Abstract
Staphylococcus aureus is a common bacterial colonizer of humans and a variety of animal species. Many strains have zoonotic potential, moving between humans and animals, including livestock, pets, and wildlife. We examined publications reporting on S. aureus presence in a variety of wildlife species in order to more cohesively review distribution of strains and antibiotic resistance in wildlife. Fifty-one studies were included in the final qualitative synthesis. The most common types documented included ST398, ST425, ST1, ST133, ST130, and ST15. A mix of methicillin-resistant and methicillin-susceptible strains were noted. A number of molecular types were identified that were likely to be found in wildlife species, including those that are commonly found in humans or other animal species (including livestock). Additional research should include follow-up in geographic areas that are under-sampled in this study, which is dominated by European studies.
1. Introduction
Staphylococcus aureus is a common commensal bacterium that lives within the nares, throat, and on the skin of humans and a wide variety of animal species. S. aureus can spread via person-to-person contact (directly or mediated by fomites) and can be transmitted zoonotically via direct contact with animals or animal products, including raw meats [1]. It can also be maintained in the environment in manure, water, or the air [2].
Because of its frequency in various environments and species, it is critical to understand movement within and between communities. S. aureus is frequently resistant to one or more classes of antibiotics, and the continued spread of methicillin-resistant S. aureus (MRSA) over the past several decades in both human and animal species has increased the risk of acquiring a resistant infection that makes treatment more difficult and costly [3].
The epidemiology of MRSA in particular has changed over the past 30 years [4]. Once primarily a hospital-associated pathogen, the rise of novel strains of MRSA in the 1990s outside of the nosocomial environment led to the recognition of “community-associated MRSA” (CA-MRSA), in contrast with the historic hospital-associated (HA-MRSA) strains [5]. In the mid-2000s, a third genre of MRSA was recognized, as colonization and infection of livestock and livestock workers led to the designation of livestock-associated MRSA (LA-MRSA) [6]. It should be noted that we know less about the changes in methicillin-susceptible S. aureus during this period (MSSA), as the bulk of surveillance is dedicated to MRSA and does not always capture MSSA epidemiology.
Wildlife are a special case and often under-studied in the epidemiology of antibiotic resistance in the community and environment. Wildlife can act as reservoirs for intrinsic resistance elements or organisms (those that are naturally occurring in the environment, including environmental bacteria and fungi living in soil and water) and may also be exposed to resistant organisms or resistance genes in the environment amplified via human activity. This may be via treated humans who excrete resistant bacteria, antimicrobials that eventually end up in sewage effluent dispersed into the environment, or from sludge or waste from humans or livestock dispersed on fields as fertilizer. This may lead to further dissemination into streams, rivers, and larger waterways and also allow for airborne transmission of dried materials. Resistance may also be generated in the environment due to spraying of antibiotics on citrus groves and other plants [7] as well as by use in aquaculture [8].
S. aureus is a commensal organism that is able to effectively colonize a wide variety of host species, including many mammalian species but also birds and fish. As such, animals besides human have the potential to harbor novel strains of S. aureus, which could enter the human population, or conversely, humans may also transmit strains of S. aureus to other animal species [9], which can then acquire additional resistance genes.
The clearest evidence of zoonotic transmission of S. aureus has been in livestock. Isolates of clonal complex 398 appear to have originated as a human-adapted lineage but were transmitted to livestock including pigs and cattle and have become both more antibiotic-resistant (including MRSA strains) and have also typically lost some human virulence factors [9]. A similar situation appears to have happened with CC5 in poultry [10]. Recent research also suggests an emerging lineage in humans, strains of CC130, originated in cattle, and typically carry a novel methicillin resistance gene (mecC, originally called mecALGA251) [11]. The role other animals may play in such cross-species transmission is less defined. Systematic collection and molecular typing of S. aureus from animal species has not been a priority; as such, cross-species transmission events from such animals to humans or wildlife to better-studied animal populations (including livestock and poultry) have likely been missed. This review examines the epidemiology of S. aureus in wildlife, including molecular typing and antibiotic resistance data where available.
2. Results
2.1. Search Results
Searching within Pubmed resulted in 856 hits, Web of Science in 58, and peer-reviewed materials within ProQuest in 918, for a total of 1832 publications. Upon searching references for additional studies that had been missed by our search terms, another nine were added. Titles and abstracts were examined to exclude duplicates (956); this left 885 remaining. Additional publications were excluded if they used animals only as an experimental model rather than examined epidemiology in wild species (such as rats, guinea pigs, and rabbits) and those that only mentioned S. aureus within the discussion or otherwise rather than consisted of a study focused on S. aureus epidemiology in wildlife. This left 69 for analysis (Figure 1).
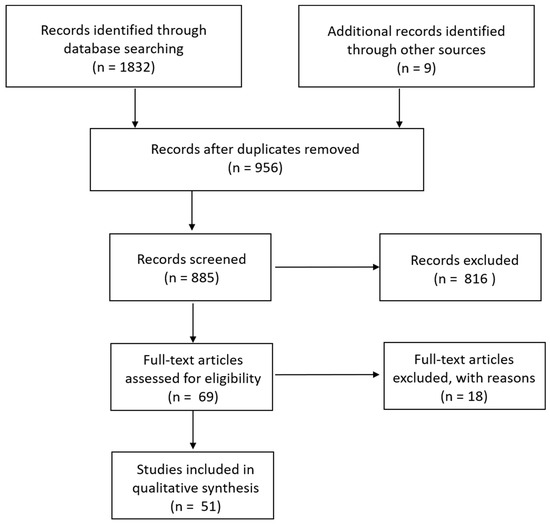
Figure 1.
