Genetic Subtyping, Biofilm-Forming Ability and Biocide Susceptibility of Listeria monocytogenes Strains Isolated from a Ready-to-Eat Food Industry
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of L. monocytogenes Isolates Collection
2.2. L. monocytogenes Confirmation and Serogrouping by PCR
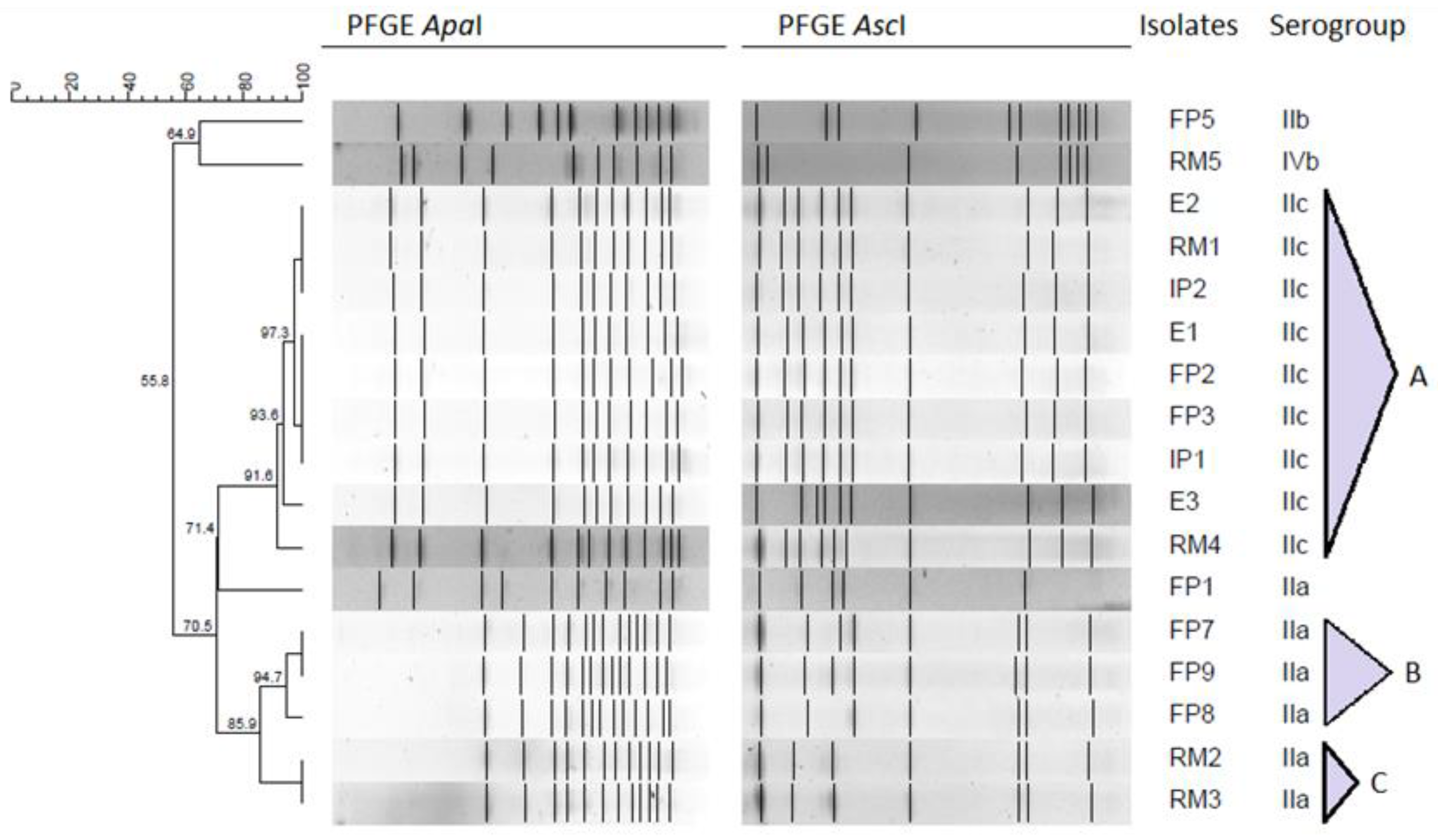
2.3. Pulsed-Field Gel Electrophoresis Typing
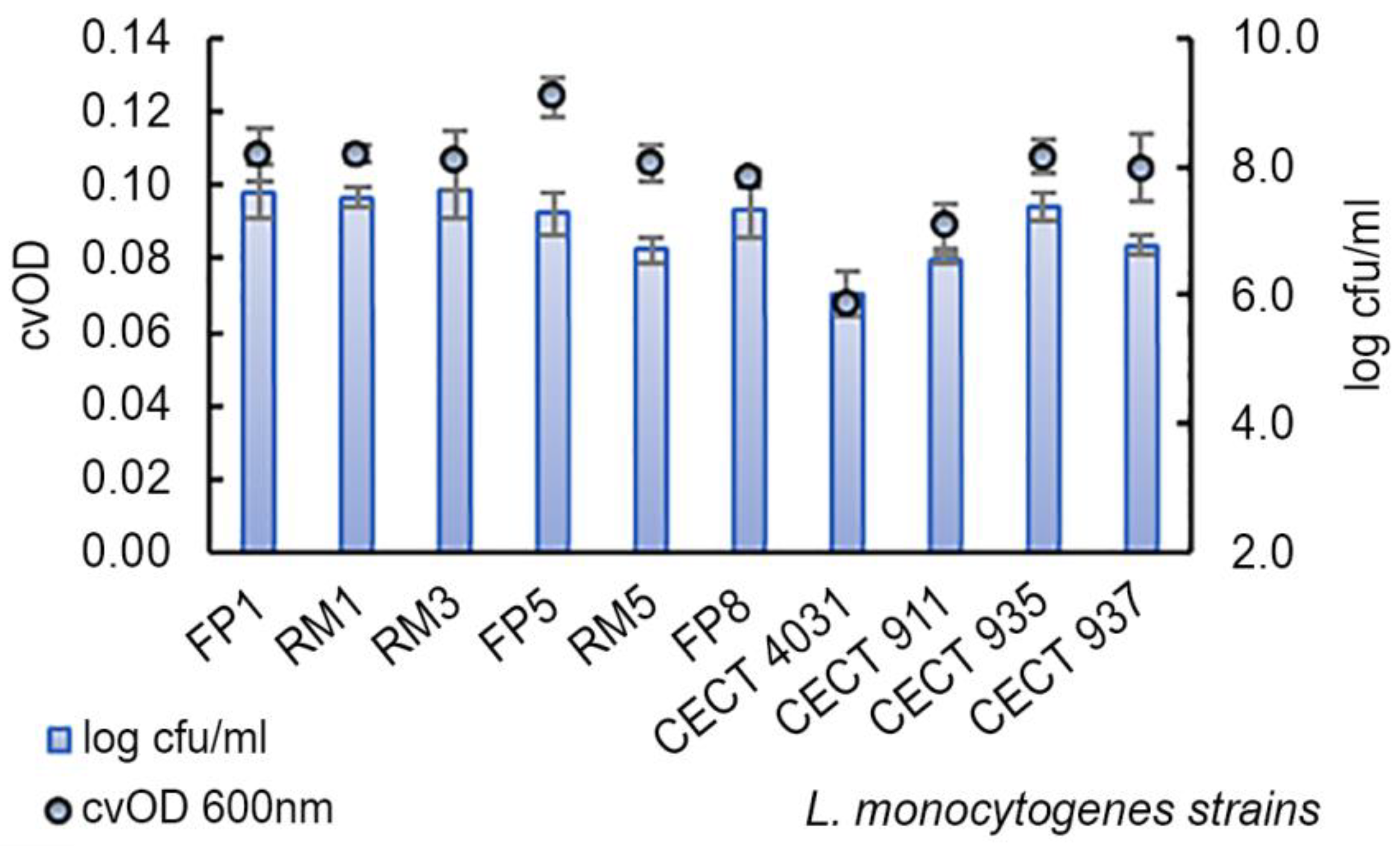
2.4. Biofilm Formation Assay
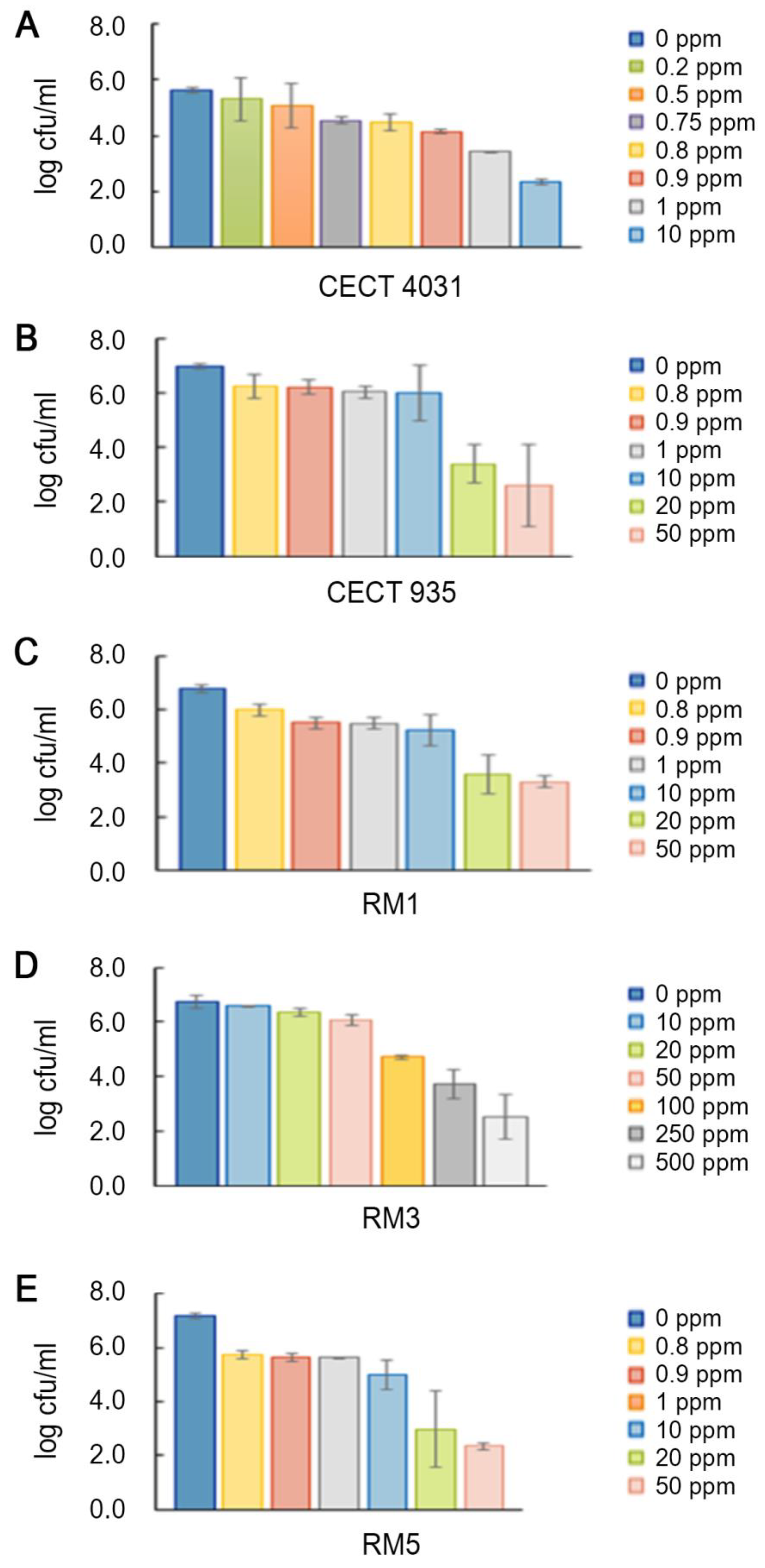
2.5. Biocides Activity Testing Assay
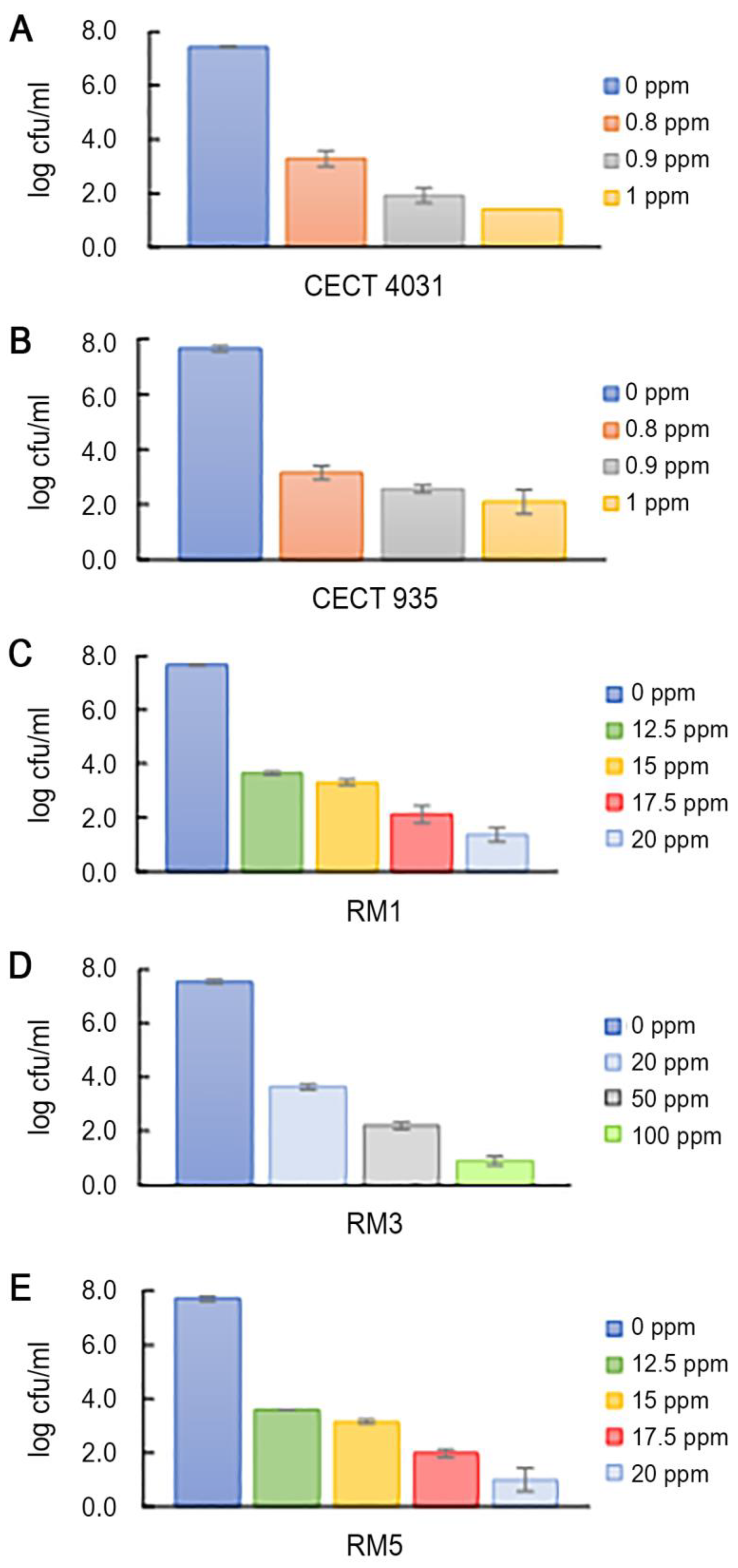
2.5.1. Activity towards L. monocytogenes Planktonic Suspension
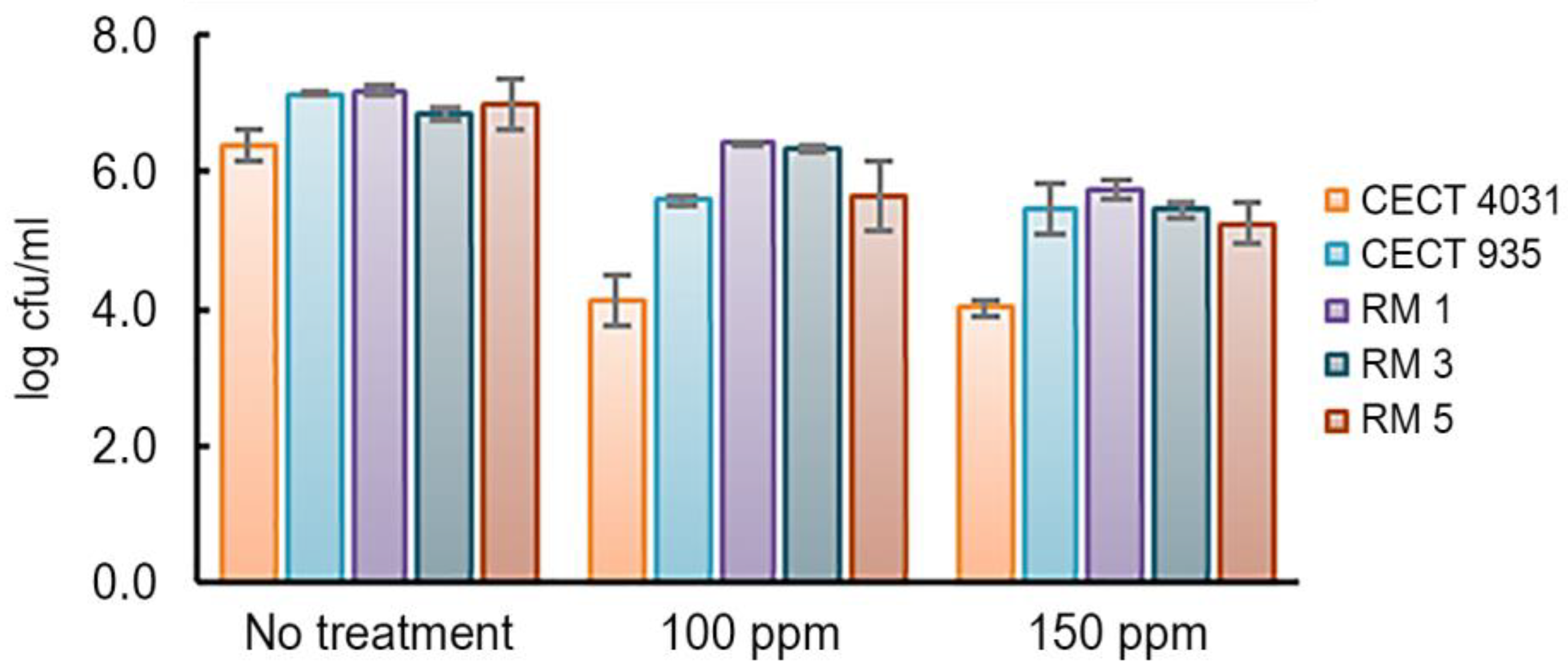
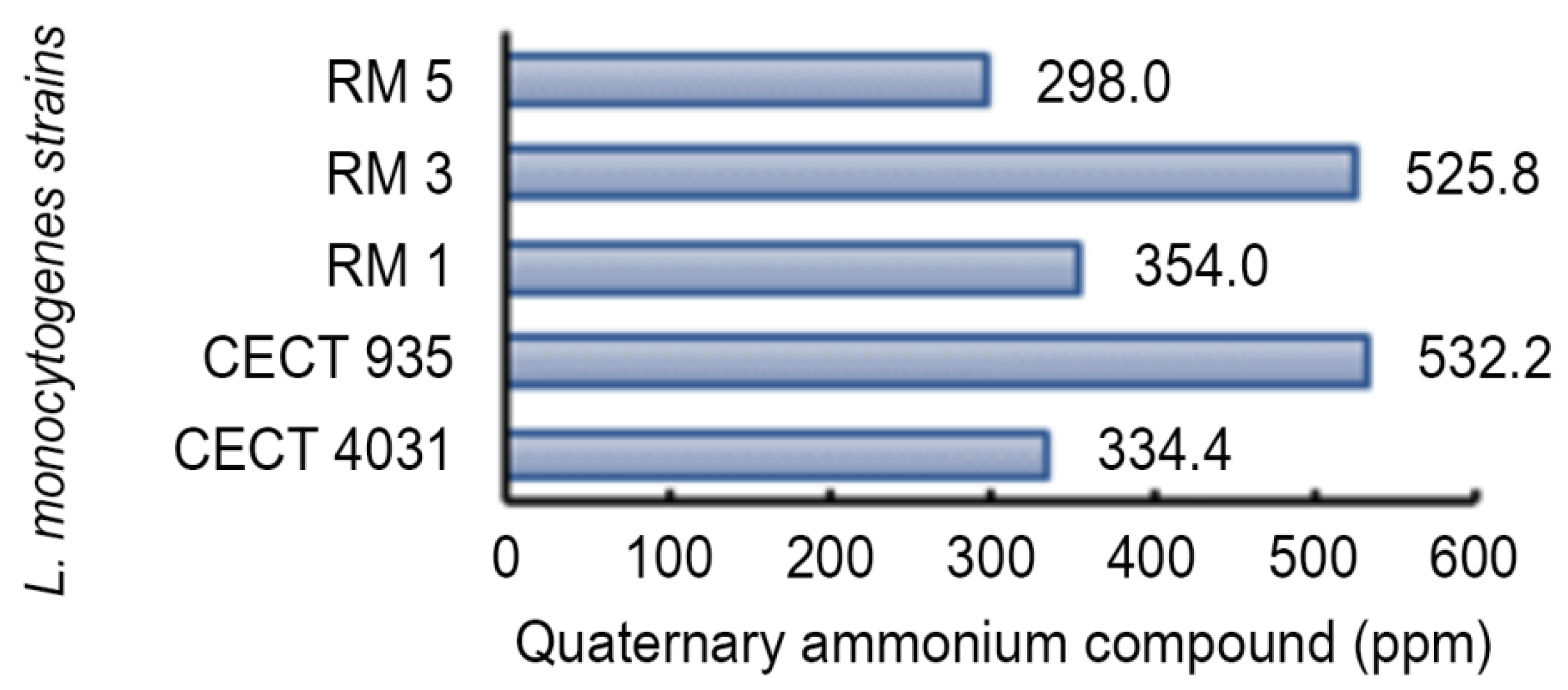
2.5.2. Activity towards L. monocytogenes 5-day-old Biofilms
3. Materials and Methods
3.1. Characterization of L. monocytogenes Isolates Collection
3.2. L. monocytogenes Confirmation and Serogrouping by PCR
3.3. Pulsed-Field Electrophoresis Typing
3.4. Biofilm-Forming Ability Assay
3.4.1. Enumeration of Viable Cells in Biofilms
3.4.2. Biofilm Assessment by Crystal Violet Staining
3.5. Biocides Activity Testing Assay
3.5.1. Activity towards L. monocytogenes Planktonic Suspension
3.5.2. Activity towards L. monocytogenes 5-day-old Biofilms
3.6. Data Analyses
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Gandhi, M.; Chikindas, M.L. Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar] [CrossRef]
- Russo, P.; Hadjilouka, A.; Beneduce, L.; Capozzi, V.; Paramithiotis, S.; Drosinos, E.; Spano, G. Effect of different conditions on Listeria monocytogenes biofilm formation and removal. Czech J. Food Sci. 2018, 36, 208–214. [Google Scholar] [CrossRef]
- Paudyal, R.; Barnes, R.H.; Karatzas, K.A.G. A novel approach in acidic disinfection through inhibition of acid resistance mechanisms; Maleic acid-mediated inhibition of glutamate decarboxylase activity enhances acid sensitivity of Listeria monocytogenes. Food Microbiol. 2018, 69, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernandes, J.A.; Araújo Junior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control. 2014, 35, 386–391. