International endodontic journal | Root-filled teeth with apical periodontitis | 60 | 9 | A. naeslundii: 4/60 A. viscosus: 3/60 A. odontolyticus: 3/60 | Culture |
| Sjogren, 1997, International endodontic journal | Apical periodontitis | 20 | 3 | Actinomyces israelii: 2/20 Actinomyces odontolyticus: 1/20 Actinomyces naeslundii: 1/20 | Culture |
| Fukushima, 1990, Journal of endodontics | Untreated cases | 21 | 4 | Actinomyces israelii: 2/21 Actinomyces viscosus: 2/21 A. meyeri: 1/21 | Culture |
| Debelian et al., 1995, Endodontics & Dental Traumatology | Teeth with asymptomatic apical periodontitis | 26 | 5 | Actinomyces israelii: 3/26 Actinomyces naeslundii: 1/26 Actinomyces odontolyticus: 1/26 | Culture |
| Bacterial Genus | Prevalence in Samples That Were Associated with Actinomyces, Given the Total Number of Samples for All Articles Selected for This Review | Prevalence in Samples, Given the Total Number of Samples for All Articles Selected for This Review | Number of Articles Reporting This Genus |
|---|---|---|---|
| Clostridium | 7/58 | 12/331(3.6%) | 4 |
| Propionibacterium | 20/58 | 51/331(15.4%) | 7 |
| Gemella | 8/58 | 12/331(3.6%) | 4 |
| Peptostreptococcus | 14/58 | 43/331(13%) | 7 |
| Vibrio | 1/58 | 1/331(0.3%) | 1 |
| Leptotrichia | 1/58 | 1/331(0.3%) | 1 |
| Staphylococcus | 14/58 | 33/331(10%) | 5 |
| Streptococcus | 26/58 | 65/331(19.6%) | 8 |
| Pseudoramibacter | 4/58 | 16/331(4.8%) | 2 |
| Enterococcus | 4/58 | 48/331(14.5%) | 3 |
| Bifidobacterium | 7/58 | 20/331(6%) | 3 |
| Veillonella | 8/58 | 12/331(3.6%) | 4 |
| Lactobacillus | 7/58 | 19/331(5.7%) | 4 |
| Bacteroides | 6/58 | 23/331(6.9%) | 2 |
| Wolinella | 1/58 | 6/331(1.8%) | 1 |
| Eubacterium | 13/58 | 44/331(13.3%) | 5 |
| Fusobacterium | 7/58 | 29/331(8:8%) | 5 |
| Gram-negative enteric rods | 1/58 | 1/331(0.3%) | 1 |
| Porphyromonas | 4/58 | 5/331(1.5%) | 3 |
| Capnocytophaga | 3/58 | 3/331(0.9%) | 1 |
| Prevotella | 10/58 | 25/331(7.5%) | 5 |
| Selenomonas | 3/58 | 3/331(0.9%) | 1 |
| Olsenella | 4/58 | 5/331(1.5%) | 1 |
| Abiotrophia | 1/58 | 1/331(0.3%) | 1 |
| Granulicatella | 2/58 | 2/331(0.6%) | 1 |
| Kocuria | 1/58 | 1/331(0.3%) | 1 |
| Micrococcus | 1/58 | 2/331(0.6%) | 1 |
| Rothia | 2/58 | 2/331(0.6%) | 1 |
| Parvimonas | 2/58 | 2/331(0.6%) | 1 |
| Solobacterium | 2/58 | 3/331(0.9%) | 1 |
| Desulfovibrio | 1/58 | 1/331(0.3%) | 1 |
| Lautropia | 1/58 | 1/331(0.3%) | 1 |
| Phascolarctobacterium | 1/58 | 1/331(0.3%) | 1 |
| Catonella | 1/58 | 1/331(0.3%) | 1 |
| Oribacterium | 1/58 | 1/331(0.3%) | 1 |
| Slackia | 3/58 | 5/331(1.5%) | 2 |
| Mogibacterium | 3/58 | 6/331(1.8%) | 1 |
| Atopobium | 2/58 | 2/331(0.6%) | 1 |
| Dialister | 3/58 | 5/331(1.5%) | 1 |
| Tanerella | 1/58 | 4/331(1.2%) | 1 |
| Campylobacter | 3/58 | 7/331(2.1%) | 3 |
| Stenotrophomonas | 1/58 | 1/331(0.3%) | 1 |
| Candida | 1/58 | 2/331(0.6%) | 1 |
| Haemophilus | 1/58 | 1/331(0.3%) | 1 |
| Peptococcus | 1/58 | 2/331(0.6%) | 1 |
| Saccharomyces | 1/58 | 1/331(0.3%) | 1 |
