Ib-M6 Antimicrobial Peptide: Antibacterial Activity against Clinical Isolates of Escherichia coli and Molecular Docking
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity
2.2. Circular Dicrhroism and 3D Structure
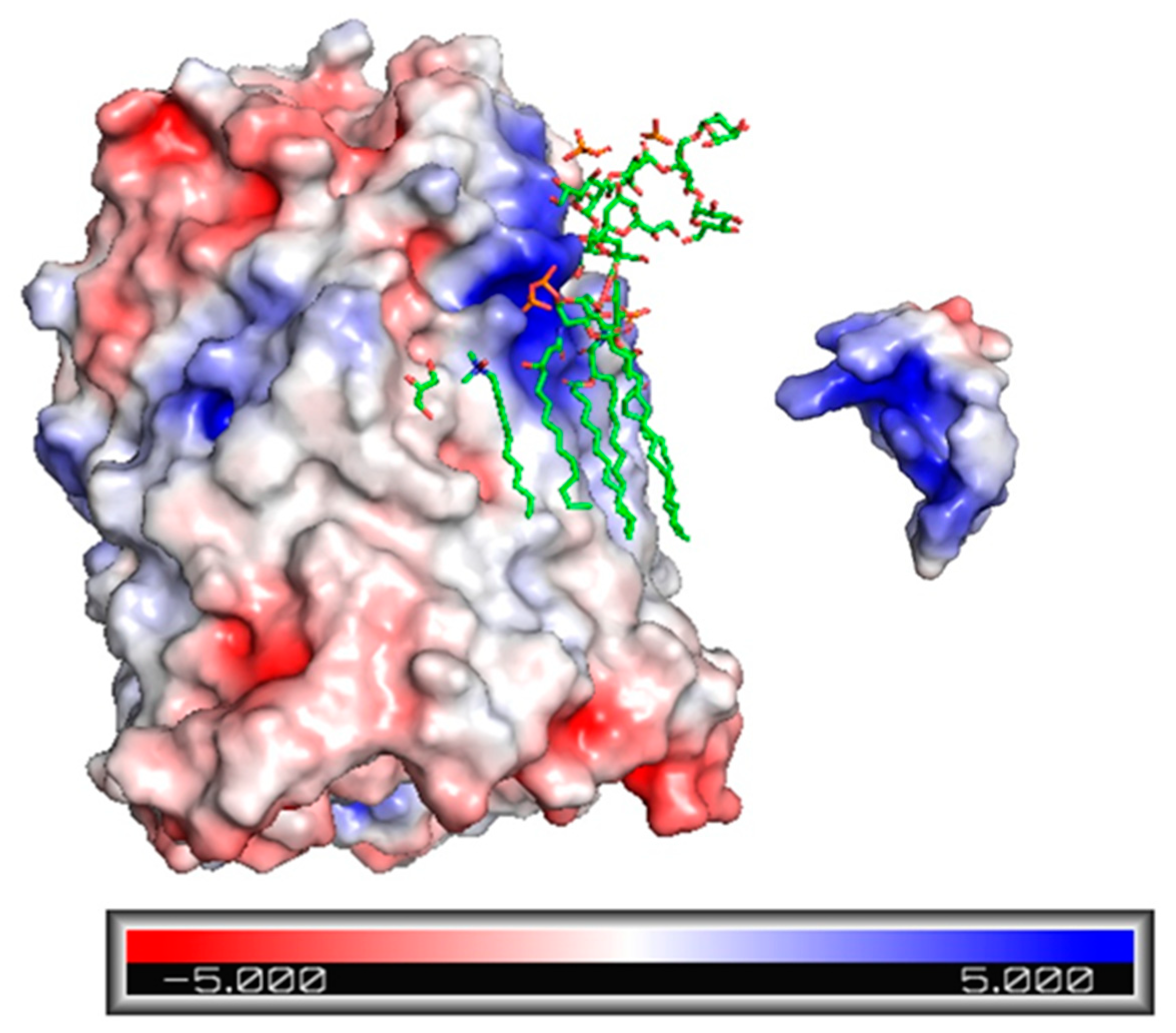
2.3. Electrostatic Potential Map of Ib-M6 Peptide and FhuA Protein
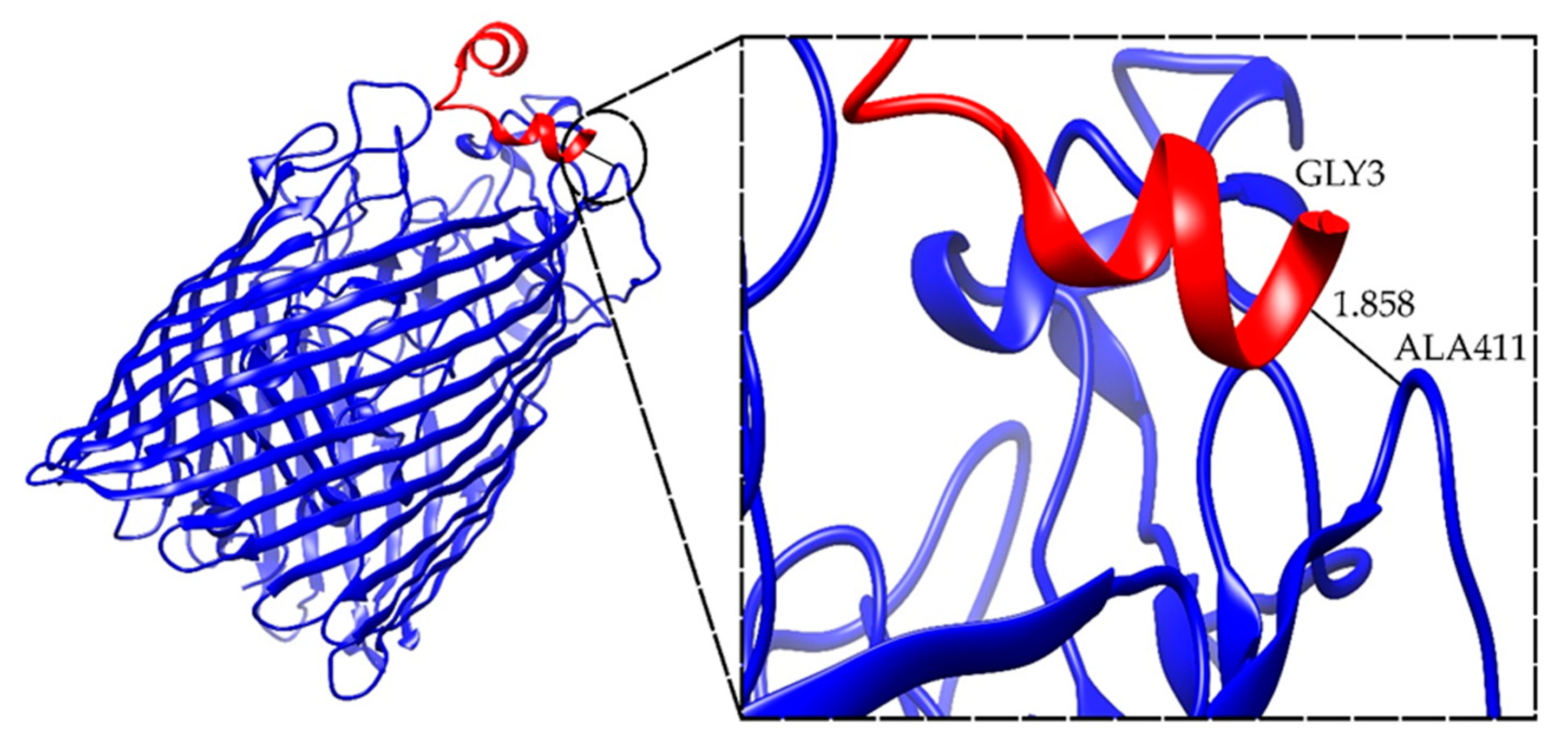
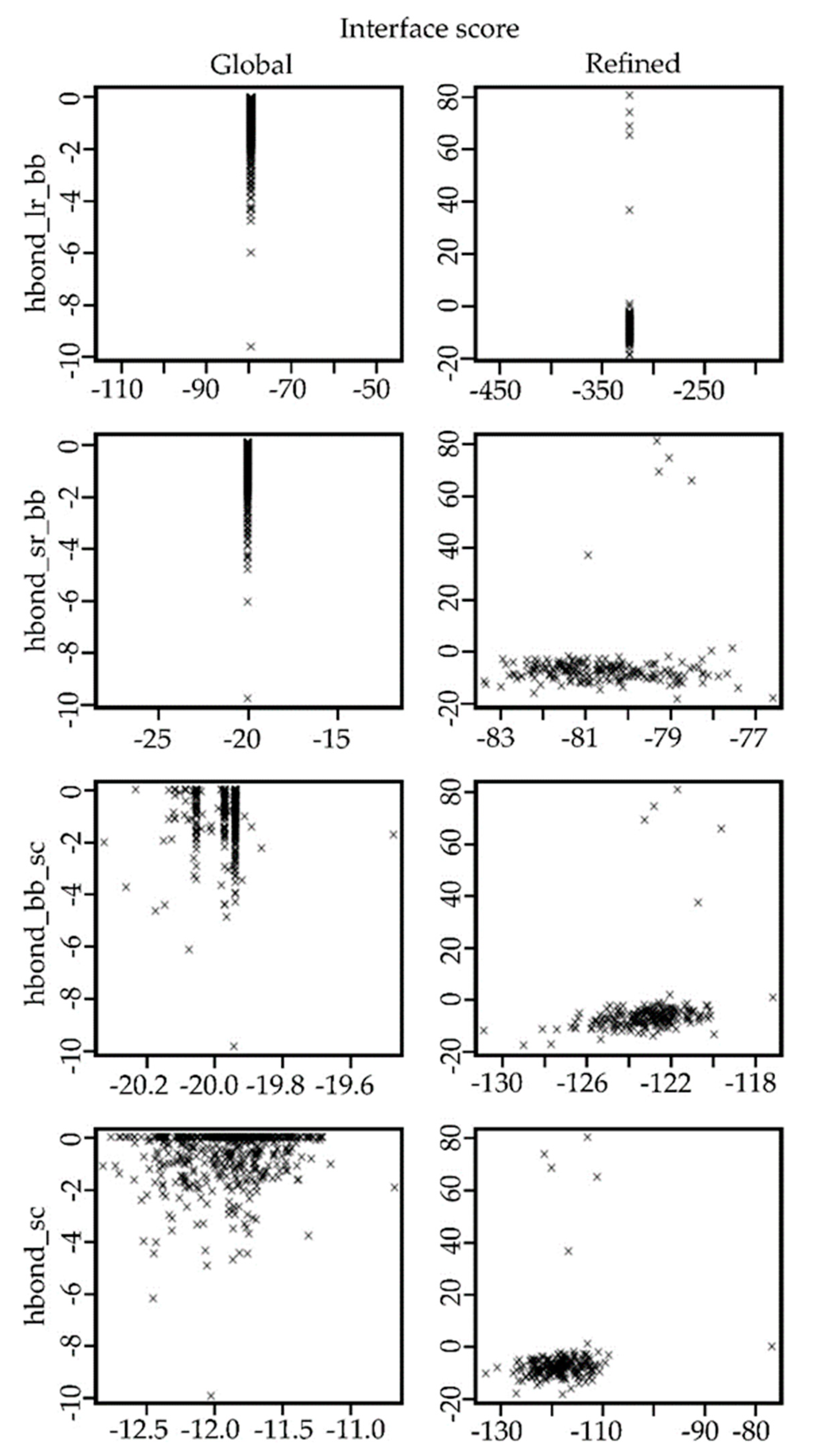