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Nicolau, A.I.; Alvarez-Ordóñez, A.; Borda, D.; Oniciuc, E.A.; Stessl, B.; Gurgu, L.; Wagner, M.; Jordan, K. Dynamics of Listeria monocytogenes colonisation in a newly-opened meat processing facility. Meat Sci. 2016, 113, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Amajoud, N.; Leclercq, A.; Soriano, J.M.; Bracq-Dieye, H.; Maadoudi, M.E.; Senhaji, N.S.; Kounnoun, A.; Moura, A.; Lecuit, M.; Abrini, J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control. 2018, 84, 436–441. [Google Scholar] [CrossRef]
- Henriques, A.R.; Fraqueza, M.J. Listeria monocytogenes and ready-to-eat meat-based food products: Incidence and control. In Listeria monocytogenes: Incidence, Growth Behavior and Control, 1st ed.; Viccario, T., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2015; pp. 71–103. [Google Scholar]
- Olszewska, M.A.; Zhao, T.; Doyle, M.P. Inactivation and induction of sublethal injury of Listeria monocytogenes in biofilm treated with various sanitizers. Food Control. 2016, 70, 371–379. [Google Scholar] [CrossRef]
- Ortiz, S.; López, V.; Martínez-Suárez, J.V. Control of Listeria monocytogenes contamination in an Iberian pork processing plant and selection of benzalkonium chloride-resistant strains. Food Microbiol. 2014, 39, 81–88. [Google Scholar] [CrossRef]
- Ibusquiza, P.S.; Herrera, J.J.; Vázquez-Sánchez, D.; Parada, A.; Cabo, M.L. A new and efficient method to obtain benzalkonium chloride adapted cells of Listeria monocytogenes. J. Microbiol. Methods 2012, 91, 57–61. [Google Scholar] [CrossRef]
- Nilsson, R.E.; Ross, T.; Bowman, J.P. Variability in biofilm production by Listeria monocytogenes correlated to strain origin and growth conditions. Int. J. Food Microbiol. 2011, 150, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, E.; Giaouris, E.D.; Berillis, P.; Boziaris, I.S. Dynamics of biofilm formation by Listeria monocytogenes on stainless steel under mono-species and mixed-culture simulated fish processing conditions and chemical disinfection challenges. Int. J. Food Microbiol. 2018, 267, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, X.; Suo, Y.; Deng, X.; Cheng, M.; Shi, C.; Shi, X. Prevalence, serotype diversity, biofilm-forming ability and eradication of Listeria monocytogenes isolated from diverse foods in Shanghai, China. Food Control. 2017, 73, 1068–1073. [Google Scholar] [CrossRef]