Schematic of search strategy.
2.2. Full-Text Articles Excluded
Eighteen studies were included within the initial analysis but excluded from further analysis Table 1 due to lack of detail reported regarding the S. aureus detected. These studies included the identification of S. aureus in a white ibis in Egypt [12], captive bustards in the United Arab Emirates [13], a peregrine falcon (Falco peregrinus) in Spain [14], from “free living insectivores” including the common shrew, lesser shrew, bank vole, root vole, and field mouse [15]; S. aureus was reported in this study but were not typed nor reported which species were positive. In Brazil, an opossum with mastitis was described but neither molecular typing nor antibiotic resistance phenotype was provided [16]. Similarly, a systemic S. aureus infection in a raccoon was also reported but not further characterized [17]. A 2013 study suggests that S. aureus infection is an important skin disease of red squirrels (Sciurus vulgaris) in Great Britain [18], and a Canadian publication determined that S. aureus was a common organism found in bite wounds from Norway and Black rats (Rattus norvegicus and Rattus rattus, respectively) [19], but no details were provided in either paper. S. aureus was also found along with other mecA-positive staphylococci in foxes in the United Kingdom, but samples were not typed [20]. S. aureus-positive Spanish Ibex (Capra pyrenaica hispanica) were identified in Spain but not typed or examined phenotypically [21]. A black rhinocerous (Diceros bicornis) in Kenya was reported to have an S. aureus infection (a possible cause of mortality) but also lacks details [22]. Bighorn sheep (Ovis canadensis nelson and Ovis canadensis mexicana) in Arizona were found to be colonized with S. aureus [23], but it was not characterized. S. aureus was identified in fecal samples taken from red deer in Poland [24] and from fecal samples from slaughtered reindeer in Finland and Norway [25] but was not further characterized. Finally, S. aureus was isolated from bottlenose dolphins (Tursiops truncatus) in the southeastern United States, but it was not further characterized [26,27]. Multiple zoo animals in Belgium were tested for MRSA, but no positive samples were reported [28]. Schaumburg [29] was not included in Table 1 because species are not specific (monkey, goat, etc. rather than exact species types) but demonstrates some sharing of spa types between humans, domestic animals, and wildlife (more for the former than the latter).

Table 1.
Summary of wildlife data.
2.3. Molecular Types
An examination of the molecular types found in wildlife demonstrates an extensive diversity of types of S. aureus. However, some broad conclusions can be suggested. Though comparisons across publications are difficult due to divergent methodology of sampling, testing, and geography, Figure 2 illustrates the most common molecular types, according to the total count publications identifying them. These molecular types include a mix of human pandemic types (ST5, ST8, ST1, ST30, ST22) [82] and molecular types that have been more commonly described in animals or at the animal–human interface (ST398, ST130, ST133, ST425) [83,84].

Figure 2.
Molecular types identified in multiple papers and associated species. Sources of photos are included in Appendix A.
3. Discussion
This review demonstrates a significant amount of diversity in Staphylococcus aureus sampled from a wide variety of wildlife species across several continents. Populations of S. aureus present in wildlife may serve as reservoirs that could be transmitted to nearby domestic livestock or poultry or directly or indirectly to humans. Such a reservoir of S. aureus in the environment may also contribute to the exchange of antibiotic resistance or virulence genes among human or animal S. aureus, potentially leading to novel strains.
The continuing encroachment of humans into animal spaces due to agriculture, deforestation, climate change can lead to “spillovers” of pathogens from one species to another [85]. Most commonly we examine this with wildlife as a reservoir and humans as the affected species (e.g., Ebola, Nipah, MERS, SARS). However, transmission may also occur in reverse, with humans seeding wildlife with pathogens [86,87]. In the case of S. aureus, it appears both may be occurring, as has been previously documented among livestock [9]. In the case of antibiotic-resistant pathogens, such bidirectional transmission may be direct, via contact between human and animal species. More likely in the case of wildlife species, transmission may be indirect, such as via environmental reservoirs of pathogens including water sources, soil, exposure to manure, air, and contact with contaminated fomites [88,89]. Transmission may also occur due to consumption of meat products contaminated with S. aureus, but sampling wildlife meat products is exceedingly difficult and has not been done in a systematic manner. Meat products from livestock are a potential way that livestock-associated strains of S. aureus may spread from farms to communities [1,89], but the impact of meat from wildlife sources (including various deer species and wild boar) which may play a role in transmission of S. aureus bacteria or resistant genes is less clear.
While few studies reviewed here examine the environment and wildlife at the same time, a study by Porrero et al. [90] found mecC-positive S. aureus in river water after the area had been found to be positive for ST425-mecC in wild boar and fallow deer at the same location [54], suggesting a shared source of exposure or transmission between the various animal species and/or the environment.
