3. Discussion
Cancer patients are known to be susceptible to various nosocomial infections due to the destructive complications of cancer treatment on their immune system [
4]. Urinary tract infection (UTI) is one of the major causes of morbidity in cancer patients.
E. coli was the most common organism isolated in cancer patients with UTI [
3].
In this study, the urine culture was taken from cancer patients, which showed that 71.7% were positive. This result is similar to that reported by Tancheva et al. [
9] in Varna where the rate was 68%. However, it is higher than that reported by Yakovlev et al. [
10] in India, where the rate was 33.4% and Raad et al. [
11] in Finland found that UTI was present in 12.5% of cancer patients.
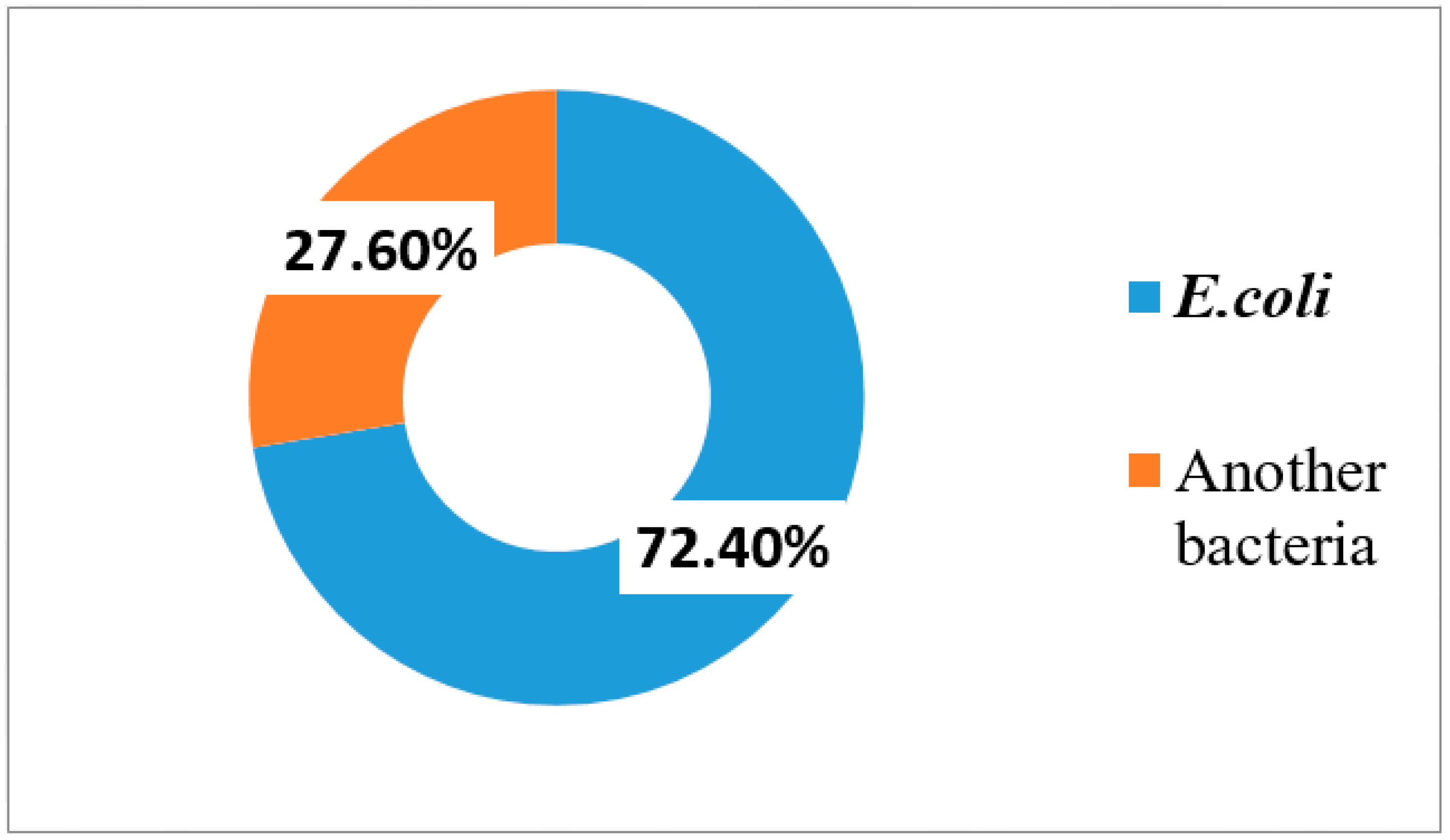
This study revealed that the main isolated organisms from urine culture taken from cancer patients were
Escherichia coli (72.4%). This is comparable with previous studies reported by Bhusa et al. [
12] in the USA, Tancheva et al. [
9] in Varna, Mukta et al. [
13] in India, and Chandra et al. [
14] in India but with lower rates than our study, where the rates were 69.5%, 64.7%, 38.09%, 37.5%, respectively.