- Doijad, S.P.; Barbuddhe, S.B.; Garg, S.; Poharkar, K.V.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Chakraborty, T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE 2015, 10, e0137046. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, T.; Ripolles-Avila, C.; Hascoët, A.S.; Rodríguez-Jerez, J.J. Effect of an enzymatic treatment on the removal of mature Listeria monocytogenes biofilms: A quantitative and qualitative study. Food Control. 2020, 114, 107266. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Mizan, M.F.R.; Kim, H.S.; Yang, S.; Ha, S.D. Activity of thyme and tea tree essential oils against selected foodborne pathogens in biofilms on abiotic surfaces. LWT 2018, 89, 134–139. [Google Scholar] [CrossRef]
- Puga, C.H.; Orgaz, B.; SanJose, C. Listeria monocytogenes impact on mature or old Pseudomonas fluorescens biofilms during growth at 4 and 20 °C. Front. Microbiol. 2016, 7, 134. [Google Scholar] [CrossRef]
- Tamburro, M.; Ripabelli, G.; Vitullo, M.; Dallman, T.J.; Pontello, M.; Amar, C.F.; Sammarco, M.L. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microbiol. Infect. Dis. 2015, 40, 31–39. [Google Scholar] [CrossRef]
- Henriques, A.R.; Fraqueza, M.J. Biofilm-forming ability and biocide susceptibility of Listeria monocytogenes strains isolated from the ready-to-eat meat-based food products food chain. LWT 2017, 81, 180–187. [Google Scholar] [CrossRef]
- Kocot, A.M.; Olszewska, M.A. Biofilm formation and microscopic analysis of biofilms formed by Listeria monocytogenes in a food processing context. LWT 2017, 84, 47–57. [Google Scholar] [CrossRef]
- Piercey, M.J.; Elis, T.C.; Macintosh, A.J.; Truelstrup, H.L. Variations in biofilm formation, desiccation resistance and benzalkonium chloride susceptibility among Listeria monocytogenes strains isolated in Canada. Int. J. Food Microbiol. 2017, 257, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, Y.; Zahid, M.S.; Yamasaki, S.; Shi, L.; Li, J.R.; Yan, H. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 2014, 190, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Barroso, I.; Maia, V.; Cabrita, P.; Martínez-Suárez, J.V.; Brito, L. The benzalkonium chloride resistant or sensitive phenotype of Listeria monocytogenes planktonic cells did not dictate the susceptibility of its biofilm counterparts. Food Res. Int. 2019, 123, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Mena, C.; Almeida, G.; Sá-Carneiro, L.; Teixeira, P.; Hogg, T.; Gibbs, P.A. Incidence of Listeria monocytogenes in different food products commercialized in Portugal. Food Microbiol. 2004, 21, 213–216. [Google Scholar] [CrossRef]
- Kérouanton, A.; Marault, M.; Petit, L.; Grout, J.; Dao, T.T.; Brisabois, A. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. J. Microbiol. Methods 2010, 80, 134–137. [Google Scholar] [CrossRef]
- Lotfollahi, L.; Chaharbalesh, A.; Ahangarzadeh, R.M.; Hasani, A. Prevalence, antimicrobial susceptibility and multiplex PCR-serotyping of Listeria monocytogenes isolated from humans, foods and livestock in Iran. Microb. Pathog. 2017, 107, 425–429. [Google Scholar] [CrossRef]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef]
- Sheng, J.; Tao, T.; Zhu, X.; Bie, X.; Lv, F.; Zhao, H.; Lu, Z. A multiplex PCR detection method for milk based on novel primers specific for Listeria monocytogenes 1/2a serotype. Food Control. 2018, 86, 183–190. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Ibusquiza, P.S.; Mosquera-Fernández, M.; López-Cabo, M. Listeria monocytogenes-carrying consortia in food industry. Composition, subtyping and numerical characterisation of mono-species biofilm dynamics on stainless steel. Int. J. Food Microbiol. 2015, 206, 84–95. [Google Scholar] [CrossRef]
- Torresi, M.; Acciari, V.A.; Zennaro, G.; Prencipe, V.; Migliorati, G. Comparison of Multiple-locus variable number tandem repeat analysis and pulsed-field gel electrophoresis in molecular subtyping of Listeria monocytogenes isolates from Italian cheese. Vet. Ital. 2015, 51, 191–198. [Google Scholar]
- Nyarko, E.B.; Donnelly, C.W. Listeria monocytogenes: Strain heterogeneity, methods, and challenges of subtyping. J. Food Sci. 2015, 80, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Kurpas, M.; Wieczorek, K.; Osek, J. Ready-to-eat meat products as a source of Listeria monocytogenes. J. Vet. Res. 2018, 62, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Malley, T.J.V.; Butts, J.; Wiedmann, M. Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J. Food Prot. 2015, 78, 436–445. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Datta, A.R.; Burall, L.S. Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Food Microbiol. 2018, 75, 18–27. [Google Scholar] [CrossRef]
