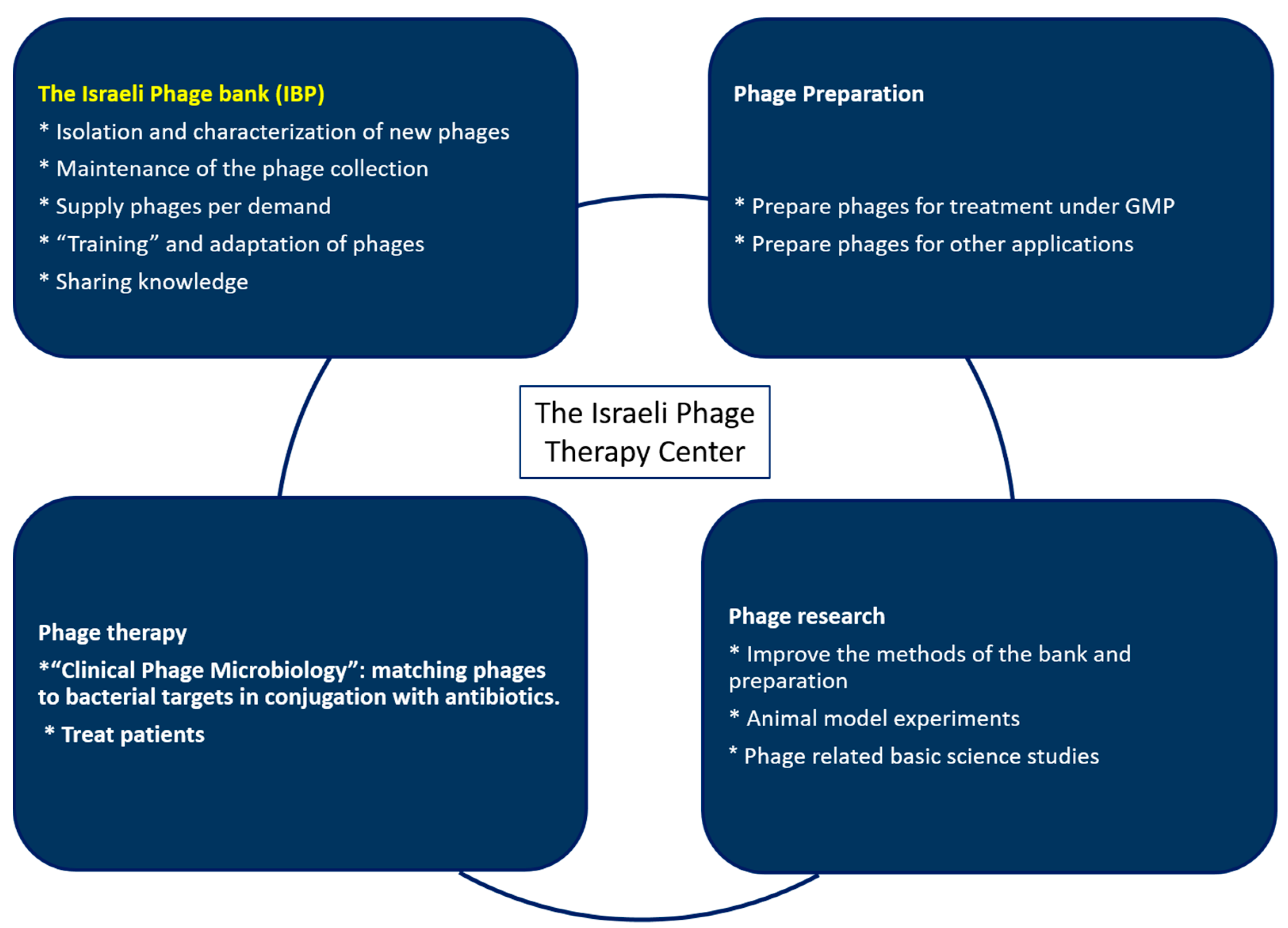
The Israeli Phage Bank (IPB)
Abstract
1. Introduction
2. Methods and Results
2.1. Bacteria Collection
2.2. Phage Isolation
2.3. Phage Characterization
2.4. IBP Phage Collection
2.5. The Phage Database
2.6. Bank Activities
3. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paule, A.; Frezza, D.; Edeas, M. Microbiota and Phage Therapy: Future Challenges in Medicine. Med. Sci. 2018, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, G.W. Bacteriophages: An Appraisal of Their Role in the Treatment of Bacterial Infections. Int. J. Antimicrob. Agents 2007, 30, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An Alternative to Antibiotics in The Age of Multi-Drug Resistance. World J. Gastrointest Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and Tolerability of a Cocktail of Bacteriophages to Treat Burn Wounds Infected by Pseudomonas Aeruginosa (Phagoburn): A Randomised, Controlled, Double-Blind Phase 1/2 Trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Building a Sustainable Phage Biobank—Trials and Tribulations | Nature Research Microbiology Community. Available online: https://naturemicrobiologycommunity.nature.com/users/342533-ruby-cy-lin/posts/59536-building-a-sustainable-phage-biobank-trials-and-tribulations (accessed on 6 March 2020).