Indeed, ST425 is a dominant molecular type that was found in wildlife papers. It does not appear to have a particular host specificity, with isolation reported from mammals including rabbits [35], boar [35,40,53,55,57,58], red deer [35,40,54], and roe deer [40,44] and from vultures [75] (see also Table 1 and Figure 1); these were found exclusively in European countries. The significance of this finding is currently unknown. ST425 isolates are known to be zoonotic, and have been described as a human colonizer as well, and its ability to cross species barriers may facilitate transmission of resistance genes, including mecC [11]. Other key molecular types present in a wide variety of species included ST398 in Norway rats [30,31,32,33,34], brown hare [40], boar [53,54,56], red deer [53,54], Iberian ibex [54], vulture [75], white stork [76], Eurasian griffon vulture [54], and Canada goose [79] and ST130/CC130, found in a rabbit [35], hedgehog [37,38,40], wood mouse [41], brown rat [40], yellow-necked mouse [42], house mouse [42], brown hare [37,40], mara [48,49], red fox [40], boar [35,55], red deer [35,59], Iberian ibex [53], fallow deer [40], and blue-winged teal [44]. The latter includes a large number of small animals and rodents, suggesting these may be an important reservoir in addition to livestock [91], and ST398 is a known colonizer of humans, particularly those with livestock contact [84]. Colonization may result in transmission of antibiotic resistance genes between species, while ST398 is also capable of causing a wide range of infections in humans [92].
Interestingly, bats and non-human primates seem to have little overlap with other animal strains. Bat molecular types consisted primarily of newly identified spa and/or MLST, though ST15 was reported twice—in a straw-colored fruit bat in Nigeria, and a captive Egyptian fruit bat sampled in Denmark [49,50].
For primate S. aureus, the papers reviewed here represent a mix of primates raised in captivity (including zoos and research facilities) and those sampled in sanctuaries and parks. As such, intensity of contact with humans who may be carrying typical human strains of S. aureus varies widely, and the importance of cross-species transmission remains in debate. Human-to-primate transmission was suggested in a study of wild primates MRSA in Nepal [63] and primates in Gabon [70]. The reverse was suggested by examination of an ST3268 strain found in macaques in primate research facilities in Singapore [68] and the United States [66,67]; this molecular type was also found in macaques in a New York research facility [65], suggesting the need for screening of animals prior to export/import. While most reports suggest preponderance of primate-associated strains, testing in a Texas facility found that their animals were colonized primarily with USA300/ST8 strains, which are common in humans and suggestive of human-to-animal transmission. However, workers at the facility were not tested for carriage [72].
Though S. aureus strains were typically taken as colonizers from healthy animals, several primates were actively infected with S. aureus. A gorilla in a primate center in Gabon was found to have a large lesion on his back; the gorilla died suddenly, and autopsy also found S. aureus in tissue samples; all were spa type t148 [70]. Though this is a human-associated strain, sampling of caretakers did not show any colonized humans involved in the animal’s care. In the Washington state facility, S. aureus was cultured from the wounds of two macaques, but both were likely primate strains (t15469/ST3268 and t13638/ST3268) [66]. Another publication from Korea documented a macaque with an acute necrotic lesion caused by MRSA, but molecular typing was not carried out [64].
How may exposure to human pathogens, including S. aureus, in great ape populations affect release of them back into wild from captivity? This is addressed in several publications, suggesting that primates from captivity may pose a risk to their wild brethren [74] due to carriage of organisms such as drug-resistant S. aureus. Others argue release still should be possible but caregivers should be screened, and those positive for S. aureus carriage should not have contact with infant apes, and post-release monitoring of animals should include screening for this bacterium [93]. This may be difficult given the high level of carriage found in wild primates (up to 100% of chimpanzees tested and 72% of lemurs) [73].
While most studies examined asymptomatic colonization of wildlife, in some reports, such as those from captive zoo animals ([44]), a number of clinical infections could be examined. These infections included abscesses, bacteremia, bite wounds, and dermatitis, among other conditions. Common animal-associated lineages were found, including CC130, CC133, and bacteremia caused by CC398 in an African wildcat. There was considerable diversity among the infection isolates, though a few did share spa or ST/CC types including two cases of t208/ST49/CC49 infections in red squirrels, two cases of t1166/ST3269/CC133 infections in a black swan and white-face whistling duck, and two cases of t15307/ST133/CC133 in another white-face whistling duck and a Baikal teal. This again suggests the potential for exposure to a contaminated environmental source for some of these animals, including water or other shared habitats within the facility.
Other captivity-based studies document the potential for bidirectional transmission between humans and animals in these facilities. In the San Diego zoo, a MRSA outbreak was noted in 2008, with pustules documented on both an elephant calf and three caretakers. Twenty total caretakers were infected over the next month, and the calf was euthanized. Investigation determined that the calf’s infection with MRSA type USA300 likely came from a colonized caretaker, as the other elephants tested were colonization-negative [60].
Isolates examined in collected studies include methicillin-resistant and methicillin -susceptibile S. aureus. This testing included a mix of phenotypic and molecular methods, with some studies employing both. With the discovery of mecC [11,94], some early papers examining phenotypic testing alone should be looked at with some skepticism, as mecC-positive S. aureus isolates do not always show up as MRSA phenotypically, which can hinder the detection of mecC-carrying isolates [55]. Indeed, wildlife may be a key reservoir for mecC, as its presence was noted in a number of European reports (see Table 1). Interestingly, mecC has not been reported in any isolates originating in the United States to date.
There are a number of limitations to this review. Sampling was concentrated in a small number of countries and a relatively limited number of animal species have been sampled in different geographic areas, making large-scale comparisons difficult. Sampling techniques and anatomical locations tested within animal species vary among research groups. Most studies employed some sort of live animal swabbing (of noses, throats, skin, cloaca, etc.), but several used feces or scat instead of live animal testing. The studies also differed significantly in molecular and antibiotic resistance testing reported, making generalizations across publications difficult. Access to many animal species is also likely a function of convenience rather than a systematic sampling of all organisms in a particular environment, leading to over-representation of some animals relative to their abundance and an under-representation of others. Additional sampling should be carried out in order to examine the continued evolution of S. aureus in wildlife, and to track any strains that may have an increased propensity for zoonotic spread and threat to human health.