- Su, X.; Zhang, J.; Shi, W.; Yang, X.; Li, Y.; Pan, H.; Kuang, D.; Xu, D.; Shi, X.; Meng, J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control. 2016, 70, 96–102. [Google Scholar] [CrossRef]
- Stepanović, S.; Cirković, I.; Ranin, L.; Svabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Meloni, D.; Consolati, S.G.; Mazza, R.; Mureddu, A.; Fois, F.; Piras, F.; Mazzette, R. Presence and molecular characterization of the major serovars of Listeria monocytogenes in ten Sardinian fermented sausage processing plants. Meat Sci. 2014, 97, 443–450. [Google Scholar] [CrossRef]
- Kwasny, S.M.; Opperman, T.J. Static biofilm cultures of gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr. Protoc. Pharmacol. 2010, 13, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Reis-Teixeira, F.B.; Alves, V.F.; Martinis, E.C.P. Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Braz. J. Microbiol. 2017, 48, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Norwood, D.E.; Gilmour, A. The growth and resistance to sodium hypochlorite of Listeria monocytogenes in a steady-state multispecies biofilm. J. Appl. Microbiol. 2000, 88, 512–520. [Google Scholar] [CrossRef]
- Aarnisalo, K.; Lundén, J.; Korkeala, H.; Wirtanen, G. Susceptibility of Listeria monocytogenes strains to disinfectants and chlorinated alkaline cleaners at cold temperatures. LWT 2007, 40, 1041–1048. [Google Scholar] [CrossRef]
- Ibusquiza, P.S.; Herrera, J.J.R.; Cabo, M.L. Resistance to benzalkonium chloride, peracetic acid and nisin during formation of mature biofilms by Listeria monocytogenes. Food Microbiol. 2011, 28, 418–425. [Google Scholar] [CrossRef]
- Sidhu, M.S.; Heir, E.; Sorum, H.; Holck, A. Genetic linkage between resistance to quaternary ammonium compounds and β-Lactam antibiotics in food-related Staphylococcus spp. Microb. Drug Res. 2001, 7, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Romanova, N.A.; Gawande, P.V.; Brovko, L.Y.; Griffiths, M.W. Rapid methods to assess sanitizing efficacy of benzalkonium chloride to Listeria monocytogenes biofilms. J. Microbiol. Methods 2007, 71, 231–237. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Vázquez-Shánchez, D.; López-Cabo, M. Current knowledge on Listeria monocytogenes biofilms in food-related environments: Incidence, resistance to biocides, ecology and biocontrol. Foods 2018, 7, 85. [Google Scholar] [CrossRef]