- Nir-Paz, R.; Gelman, D.; Khouri, A.; Sisson, B.M.; Fackler, J.; Alkalay-Oren, S.; Khalifa, L.; Rimon, A.; Yerushalmy, O.; Bader, R.; et al. Successful Treatment of Antibiotic-resistant, Poly-microbial Bone Infection With Bacteriophages and Antibiotics Combination. Clin. Infect. Dis. 2019, 69, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages: Methods and Protocols; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Hyman, P. Phages For Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.A.; Beyth, N.; Hazan, R. Targeting Enterococcus Faecalis Biofilms With Phage Therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmy, O.; Coppenhagen-Glazer, S.; Nir-Paz, R.; Tuomala, H.; Skurnik, M.; Kiljunen, S.; Hazan, R. Complete Genome Sequences of Two Klebsiella pneumoniae Phages Isolated as Part of an International Effort. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Shlezinger, M.; Beyth, S.; Houri-Haddad, Y.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Phage Therapy Against Enterococcus Faecalis in Dental Root Canals. J. Oral Microbiol. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Pirnay, J.P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Methods Mol. Biol. 2018, 1693, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Friman, V.P.; Soanes-Brown, D.; Sierocinski, P.; Molin, S.; Johansen, H.K.; Merabishvili, M.; Pirnay, J.P.; De Vos, D.; Buckling, A. Pre-Adapting Parasitic Phages to a Pathogen Leads to Increased Pathogen Clearance and Lowered Resistance Evolution with Pseudomonas Aeruginosa Cystic Fibrosis Bacterial Isolates. J. Evol. Biol. 2016, 29, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
| Phage Collection Name | Number of Phages | Hosts | Link |
|---|---|---|---|
| Adaptive phage therapy (APT) phage bank | ~1000 | Mainly the ESKAPE pathogens | http://www.aphage.com/the-science/ |
| The Felix d’Hérelle Reference Center for Bacterial Viruses | >400 | A few dozen hosts | https://www.phage.ulaval.ca/en/phages-catalog/ |
| The Bacteriophage Bank of Korea | ~1000 | A few dozen hosts | http://www.phagebank.or.kr/intro/eng_intro.jsp |
| Leibniz Institute—DSMZ (German Collection of Microorganisms and Cell Cultures) | ~300 | Unknown | https://www.dsmz.de/collection/collection-experts |
| ATCC Bacteriophage Collection | ~400 | A few dozen hosts | https://www.atcc.org/search#q=phage&sort=relevancy&f:productcategoryFacet=[Bacteria%20%26%20Phages]&f:listofapplicationsFacet=[Bacteriophage] |
| NCTC Bacteriophage Collection | >100 | Streptococcus ssp Staphylococcus ssp Campylobacter | https://www.phe-culturecollections.org.uk/products/bacteria/bacteriophages.aspx |
| Hatfull Lab Phage Collection | >15,000 | Species from Actinobacteria phylum | https://phagesdb.org/ http://www.hatfull.org/sea-phages |
| TUDelft | Unknown | Unknown | https://www.tudelft.nl/en/delft-university-fund/ |
| P.H.A.G.E | Unknown | Unknown | http://www.p-h-a-g-e.org/ |