4. Materials and Methods
4.1. Eligibility Criteria
Studies that reported the presence of S. aureus (methicillin-resistant or susceptible) in any species of wildlife were eligible for inclusion.
4.2. Information Sources and Search Strategies
PubMed, Web of Science, and peer-reviewed materials within ProQuest databases were searched in May 2019 to identify eligible studies. The following search terms were used “MRSA OR Methicillin Resistant Staphylococcus aureus OR Staphylococcus aureus AND wildlife.” Reference lists of the identified studies were also checked for additional studies. “Wildlife” was defined as wild animals but also captive animals who would typically be wild (such as zoo elephants) and those on nature preserves. Captive animals used as livestock or poultry or otherwise farmed or used as pets or work animals were also excluded. Articles were limited to English language only. Articles were examined and duplicate articles were removed.
Titles and abstracts were examined and articles were retained when there was evidence of S. aureus colonization or infection reported within wildlife species as defined above. Citations which included information on S. aureus antibiotic resistance and/or molecular typing were included in Table 1 and were grouped by animal species type.
Author Contributions
Conceptualization, T.C.S.; Data extraction: T.C.S., C.J.H., G.R.G., L.D.S., and V.P.; Formal analysis: T.C.S.; Original draft preparation: T.C.S.; Writing—review and editing: T.C.S., C.J.H., G.R.G., L.D.S., and V.P.; Visualization: T.C.S. and C.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Picture credits.
Table A1.
Picture credits.
References
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. BioMed Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef]
- Smith, T.C.; Moritz, E.D.; Leedom Larson, K.R.; Ferguson, D.D. The environment as a factor in methicillin-resistant Staphylococcus aureus transmission. Rev. Environ. Health 2010, 25, 121–134. [Google Scholar] [CrossRef]
- Haag, A.F.; Fitzgerald, J.R.; Penades, J.R. Staphylococcus aureus in Animals. Gram-Posit. Pathog. 2019, 7, 731–746. [Google Scholar]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178–182. [Google Scholar] [CrossRef]
- Fluit, A.C. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 2012, 18, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Sundin, G.W.; Wang, N. Antibiotic Resistance in Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. MBio 2012, 3, e00305. [Google Scholar]
- Lowder, B.V.; Guinane, C.M.; Ben Zakour, N.L.; Weinert, L.A.; Conway-Morris, A.; Cartwright, R.A.; Simpson, A.J.; Rambaut, A.; Nübel, U.; Fitzgerald, J.R. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2009, 106, 19545–19550. [Google Scholar] [CrossRef]
- Garcia-Alvarez, L.; Holden, M.T.; Lindsay, H.; Webb, C.R.; Brown, D.F.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Awad-Alla, M.E.; Abdien, H.M.; Dessouki, A.A. Prevalence of bacteria and parasites in White Ibis in Egypt. Vet. Ital. 2010, 46, 277–286. [Google Scholar] [PubMed]
- Silvanose, C.D.; Bailey, T.A.; Naldo, J.L.; Howlett, J.C. Bacterial flora of the conjunctiva and nasal cavity in normal and diseased captive bustards. Avian Dis. 2001, 45, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.; Baldoma, L.; Molina-Lopez, R.A.; Martin, M.; Darwich, L. Microbiological diagnosis and antimicrobial sensitivity profiles in diseased free-living raptors. Avian Pathol. 2017, 46, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, T.; Slizewski, P.; Masiewicz, P. Species distribution of staphylococci from small wild mammals. Syst. Appl. Microbiol. 2010, 33, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, D.B.; Aessio, F.M.; Mota, R.A.; Marvulo, M.F.; Mauffrey, J.F.; Monteiro, S.R.; Farias, R.C.; Cunha, R.C.; Oliveira, R.L.; Souza, T.C.; et al. Staphylococcus aureus mastitis in a white-eared opossum (Didelphis albiventris) in the Atlantic Forest of northeast Brazil. J. Zoo Wildl. Med. 2010, 41, 526–529. [Google Scholar] [CrossRef]
- Hamir, A.N. Systemic Staphylococcus aureus infection in a free-ranging raccoon (Procyon lotor). J. Wildl. Dis. 2010, 46, 665–668. [Google Scholar] [CrossRef][Green Version]
- Simpson, V.R.; Hargreaves, J.; Butler, H.M.; Davison, N.J.; Everest, D.J. Causes of mortality and pathological lesions observed post-mortem in red squirrels (Sciurus vulgaris) in Great Britain. BMC Vet. Res. 2013, 9, 229. [Google Scholar] [CrossRef]
- Himsworth, C.G.; Zabek, E.; Tang, P.; Parsons, K.L.; Koehn, M.; Jardine, C.M.; Patrick, D.M. Bacteria isolated from conspecific bite wounds in Norway and black rats: Implications for rat bite-associated infections in people. Vector-Borne Zoonotic Dis. 2014, 14, 94–100. [Google Scholar] [CrossRef]