- Nocker, A.; Sossa, K.E.; Camper, A.K. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 2007, 70, 252–260. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F.; Kathariou, S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 2006, 72, 7711–7717. [Google Scholar] [CrossRef] [PubMed]
- Katharios-Lanwermeyer, S.; Rakic-Martinez, M.; Elhanafi, D.; Ratani, S.; Tiedje, J.M.; Kathariou, S. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other listeriae. Appl. Environ. Microbiol. 2012, 78, 7549–7556. [Google Scholar] [CrossRef]
- Nakamura, H.; Takakura, K.; Sone, Y.; Itano, Y.; Nishikawa, Y. Biofilm formation and resistance to benzalkonium chloride in Listeria monocytogenes isolated from a fish processing plant. J. Food Prot. 2013, 76, 1179–1186. [Google Scholar] [CrossRef]
- Moltz, A.G.; Martin, S.E. Formation of biofilms by Listeria monocytogenes under various growth conditions. J. Food Prot. 2005, 68, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Kastbjerg, V.G.; Laesen, M.H.; Ingmer, H. Influence of sublethal concentrations of common disinfectants on expression of virulence genes in Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Srey, S.; Jahid, I.K.; Ha, S.D. Biofilm formation in food industries: A food safety concern. Food Control. 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Schlafer, S.; Meyer, R.L. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods 2017, 138, 50–59. [Google Scholar] [CrossRef]
- Gómez, D.; Azón, E.; Marco, N.; Carramiñana, J.J.; Rota, C.; Ariño, A.; Yanguela, J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014, 42, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Ha, S.D. The paradox of mixed-species biofilms in the context of food safety. Compr. Rev. Food Sci. Food Saf. 2014, 13, 990–1011. [Google Scholar] [CrossRef]