2.4. Docking Results for the Ib-M6 Peptide in Complex with FhuA Protein
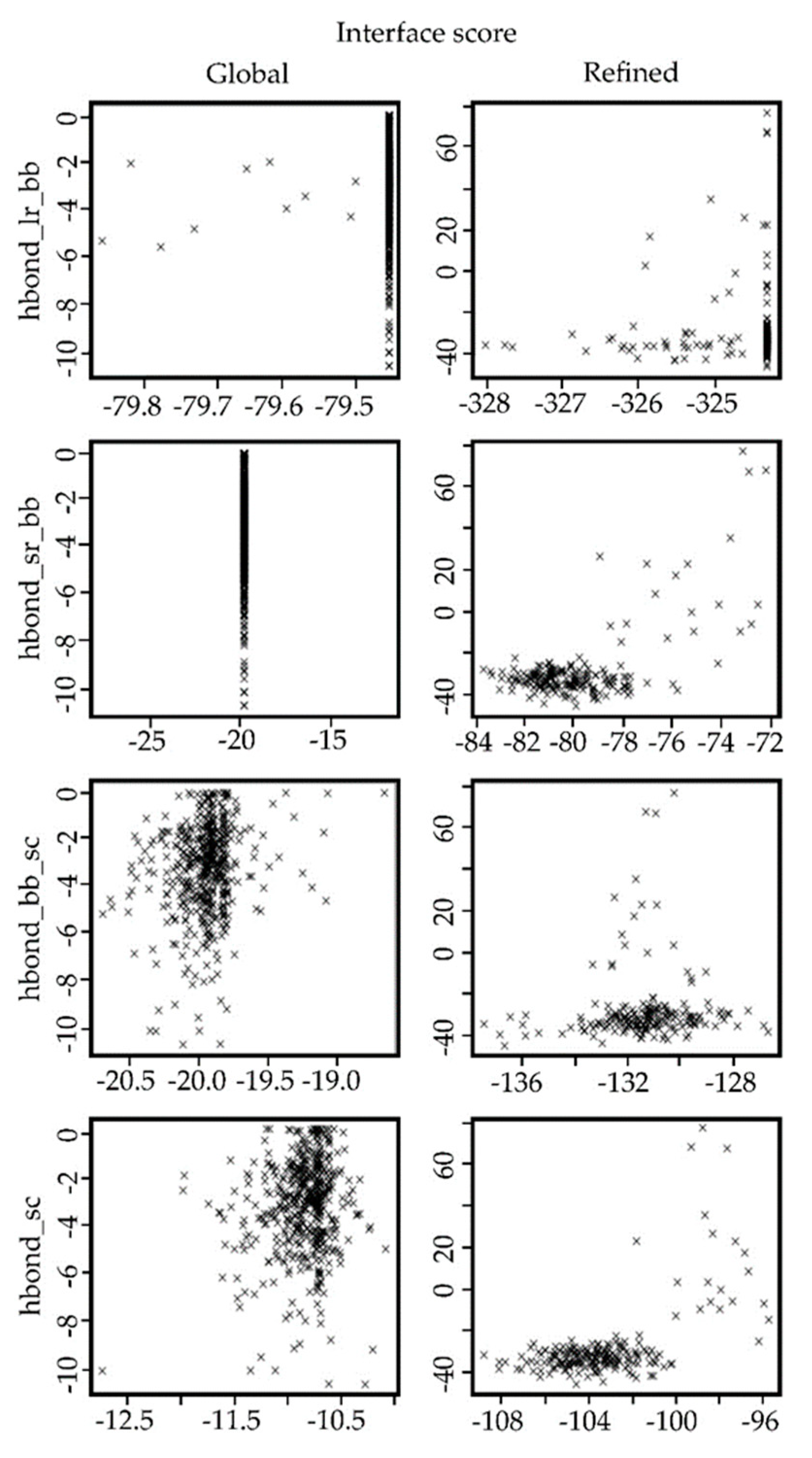
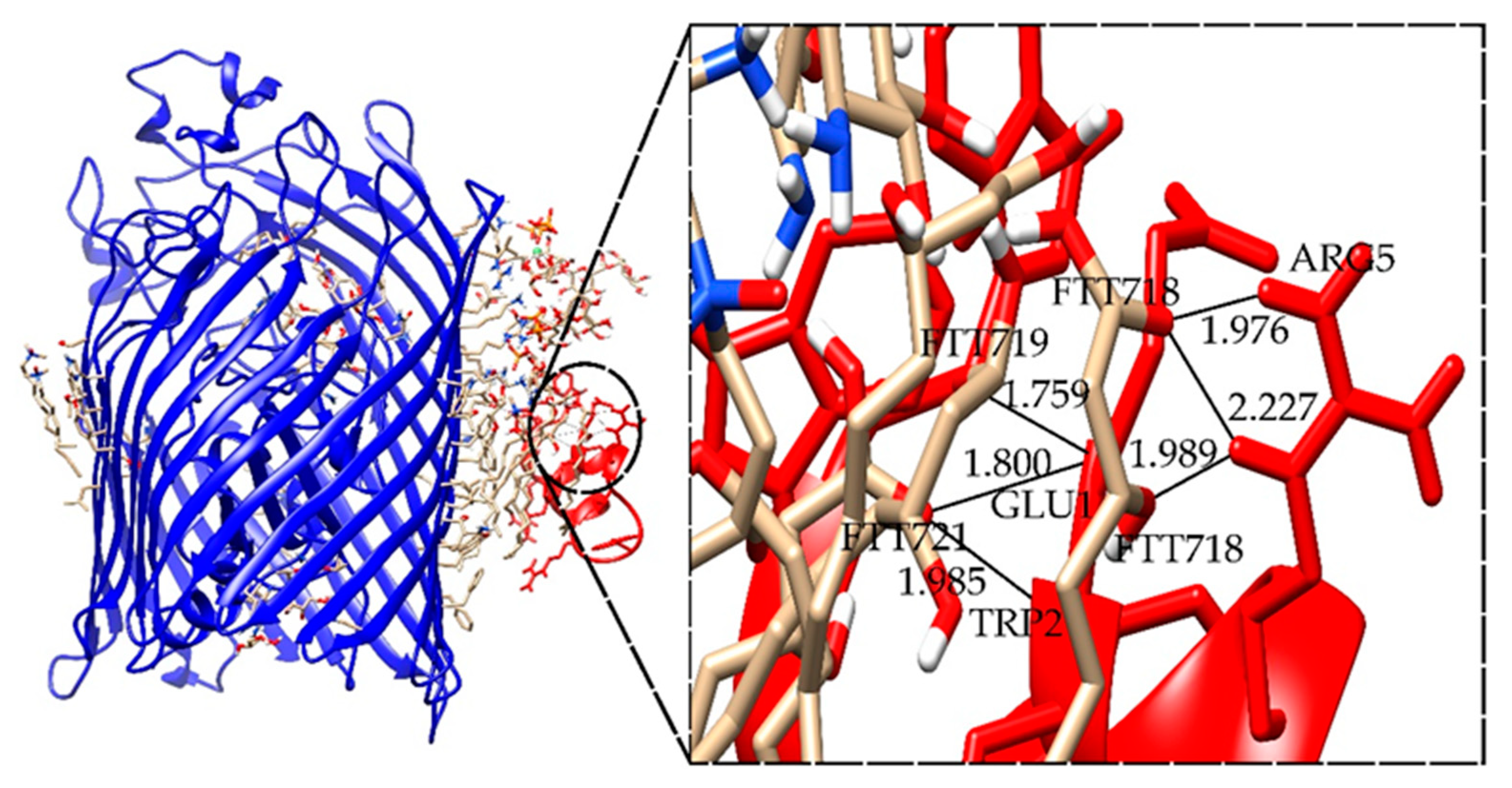
2.5. Docking Results for the Ib-M6 Peptide Interacting with Lipopolysaccharide–FhuA Complex
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Agents and Bacterial Strains
4.2. Antimicrobial Activity of Ib-M6 Peptide
4.3. Circular Dichroism and Structure 3D of Peptide