- Carson, M.; Meredith, A.L.; Shaw, D.J.; Giotis, E.S.; Lloyd, D.H.; Loeffler, A. Foxes as a potential wildlife reservoir for mecA-positive Staphylococci. Vector-Borne Zoonotic Dis. 2012, 12, 583–587. [Google Scholar] [CrossRef]
- Gonzalez-Candela, M.; Cubero-Pablo, M.J.; Martin-Atance, P.; Leon-Vizcaino, L. Potential pathogens carried by Spanish ibex (Capra pyrenaica hispanica) in southern Spain. J. Wildl. Dis. 2006, 42, 325–334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clausen, B.; Ashford, W.A. Bacteriologic survey of black rhinoceros (Diceros bicornis). J. Wildl. Dis. 1980, 16, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M.M.; Songer, J.G.; Chilelli, C.J.; deVos, J.C. Isolations of aerobic bacteria from wild desert bighorn sheep (Ovis canadensis nelsoni and, O. c. mexicana) in Arizona. J. Wildl. Dis. 1983, 19, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Gnat, S.; Troscianczyk, A.; Nowakiewicz, A.; Majer-Dziedzic, B.; Ziolkowska, G.; Dziedzic, R.; Ziółkowska, G.; Dziedzic, R.; Zięba, P.; Teodorowski, O. Experimental studies of microbial populations and incidence of zoonotic pathogens in the faeces of red deer (Cervus elaphus). Lett. Appl. Microbiol. 2015, 61, 446–452. [Google Scholar] [CrossRef]
- Laaksonen, S.; Oksanen, A.; Julmi, J.; Zweifel, C.; Fredriksson-Ahomaa, M.; Stephan, R. Presence of foodborne pathogens, extended-spectrum beta-lactamase -producing Enterobacteriaceae, and methicillin-resistant Staphylococcus aureus in slaughtered reindeer in northern Finland and Norway. Acta Vet. Scand. 2017, 59, 2. [Google Scholar] [CrossRef][Green Version]
- Stewart, J.R.; Townsend, F.I.; Lane, S.M.; Dyar, E.; Hohn, A.A.; Rowles, T.K.; Staggs, L.A.; Wells, R.S.; Balmer, B.C.; Schwacke, L.H. Survey of antibiotic-resistant bacteria isolated from bottlenose dolphins Tursiops truncatus in the southeastern USA. Dis. Aquat. Organ. 2014, 108, 91–102. [Google Scholar] [CrossRef]
- Morris, P.J.; Johnson, W.R.; Pisani, J.; Bossart, G.D.; Adams, J.; Reif, J.S.; Fair, P.A. Isolation of culturable microorganisms from free-ranging bottlenose dolphins (Tursiops truncatus) from the southeastern United States. Vet. Microbiol. 2010, 148, 440–447. [Google Scholar] [CrossRef]
- Vercammen, F.; Bauwens, L.; De Deken, R.; Brandt, J. Prevalence of methicillin-resistant Staphylococcus aureus in mammals of the Royal Zoological Society of Antwerp, Belgium. J. Zoo Wildl. Med. 2012, 43, 159–161. [Google Scholar] [CrossRef]
- Schaumburg, F.; Pauly, M.; Anoh, E.; Mossoun, A.; Wiersma, L.; Schubert, G.; Flammen, A.; Alabi, A.S.; Muyembe-Tamfum, J.J.; Grobusch, M.P.; et al. Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 2015, 21, 345. [Google Scholar] [CrossRef]
- Rothenburger, J.L.; Himsworth, C.G.; Nemeth, N.M.; Pearl, D.L.; Jardine, C.M. Environmental Factors Associated with the Carriage of Bacterial Pathogens in Norway Rats. EcoHealth 2018, 15, 82–95. [Google Scholar] [CrossRef]
- Rothenburger, J.L.; Rousseau, J.D.; Weese, J.S.; Jardine, C.M. Livestock-associated methicillin-resistant Staphylococcus aureus and Clostridium difficile in wild Norway rats (Rattus norvegicus) from Ontario swine farms. Can. J. Vet. Res. Rev. Can. Rech. Vet. 2018, 82, 66–69. [Google Scholar]
- Rothenburger, J.L.; Himsworth, C.G.; La Perle, K.M.D.; Leighton, F.A.; Nemeth, N.M.; Treuting, P.M.; Jardine, C.M. Pathology of wild Norway rats in Vancouver, Canada. J. Vet. Diagn. Investig. 2019, 31, 184–199. [Google Scholar] [CrossRef]
- Lee, M.J.; Byers, K.A.; Donovan, C.M.; Zabek, E.; Stephen, C.; Patrick, D.M.; Himsworth, C.G. Methicillin-resistant Staphylococcus aureus in urban Norway rat (Rattus norvegicus) populations: Epidemiology and the impacts of kill-trapping. Zoonoses Public Health 2019, 66, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Himsworth, C.G.; Miller, R.R.; Montoya, V.; Hoang, L.; Romney, M.G.; Al-Rawahi, G.N.; Kerr, T.; Jardine, C.M.; Patrick, D.M.; Tang, P.; et al. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS ONE 2014, 9, e87983. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ripa, L.; Alcala, L.; Simon, C.; Gomez, P.; Mama, O.M.; Rezusta, A.; Zarazaga, M.; Torres, C. Diversity of Staphylococcus aureus clones in wild mammals in Aragon, Spain, with detection of MRSA ST130-mecC in wild rabbits. J. Appl. Microbiol. 2019, 127, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Vancraeynest, D.; Haesebrouck, F.; Deplano, A.; Denis, O.; Godard, C.; Wildemauwe, C.; Hermans, K. International dissemination of a high virulence rabbit Staphylococcus aureus clone. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Loncaric, I.; Kubber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 2013, 68, 2222–2225. [Google Scholar] [CrossRef]