- Metselaar, K.; Ibusquiza, P.S.; Ortiz, C.A.R.; Krieg, M.; Zwietering, M.H.; den Besten, H.M.; Abee, T. Performance of stress resistant variants of Listeria monocytogenes in mixed species biofilms with Lactobacillus plantarum. Int. J. Food Microbiol. 2015, 213, 24–30. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO). 1290-1:1996. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes; Part 1: Detection Method; International Organization for Standardization: Geneva, Switzerland, 1996. [Google Scholar]
- Graves, L.M.; Swaminathan, B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- European standard (EN). 1276:2009. Chemical Disinfectants and Antiseptics—Quantitative Suspension Test for the Evaluation of Bactericidal Activity of Chemical Disinfectants and Antiseptics Used in Food, Industrial, Domestic and Institutional Areas—Test Method and Requirements (Phase 2, Step 1); International Organization for Standardization: Geneva, Switzerland, 2009. [Google Scholar]

| Date of Collection | Sample Description/Type | Presumptive L. monocytogenes Isolate Code |
|---|---|---|
| July 2010 | Chourição/Final product | FP1 |
| February 2013 | Chouriço/Final product | FP2 |
| March 2013 | Seasoned ham meat/Intermediate product | IP1 |
| March 2013 | In-use meat mincing machine/Equipment | E1 |
| March 2013 | Meat sausage/Final product | FP3 |
| April 2013 | Unseasoned ham meat/Intermediate product | IP2 |
| April 2013 | Seasoned ham meat/Intermediate product | IP3 |
| April 2013 | Pork meat/Raw material | RM1 |
| April 2013 | In-use meat mincing machine/Equipment | E2 |
| April 2013 | Raw meat transport box/Equipment | E3 |
| July 2013 | Pork meat/Raw material | RM2 |
| October 2013 | Chouriço/Final product | FP4 |
| February 2014 | Lard for chouriço/Raw material | RM3 |
| February 2014 | Chouriço/Final product | FP5 |
| April 2014 | Boneless pork shoulder/Raw material | RM4 |
| May 2014 | Chouriço/Final product | FP6 |
| May 2014 | Alheira/Final product | FP7 |
| January 2015 | Boneless pork shoulder/Raw material | RM5 |
| February 2015 | Farinheira/Final product | FP8 |
| April 2015 | Chouriço/Final product | FP9 |
| Serogroup | Proportion | Isolate Code 1 |
|---|---|---|
| IIa | 6 (35.3%) | FP1, RM2, RM3, FP7, FP8, FP9 |
| IIb | 1 (5.9%) | FP5 |
| IIc | 9 (52.9%) | FP2, IP1, E1, FP3, IP2, RM1, E2, E3, RM4 |
| IVb | 1 (5.9%) | RM5 |