Antibiotic resistance is a major clinical problem when treating UTI in cancer patients caused by UPEC. The resistance to imipenem seen in our study, which is more than any other previous study, perhaps finds a logical explanation due to the frequent use of imipenem as routine treatment for resistant strains. Other studies reported lower rates of resistance to imipenem, such as Sedighi et al. [
15] in Iran, who reported it to be 3.3% and Mukta et al. [
13] in Bulgaria, who found the resistance to be 9%. In addition, Elsayed et al. [
16], in Egypt, reported that the resistance rate was 2%. The lower resistant rates for imipenem are probablybecause it is a very powerful drug used only in hospital settings and not as first-line therapy in out-patients clinics [
17].
In our study,
E. coli isolates exhibited maximal resistance against ceftriaxone. This finding is quite challenging because ceftriaxone is a commonly-used empirical therapy in most hospitals. This result is similar to that reported by Mahgoub et al. [
18] in Egypt, where the rate was 79.6%. However, the result in this study is higher than that reported by Abdel-Moaty et al. [
19] in Egypt and Khan et al. [
20] in Bangladesh, where the rates were 61% and 41.9%, respectively.
The regional variations of resistance to antibiotics may be explained by different local antibiotic practices. The influence of inappropriate antibiotic use on the event of antibiotic-resistant strains, especially broad-spectrum agents, has been proven through empirical observation [
21].
Extended-spectrum β-lactamase (ESBL) production is an important resistance mechanism that inhibits the antimicrobial actions against infections caused by Enterobacteriaceae. ESBLs are considered a serious threat to the currently available antimicrobial agents [
22]. The prevalence of bacteria producing ESBLs varies worldwide, with reports from North America, Europe, South America, Africa and Asia [
23].
Preliminary detection of ESBL-producing
E. coli isolates was done by screening tests according to CLSI (2016) that depend on reduced susceptibility to one or more of cefotaxime, ceftazidime, aztreonam, cefotaxime or ceftriaxone. Accordingly, 46.63% (51/110) of UPEC isolates were considered as potential ESBL producers. This is comparable with Shakya et al. [
24] in Nepal who reported that 43.8% of UPEC isolates were potential ESBL producers by screening tests.
In addition, a similar result was obtained by Alqasim et al. [
25] in Saudi Arabia who reported that 41% of UPEC isolates were potential ESBL producers, but a higher rate of potential ESBL-producing UPEC was reported by Al-Mayahie et al. [
26] in Iraq, Thabit et al. [
27] in Egypt and Mukherjee et al. [
28] in India, where the rates were 80.2%, 76.47, 70%, respectively. For the ESBL confirmatory double-disk synergy test, DDST detected ESBLs in 42/110 (38.18%). This percentage is similar to the result by Alqasim et al. [
25] in Saudi Arabia who reported that 33.3% of UPEC isolates were confirmed ESBL producers by DDST. Furthermore, Islam et al. [
29] in Bangladesh reported a comparable result where 32% of UPEC isolates were confirmed ESBL producers by the same test.
Higher levels of ESBL production were reported by Al-Mayahie et al. [
26] in Iraq, Chandra et al. [
14] in India, Al-Agamy et al. [
30] in Egypt, Mekki et al. [
31] in Sudan, and Abayneh et al. [
32] in Southwest Ethiopia. The rates reported for each study were 64.8%, 62.5%, 60.9%, 53% and 76.5%, respectively. In contrast, lower results were mentioned by Sedighi et al. [
15] in Iran (27.3%) and Villanueva et al. [
33] in the Philippines (12.4%).
In our study, a higher degree of resistance was shown by ESBL producers than ESBL non-producers. The obtained results revealed that the resistance level to all cephalosporins (cefotaxime, ceftazidime, ceftriaxone, and cefepime) and aztreonam was significantly higher in ESBL-producing
E. coli in comparison with non-ESBL-producing isolates (
p < 0.001). This finding is in accordance with other reports, such as Islam et al. [
29] in Bangladesh, and Abdel-Moaty et al. [
19] in Egypt.
In the current study, ESBL-producing isolates exhibited significantly higher resistant rates to non-β-lactamase antimicrobials agents including fluoroquinolone, aminoglycosides, tetracycline and trimethoprim/sulfamethoxazole, compared to non-ESBL-producing isolates. The possible explanation for this observation may be that ESBLs are encoded on plasmids and can be mobile and therefore, easily transmissible as resistance gene elements for other antimicrobials from one organism to another.