| Bacterial Genus | Sub-Group Culture | Sub-Group PCR | ||
|---|---|---|---|---|
| Prevalence in Samples That Were Associated with Actinomyces | Prevalence in Samples, Given the Total of Number of Samples | Prevalence in Samples That Were Associated with Actinomyces | Prevalence in Samples, Given the Total of Number of Samples | |
| Clostridium | 6/16 | 10/83 | 1/12 | 2/20 |
| Propionibacterium | 9/29 | 22/156 | 11/13 | 29/42 |
| Streptococcus | 13/36 | 43/228 | 13/14 | 20/40 |
| Peptostreptococcus | 14/37 | 43/212 | - | - |
| Staphylococcus | 13/34 | 25/129 | 1/2 | 8/20 |
| Eubacterium | 9/17 | 38/139 | 4/12 | 6/20 |
| Fusobacterium | 6/25 | 24/171 | 1/2 | 5/20 |
| Prevotella | 9/29 | 21/126 | 1/2 | 4/20 |
| Veillonella | 5/20 | 8/107 | 3/9 | 4/20 |
| Lactobacillus | 6/14 | 18/119 | 1/12 | 1/20 |
| enterococcus | 2/9 | 28/60 | 2/13 | 20/42 |
| Porphyromonas | 2/11 | 3/39 | 2/12 | 2/20 |
| Campylobacter | 2/8 | 5/46 | 1/12 | 2/20 |
| Bifidobacterium | 6/15 | 18/81 | 1/12 | 2/20 |
| Species of the Genus Actinomyces | Prevalence of the Actinomyces Species in Samples for the Total Number of Analyzed Samples for All Articles | Number of Articles Reporting This Species |
|---|---|---|
| Actinomyces israelii | 15/331(4.5%) | 6 |
| Actinomyces viscosus | 7/331(2.1%) | 3 |
| Actinomyces meyeri | 3/331 (0.9%) | 3 |
| Actinomyces naeslundii | 11/331(3.3%) | 6 |
| Actinomyces radicidentis | 2/331(0.6%) | 2 |
| Actinomyces species | 10/331(3%) | 2 |
| Actinomyces sp. I | 1/331(0.3%) | 1 |
| Actinomyces sp. II | 1/331(0.3%) | 1 |
| Actinomyces gerencseriae oral taxon 618 | 1/331(0.3%) | 1 |
| Actinomyces sp. oral clone CT047 | 1/331(0.3%) | 1 |
| Actinomyces massiliensis | 1/331(0.3%) | 1 |
| Actinomyces sp. oral taxon 169 clone AG004 | 3/331(0.9%) | 1 |
| Actinomyces sp. oral clone JA063 | 1/331(0.3%) | 1 |
| Actinomyces sp. oral taxon 181 strain Hal1065 | 1/331(0.3%) | 1 |
| Actinomyces strain B27SC | 2/331(0.6%) | 1 |
| Actinomyces strain C29KA | 1/331(0.3%) | 1 |
| Actinomyces odontolyticus | 5/331(1.5%) | 3 |
| Selection | Comparability | Exposure | Score | Sub-Group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Definition of Cases | Representativeness of Cases | Selection of Controls | Definition of Controls | Comparability of Cases and Controls on the Basis of the Design or Analysis | Ascertainment of Exposure | Same Method of Ascertainment for Cases and Controls | Non-Response Rate | ||
| [34] Ledezma-Rasillo et al., 2010 The Journal of clinical pediatric dentistry | 3 | 1 | 2 | 2 | 2 | 2 | 3 | 0 | 15 | Streptococcus, Propionibacterium, |
| [37] Niazi et al., 2010 Journal of endodontics | 3 | 1 | 3 | 3 | 2 | 1 | 3 | 0 | 16 | Streptococcus, Propionibacterium, Peptostreptococcus, Staphylococcus, Eubacterium |
| [38] Fujii et al., 2009 Oral microbiology and immunology | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 0 | 11 | Streptococcus, Propionibacterium, Peptostreptococcus, Staphylococcus |