- Monecke, S.; Gavier-Widen, D.; Mattsson, R.; Rangstrup-Christensen, L.; Lazaris, A.; Coleman, D.C.; Shore, A.C.; Ehricht, R. Detection of mecC-positive Staphylococcus aureus (CC130-MRSA-XI) in diseased European hedgehogs (Erinaceus europaeus) in Sweden. PLoS ONE 2013, 8, e66166. [Google Scholar] [CrossRef]
- Bengtsson, B.; Persson, L.; Ekstrom, K.; Unnerstad, H.E.; Uhlhorn, H.; Borjesson, S. High occurrence of mecC-MRSA in wild hedgehogs (Erinaceus europaeus) in Sweden. Vet. Microbiol. 2017, 207, 103–107. [Google Scholar] [CrossRef]
- Monecke, S.; Gavier-Widen, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Muller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- Gomez, P.; Gonzalez-Barrio, D.; Benito, D.; Garcia, J.T.; Vinuela, J.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014, 69, 2061–2064. [Google Scholar] [CrossRef] [PubMed]
- Mrochen, D.M.; Schulz, D.; Fischer, S.; Jeske, K.; El Gohary, H.; Reil, D.; Imholt, C.; Trube, P.; Suchomel, J.; Tricaud, E.; et al. Wild rodents and shrews are natural hosts of Staphylococcus aureus. Int. J. Med. Microbiol. 2018, 308, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Kmet, V.; Cuvalova, A.; Stanko, M. Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol. 2018, 63, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Fessler, A.T.; Thomas, P.; Muhldorfer, K.; Grobbel, M.; Brombach, J.; Eichhorn, I.; Monecke, S.; Ehricht, R.; Shwartz, S. Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from zoo and wild animals. Vet. Microbiol. 2018, 218, 98–103. [Google Scholar] [CrossRef]
- Nowakiewicz, A.; Ziolkowska, G.; Zieba, P.; Gnat, S.; Wojtanowicz-Markiewicz, K.; Troscianczyk, A. Coagulase-positive Staphylococcus isolated from wildlife: Identification, molecular characterization and evaluation of resistance profiles with focus on a methicillin-resistant strain. Comp. Immunol. Microbiol. Infect. Dis. 2016, 44, 21–28. [Google Scholar] [CrossRef]
- Wardyn, S.E.; Kauffman, L.K.; Smith, T.C. Methicillin-resistant Staphylococcus aureus in central Iowa wildlife. J. Wildl. Dis. 2012, 48, 1069–1073. [Google Scholar] [CrossRef]
- Chen, M.M.S.; Monecke, S.; Brown, M.H. Clonal diversity of methicillin-sensitive Staphylococcus aureus from South Australian wallabies. One Health 2016, 2, 31–32. [Google Scholar] [CrossRef]
- Espinosa-Gongora, C.; Harrison, E.M.; Moodley, A.; Guardabassi, L.; Holmes, M.A. MRSA carrying mecC in captive mara. J. Antimicrob. Chemother. 2015, 70, 1622–1624. [Google Scholar] [CrossRef]
- Espinosa-Gongora, C.; Chrobak, D.; Moodley, A.; Bertelsen, M.F.; Guardabassi, L. Occurrence and distribution of Staphylococcus aureus lineages among zoo animals. Vet. Microbiol. 2012, 158, 228–231. [Google Scholar] [CrossRef]
- Akobi, B.; Aboderin, O.; Sasaki, T.; Shittu, A. Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 2012, 12, 279. [Google Scholar] [CrossRef]
- Olatimehin, A.; Shittu, A.O.; Onwugamba, F.C.; Mellmann, A.; Becker, K.; Schaumburg, F. Staphylococcus aureus Complex in the Straw-Colored Fruit Bat (Eidolon helvum) in Nigeria. Front. Microbiol. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Held, J.; Gmeiner, M.; Mordmuller, B.; Matsiegui, P.B.; Schaer, J.; Eckerle, I.; Weber, N.; Matuschewski, K.; Bletz, S.; Schaumburg, F. Bats are rare reservoirs of Staphylococcus aureus complex in Gabon. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017, 47, 118–120. [Google Scholar] [CrossRef]
- Porrero, M.C.; Mentaberre, G.; Sanchez, S.; Fernandez-Llario, P.; Casas-Diaz, E.; Mateos, A.; Vidal, D.; Lavin, S.; Fernández-Garayzábal, J.-F.; Dominguez, L. Carriage of Staphylococcus aureus by free-living wild animals in Spain. Appl. Environ. Microbiol. 2014, 80, 4865–4870. [Google Scholar] [CrossRef] [PubMed]