| L. monocytogenes Serogroup | n | Log cfu/mL (Mean ± SD) | cvOD (Mean ± SD) |
|---|---|---|---|
| IIa | 4 | 7.2 ± 0.8 | 0.096 ± 0.019 |
| IIb | 2 | 7.0 ± 0.4 | 0.114 ± 0.002 |
| IIc | 2 | 7.0 ± 0.7 | 0.099 ± 0.003 |
| IVb | 2 | 7.1 ± 0.5 | 0.107 ± 0.001 |
| LD90 Values | Log cfu/mL | QaC LD90 |
|---|---|---|
| QaC | 0.652 | 1 |
| BaC | 0.554 | 0.607 |
| Biocide | Tested Concentrations | Neutralizer | ||
|---|---|---|---|---|
| Oxidizing compound (OxC) | 50 ppm | 100 ppm | 150 ppm | Polysorbate 80, 30 g/L + lecithin, 3 g/L + sodium thiosulphate 10 g/L |
| Quaternary ammonium compound (QaC) | 50 ppm | 100 ppm | 150 ppm | Polysorbate 80, 30 g/L + sodium dodecyl sulphate, 4 g/L + lecithin, 3 g/L |
| Ethanol-based compound (EthC) | 50% | 70% | 100% | Polysorbate 80, 30 g/L + saponin, 30 g/L + lecithin, 3 g/L |
| Benzalkonium chloride (BaC) | Planktonic cells | Biofilm | ||
| 0.8–150 ppm | 0.2–500 ppm | Polysorbate 80, 30 g/L + sodium dodecyl sulphate, 4 g/L + lecithin, 3 g/L | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, J.C.; João, A.L.; Alonso, C.d.S.; Barreto, A.S.; Henriques, A.R. Genetic Subtyping, Biofilm-Forming Ability and Biocide Susceptibility of Listeria monocytogenes Strains Isolated from a Ready-to-Eat Food Industry. Antibiotics 2020, 9, 416. https://doi.org/10.3390/antibiotics9070416
Andrade JC, João AL, Alonso CdS, Barreto AS, Henriques AR. Genetic Subtyping, Biofilm-Forming Ability and Biocide Susceptibility of Listeria monocytogenes Strains Isolated from a Ready-to-Eat Food Industry. Antibiotics. 2020; 9(7):416. https://doi.org/10.3390/antibiotics9070416
Chicago/Turabian StyleAndrade, Joana Catarina, António Lopes João, Carlos de Sousa Alonso, António Salvador Barreto, and Ana Rita Henriques. 2020. "Genetic Subtyping, Biofilm-Forming Ability and Biocide Susceptibility of Listeria monocytogenes Strains Isolated from a Ready-to-Eat Food Industry" Antibiotics 9, no. 7: 416. https://doi.org/10.3390/antibiotics9070416
APA StyleAndrade, J. C., João, A. L., Alonso, C. d. S., Barreto, A. S., & Henriques, A. R. (2020). Genetic Subtyping, Biofilm-Forming Ability and Biocide Susceptibility of Listeria monocytogenes Strains Isolated from a Ready-to-Eat Food Industry. Antibiotics, 9(7), 416. https://doi.org/10.3390/antibiotics9070416