| [33] Siqueira et al., 2004 Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics | 3 | 1 | 1 | 1 | 1 | 3 | 2 | 0 | 12 | Streptococcus, Propionibacterium, Peptostreptococcus |
| [14] Pinheiro et al., 2003 International endodontic journal | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 0 | 17 | Streptococcus, Propionibacterium, Peptostreptococcus, Staphylococcus, Eubacterium |
| [32] Sunde et al., 2002 Journal of endodontics | 2 | 2 | 2 | 2 | 3 | 2 | 3 | 0 | 16 | Streptococcus, Propionibacterium, Peptostreptococcus, Staphylococcus |
| [3] Sjogren et al., 1997 International endodontic journal | 2 | 2 | 2 | 2 | 3 | 2 | 3 | 0 | 16 | Streptococcus, Propionibacterium, Peptostreptococcus, Eubacterium |
| [40] Debelian, et al. 1995 Endodontics & dental traumatology | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 0 | 15 | Streptococcus, Propionibacterium, Eubacterium |
| [39] Fukushima et al., 1990 Journal of endodontics | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | 10 | Streptococcus, Propionibacterium, Peptostreptococcus, Eubacterium |
| [35] Sundqvist et al., 1989 Journal of endodontics | 3 | 3 | 3 | 1 | 1 | 1 | 2 | 0 | 14 | Streptococcus, Peptostreptococcus, Eubacterium |
| [36] Abou-Rass et al., 1998 International endodontic journal | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 14 | Streptococcus, Propionibacterium, Peptostreptococcus, Staphylococcus |
| Covariate | Coefficients | Lower Bound | Upper Bound | Std. Error | Z-Value | p-Value |
|---|---|---|---|---|---|---|
| Intercept | 2.226 | −6.222 | 10.675 | 4.310 | 0.5164 | 0.605 |
| Risk of bias | −0.140 | −0.724 | 0.444 | 0.298 | −0.4697 | 0.639 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dioguardi, M.; Quarta, C.; Alovisi, M.; Crincoli, V.; Aiuto, R.; Crippa, R.; Angiero, F.; Laneve, E.; Sovereto, D.; De Lillo, A.; et al. Microbial Association with Genus Actinomyces in Primary and Secondary Endodontic Lesions, Review. Antibiotics 2020, 9, 433. https://doi.org/10.3390/antibiotics9080433
Dioguardi M, Quarta C, Alovisi M, Crincoli V, Aiuto R, Crippa R, Angiero F, Laneve E, Sovereto D, De Lillo A, et al. Microbial Association with Genus Actinomyces in Primary and Secondary Endodontic Lesions, Review. Antibiotics. 2020; 9(8):433. https://doi.org/10.3390/antibiotics9080433
Chicago/Turabian StyleDioguardi, Mario, Cristian Quarta, Mario Alovisi, Vito Crincoli, Riccardo Aiuto, Rolando Crippa, Francesca Angiero, Enrica Laneve, Diego Sovereto, Alfredo De Lillo, and et al. 2020. "Microbial Association with Genus Actinomyces in Primary and Secondary Endodontic Lesions, Review" Antibiotics 9, no. 8: 433. https://doi.org/10.3390/antibiotics9080433
APA StyleDioguardi, M., Quarta, C., Alovisi, M., Crincoli, V., Aiuto, R., Crippa, R., Angiero, F., Laneve, E., Sovereto, D., De Lillo, A., Troiano, G., & Lo Muzio, L. (2020). Microbial Association with Genus Actinomyces in Primary and Secondary Endodontic Lesions, Review. Antibiotics, 9(8), 433. https://doi.org/10.3390/antibiotics9080433