- Porrero, M.C.; Mentaberre, G.; Sanchez, S.; Fernandez-Llario, P.; Gomez-Barrero, S.; Navarro-Gonzalez, N.; Serrano, E.; Casas-Diaz, E.; Marco, I.; Fernández-Garayzabal, J.-F.; et al. Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 2013, 198, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Porrero, M.C.; Valverde, A.; Fernandez-Llario, P.; Diez-Guerrier, A.; Mateos, A.; Lavin, S.; Canton, R.; Fernández-Garayzabal, J.-F.; Dominguez, L. Staphylococcus aureus carrying mecC gene in animals and urban wastewater, Spain. Emerg. Infect. Dis. 2014, 20, 899–901. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Silva, N.; Manageiro, V.; Ramos, S.; Coelho, A.; Goncalves, D.; Caniça, M.; Torres, C.; Igrejas, G.; Poeta, P. First report on MRSA CC398 recovered from wild boars in the north of Portugal. Are we facing a problem? Sci. Total Environ. 2017, 596–597, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Seinige, D.; Von Altrock, A.; Kehrenberg, C. Genetic diversity and antibiotic susceptibility of Staphylococcus aureus isolates from wild boars. Comp. Immunol. Microbiol. Infect. Dis. 2017, 54, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Meemken, D.; Blaha, T.; Hotzel, H.; Strommenger, B.; Klein, G.; Ehricht, R.; Monecke, S.; Kehrenberg. Genotypic and phenotypic characterization of Staphylococcus aureus isolates from wild boars. Appl. Environ. Microbiol. 2013, 79, 1739–1742. [Google Scholar] [CrossRef]
- Gomez, P.; Lozano, C.; Gonzalez-Barrio, D.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet. Microbiol. 2015, 177, 326–331. [Google Scholar] [CrossRef]
- CDC. Prevention. Methicillin-resistant Staphylococcus aureus skin infections from an elephant calf-San Diego, California, 2008. Morb. Mortal. Wkly. Rep. 2009, 58, 194–198. [Google Scholar]
- Luzzago, C.; Locatelli, C.; Franco, A.; Scaccabarozzi, L.; Gualdi, V.; Vigano, R.; Sironi, G.; Besozzi, M.; Castiglioni, B.; Lanfranchi, P.; et al. Clonal diversity, virulence-associated genes and antimicrobial resistance profile of Staphylococcus aureus isolates from nasal cavities and soft tissue infections in wild ruminants in Italian Alps. Vet. Microbiol. 2014, 170, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Ehricht, R.; Slickers, P.; Wernery, R.; Johnson, B.; Jose, S.; Wernery, U. Microarray-based genotyping of Staphylococcus aureus isolates from camels. Vet. Microbiol. 2011, 150, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Joshi, P.R.; Greninger, A.L.; Melendez, D.; Paudel, S.; Acharya, M.; Kishor Bimali, N.; Koju, N.P.; No, D.; Chalise, M.; et al. The human clone ST22 SCCmec IV methicillin-resistant Staphylococcus aureus isolated from swine herds and wild primates in Nepal: Is man the common source? FEMS Microbiol. Ecol. 2018, 94, fiy052. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.I.; Kim, K.S.; Oh, B.C.; Kim, N.A.; Kim, I.H.; Park, C.G.; Kim, S.J. Acute necrotic stomatitis (noma) associated with methicillin-resistant Staphylococcus aureus infection in a newly acquired rhesus macaque (Macaca mulatta). J. Med. Primatol. 2011, 40, 188–193. [Google Scholar] [CrossRef]
- Pardos de la Gandara, M.; Diaz, L.; Euler, C.W.; Chung, M.; Gonzalez, A.; Cheleuitte, C.; Freiwald, W.; Tomasz, A.; Fischetti, V.A.; de Lencastre, H.; et al. Staphylococcus aureus Infecting and Colonizing Experimental Animals, Macaques, in a Research Animal Facility. Microb. Drug Resist. 2019, 25, 54–62. [Google Scholar] [CrossRef]
- Roberts, M.C.; Fessler, A.T.; Monecke, S.; Ehricht, R.; No, D.; Schwarz, S. Molecular Analysis of Two Different MRSA Clones ST188 and ST3268 From Primates (Macaca spp.) in a United States Primate Center. Front. Microbiol. 2018, 9, 2199. [Google Scholar] [CrossRef]
- Soge, O.O.; No, D.; Michael, K.E.; Dankoff, J.; Lane, J.; Vogel, K.; Smedley, J.; Roberts, M.C. Transmission of MDR MRSA between primates, their environment and personnel at a United States primate centre. J. Antimicrob. Chemother. 2016, 71, 2798–2803. [Google Scholar] [CrossRef]
- Hsu, L.Y.; Holden, M.T.G.; Koh, T.H.; Pettigrew, K.A.; Cao, D.; Hon, P.Y.; Sergio, D.M.; Pena, E.; Ogden, B.E. ST3268: A geographically widespread primate MRSA clone. J. Antimicrob. Chemother. 2017, 72, 2401–2403. [Google Scholar] [CrossRef][Green Version]
- Bittar, F.; Keita, M.B.; Lagier, J.C.; Peeters, M.; Delaporte, E.; Raoult, D. Gorilla gorilla gorilla gut: A potential reservoir of pathogenic bacteria as revealed using culturomics and molecular tools. Sci. Rep. 2014, 4, 7174. [Google Scholar] [CrossRef]
- Nagel, M.; Dischinger, J.; Turck, M.; Verrier, D.; Oedenkoven, M.; Ngoubangoye, B.; Le Flohic, G.; Drexler, J.F.; Bierbaum, G.; Goonzalez, J.P. Human-associated Staphylococcus aureus strains within great ape populations in Central Africa (Gabon). Clin. Microbiol. Infect. 2013, 19, 1072–1077. [Google Scholar] [CrossRef]
- Schaumburg, F.; Alabi, A.S.; Kock, R.; Mellmann, A.; Kremsner, P.G.; Boesch, C.; Becker, K.; Leendertz, F.H.; Peters, G. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ. Microbiol. Rep. 2012, 4, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Hanley, P.W.; Barnhart, K.F.; Abee, C.R.; Lambeth, S.P.; Weese, J.S. Methicillin-Resistant Staphylococcus aureus Prevalence among Captive Chimpanzees. Emerg. Infect. Dis. 2015, 21, 2158–2160. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, F.; Mugisha, L.; Kappeller, P.; Fichtel, C.; Kock, R.; Kondgen, S.; Becker, K.; Boesch, C.; Peters, G.; Leendertz, F. Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS ONE 2013, 8, e78046. [Google Scholar] [CrossRef]