In the present study, the genotyping of ESBL-producing UPEC isolates was done by PCR to determine the most common ESBL genes responsible for resistance. We reported that CTX-M was the main ESBL type (52%), followed by TEM (40%), then SHV (17%). Twelve isolates had more than one type of ESBL, where CTX-M + TEM were found in 7 (16.67%) isolates, CTX-M + SHV were observed in 11.9% of isolates and 4.76% of isolates had TEM + SHV genes. The same order of gene type presence, but with different percentages, was reported by Chakraborty et al. [
34] in India, where the CTX-M gene was detected in 88% of
E. coli, followed by TEM (19%) and SHV (2%). In addition, the same pattern was mentioned by Zhao et al. [
35] in China, who reported that the rate of CTX-M, TEM, and SHV among
E. coli isolates was 42.5%, 4.2%, and 0.8%, respectively.
The increase of consumption of cefotaxime and ceftazidime could have contributed to the emergence of CTX-M enzymes encoding genes among
E. coli strains in Egyptian hospitals [
36].
CTX-M β-lactamases constitute a novel and rapidly growing family of plasmid-mediated ESBLs, which are currently replacing mutant TEM or SHV ESBL families [
6].
In contrast, Azargun et al. [
37] in Iran, reported that the TEM gene was the major ESBL gene in UPEC isolates, followed by the CTX-M gene and the SHV gene, where the rates for TEM, CTX-M, and SHV were 75.6%, 78.6% and 33.3%, respectively.
In our study, Triplex PCR-based phylogenetic analysis was carried out for EP-UPEC isolates according to the method described by Clermont et al. [
7]. Phylogenetic grouping revealed that most of the isolates belonged to the B2 group (
n = 18, 43%), followed by group D (
n = 15, 36%), group A (
n= 8, 19%) and group B1 (2%). The majority of studies concerning the phylogenetic grouping among UPEC have reported a similar distribution, such as Zhao et al. [
35] in china, Abdi et al. [
38] in Iran, Lee et al. [
39] in Korea, Johnson, and Stell [
40] in Minnesota, Picard et al. [
41] in France, Ejrnæs et al. [
42] in Denmark, Kanamaru et al. [
43] in Japan, and Alghoribi et al. [
44] in England.
However, a few studies have reported a different distribution of phylogenetic groupings for UPEC isolates, such as Marialouis et al. [
45] in India who found that most of the isolates belonged to B2, followed by A, B1, and D, and phylogenetic group D isolates were the least frequent. In addition, Bashir et al. [
46] in Pakistan observed the same finding with their phylogenetic analysis.
In contrast, some studies observed that most of the UPEC isolates belonged to phylogenetic group D, followed by A, B1, and B2, such as Adwan et al. [
47] in Palestine and Abdallah et al. [
48] in china, where phylogenetic group B2 isolates were the least frequent phylogenetic group.
The results of drug resistance according to phylogenetic groups are shown in
Table 4. Our findings showed that group B2 was the most predominant phylogenetic group and most resistant strain to commonly used antibiotics among patients. This finding is in agreement with other studies such as Iranpour et al. [
49] in Iran. However, Bashir et al. [
46] in Pakistan reported that group B2 isolates exhibited lower levels of drug resistance than our study.
In our study, group D isolates were totally resistant to ceftriaxone, aztreonam, and ceftazidime, but highly resistant to cefotaxime (93.3%), cefepime (86.8%), sulfamethoxazole/trimethoprim (73.33%) and ciprofloxacin (66.67%), and less resistant to gentamicin (46.4%), imipenem (33.3%), amikacin (20%), nitrofurantoin (20%) and levofloxacin (40%). This result is similar to Bashir et al. [
46]. In contrast, Iranpour et al. [
49] in Iran found that group D isolates exhibited a low level of drug resistance.
In this study, Group A isolates were totally sensitive to imipenem and sulfamethoxazole/trimethoprim, less resistant to gentamicin (25.4%), amikacin (12.5%), nitrofurantoin (37.5%), levofloxacin (37.5%), colistin (12.5%) and ciprofloxacin (12.5%), and highly resistant to ceftriaxone (87.5%), ceftazidime (75%), cefotaxime (87.5%), cefepime (62.5%) and aztreonam (50%). In contrast, Bashir et al. [
46] in Pakistan found that group A isolates showed a higher level of drug resistance than our study.
In our study, we observed a significant difference between phylogenetic groups and resistance to different groups of antibiotics. This finding is similar to the study reported in china by Wang et al. [
50]. In contrast, Cristea et al. [
51] in Romania found that their statistical analyses did not reveal any statistical significance of the correlation between antibiotic resistance and
E. coli phylogenetic groups.
The frequency
of blaCTX-M,
blaTEM and
bla SHV ESBL genes in phylogenetic groups is summarized in
Table 5. Our phylogenetic analysis of the isolates revealed that strains harboring CTX-M gene were associated with the D phylogenetic group and there is a significant difference was observed in the frequency of CTX-M type between the phylogenetic groups.