| Israeli Phage Bank (IPB) | >300 | 16 different species | https://ronenhazanlab.wixsite.com/hazanlab/the-404-israeli-phage-bank |
| Bacteria | Number of Srains |
|---|---|
| Acinetobacter baumanii | 12 |
| Bacillus anthracis | 1 |
| Burkholderia cepacia | 2 |
| Burkholderia contaminans | 2 |
| Burkholderia lata | 1 |
| Others Burkholderia ssp | 20 |
| Enterobacter cloacae | 8 |
| Enterococcus faecalis | 15 |
| Enterococcus faecium | 10 |
| Escherichia coli | 8 |
| Klebsiella pneumoniae | 10 |
| Mycobacterium abscessus | 6 |
| Propionibacterium acnes | 36 |
| Providencia rettgeri | 21 |
| Providencia stuartii | 20 |
| Pseudomonas aeruginosa | 120 |
| Pseudomonas stutzeri | 2 |
| Pseudomonas syringae | 1 |
| Salmonella enterica | 2 |
| Shigella ssp | 30 |
| Staphylococcus aureus | 40 |
| Streptococcus mutans | 15 |
| Total | 382 |
| Bacteria Strain | Clinical Strains | Phages | Percentage of Coverage | Minimum Phages Require for Coverage |
|---|---|---|---|---|
| Staphylococcus aureus | 20 | 25 | 100% | 1 |
| Klebsiella pneumoniae | 8 | 30 | 100% | 2 |
| Pseudomonas aeruginosa | 100 | 54 | 100% | 9 |
| Pseudomonas stutzeri | 2 | 6 | 100% | 1 |
| Enterobacter spp | 7 | 20 | 100% | 3 |
| Burkholderia spp | 23 | 15 | 90% | 5 |
| Burkholderia lata | 1 | 2 | 100% | 1 |
| Enterococcus faecalis + Enterococcus faecium | 8 | 38 | 100% | 1 |
| Streptococcus mutans | 15 | 1 | 100% | 1 |
| Acinetobacter baumannii | 8 | 19 | 100% | 2 |
| Propionibacterium acnes | 36 | 22 | 86% | 1 |
| Providencia spp | 35 | 10 | 100% | 3 |
| Escherichia coli | 5 | 25 | 100% | 1 |
| Bacillus anthracis | 1 | 6 | 100% | 1 |
| Salmonella | 30 | 1 | 100% | 1 |
| Shigella | 2 | 12 | 100% | 1 |
| Mycobacterium abscessus | 6 | 3 | 100% | 1 |
| Total | 307 | 289 | 97% | 35 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yerushalmy, O.; Khalifa, L.; Gold, N.; Rakov, C.; Alkalay-Oren, S.; Adler, K.; Ben-Porat, S.; Kraitman, R.; Gronovich, N.; Shulamit Ginat, K.; et al. The Israeli Phage Bank (IPB). Antibiotics 2020, 9, 269. https://doi.org/10.3390/antibiotics9050269
Yerushalmy O, Khalifa L, Gold N, Rakov C, Alkalay-Oren S, Adler K, Ben-Porat S, Kraitman R, Gronovich N, Shulamit Ginat K, et al. The Israeli Phage Bank (IPB). Antibiotics. 2020; 9(5):269. https://doi.org/10.3390/antibiotics9050269
Chicago/Turabian StyleYerushalmy, Ortal, Leron Khalifa, Naama Gold, Chani Rakov, Sivan Alkalay-Oren, Karen Adler, Shira Ben-Porat, Reut Kraitman, Niv Gronovich, Kerem Shulamit Ginat, and et al. 2020. "The Israeli Phage Bank (IPB)" Antibiotics 9, no. 5: 269. https://doi.org/10.3390/antibiotics9050269
APA StyleYerushalmy, O., Khalifa, L., Gold, N., Rakov, C., Alkalay-Oren, S., Adler, K., Ben-Porat, S., Kraitman, R., Gronovich, N., Shulamit Ginat, K., Abdalrhman, M., Coppenhagen-Glazer, S., Nir-Paz, R., & Hazan, R. (2020). The Israeli Phage Bank (IPB). Antibiotics, 9(5), 269. https://doi.org/10.3390/antibiotics9050269