- Schaumburg, F.; Mugisha, L.; Peck, B.; Becker, K.; Gillespie, T.R.; Peters, G.; Leendertz, F.H. Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 2012, 74, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ripa, L.; Gomez, P.; Alonso, C.A.; Camacho, M.C.; de la Puente, J.; Fernandez-Fernandez, R.; Ramiro, Y.; Quivedo, M.A.; Blanco, J.M.; Zarazaga, M.; et al. Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in Magpies and Cinereous Vultures in Spain. Microb. Ecol. 2019, 78, 409–415. [Google Scholar] [CrossRef]
- Gomez, P.; Lozano, C.; Camacho, M.C.; Lima-Barbero, J.F.; Hernandez, J.M.; Zarazaga, M.; Hofle, U.; Torres, C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016, 71, 53–57. [Google Scholar] [CrossRef]
- Sousa, M.; Silva, N.; Igrejas, G.; Silva, F.; Sargo, R.; Alegria, N.; Benito, D.; Gomez, P.; Lozano, C.; Gomez-Sanz, E.; et al. Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet. Microbiol. 2014, 171, 436–440. [Google Scholar] [CrossRef]
- Robb, A.; Pennycott, T.; Duncan, G.; Foster, G. Staphylococcus aureus carrying divergent mecA homologue (mecA LGA251) isolated from a free-ranging wild bird. Vet. Microbiol. 2013, 162, 300–301. [Google Scholar] [CrossRef]
- Thapaliya, D.; Dalman, M.; Kadariya, J.; Little, K.; Mansell, V.; Taha, M.Y.; Grenier, D.; Smith, T.C. Characterization of Staphylococcus aureus in Goose Feces from State Parks in Northeast Ohio. Ecohealth 2017, 14, 303–309. [Google Scholar] [CrossRef]
- Atyah, M.A.; Zamri-Saad, M.; Siti-Zahrah, A. First report of methicillin-resistant Staphylococcus aureus from cage-cultured tilapia (Oreochromis niloticus). Vet. Microbiol. 2010, 144, 502–504. [Google Scholar] [CrossRef]
- Faires, M.; Gehring, E.; Mergl, J.; Weese, J. Methicillin-Resistant Staphylococcus aureus in Marine Mammals. Emerg. Infect. Dis. 2009, 15, 2071–2072. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.A.; Zadoks, R.N. Methicillin resistant, S. aureus in human and bovine mastitis. J. Mammary Gland Biol. Neoplasia 2011, 16, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C.; Pearson, N. The Emergence of Staphylococcus aureus ST398. Vector-Borne Zoonotic Dis. 2010, 11, 327–339. [Google Scholar] [CrossRef]
- Kruse, H.; kirkemo, A.M.; Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 2004, 10, 2067–2072. [Google Scholar] [CrossRef]
- Messenger, A.M.; Barnes, A.N.; Gray, G.C. Reverse zoonotic disease transmission (zooanthroponosis): A systematic review of seldom-documented human biological threats to animals. PLoS ONE 2014, 9, e89055. [Google Scholar] [CrossRef]
- Epstein, J.H.; Price, J.T. The significant but understudied impact of pathogen transmission from humans to animals. Mt. Sinai J. Med. J. Transl. Pers. Med. 2009, 76, 448–455. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Blaak, H.; de Jong, M.C.; Graat, E.A.; Vandenbroucke-Grauls, C.M.; de Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Hiltunen, T.; Virta, M.; Laine, A.L. Antibiotic resistance in the wild: An eco-evolutionary perspective. Philos. Trans. R. Soc. Biol. Sci. 2017, 372, 20160039. [Google Scholar] [CrossRef]
- Concepcion Porrero, M.; Harrison, E.M.; Fernandez-Garayzabal, J.F.; Paterson, G.K.; Diez-Guerrier, A.; Holmes, M.A.; Dominguez, L. Detection of mecC-Methicillin-resistant Staphylococcus aureus isolates in river water: A potential role for water in the environmental dissemination. Environ. Microbiol. Rep. 2014, 6, 705–708. [Google Scholar] [CrossRef]
- Harrison, E.M.; Paterson, G.K.; Holden, M.T.; Larsen, J.; Stegger, M.; Larsen, A.R.; Petersen, A.; Skov, R.L.; Christensen, J.M.; Bak Zeuthen, A.; et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 2013, 5, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C.; Wardyn, S.E. Human Infections with Staphylococcus aureus CC398. Curr. Environ. Health Rep. 2015, 2, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Unwin, S.; Robinson, I.; Schmidt, V.; Colin, C.; Ford, L.; Humle, T. Does confirmed pathogen transfer between sanctuary workers and great apes mean that reintroduction should not occur? Commentary on “Drug-resistant human Staphylococcus aureus findings in sanctuary apes and its threat to wild ape populations”. Am. J. Primatol. 2012, 74, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.C.; Deasy, E.C.; Slickers, P.; Brennan, G.; O’Connell, B.; Monecke, S.; Ehricht, R.; Coleman, D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).