4.4. Preparation of Ligand and Targets Molecules
4.5. Docking Procedure for the Ib-M6 Peptide in Complex with FhuA Protein
4.6. Docking Procedure for the Ib-M6 Peptide in Complex with Lipopolysaccharide–FhuA
4.7. Analysis of the Resulting Potential Complexes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 20, 1–16. [Google Scholar]
- UK Parliament. Antimicrobial Resistance; Health and Social Care Committee: House of Commons; UK Parliament: London, UK, 2018. [Google Scholar]
- La Organización Mundial de la Salud (OMS). Estrategia Mundial DE La OMS Para Contener La Resistencia a Los Antimicrobianos; La Organización Mundial de la Salud OMS: Geneva, Switzerland, 2001. [Google Scholar]
- WHO. Global Antimicrobial Resistance Surveillance System (GLASS) Report; Early Implementation 2016–2017; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Kaper, J.B. Pathogenic Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 355–356. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R1–R21. [Google Scholar] [CrossRef]
- Arnusch, C.J.; Pieters, R.J.; Breukink, E. Enhanced membrane pore formation through high-affinity targeted antimicrobial peptides. PLoS ONE 2012, 7, e39768. [Google Scholar] [CrossRef]
- Almaaytah, A.; Qaoud, M.T.; Mohammed, G.K.; Abualhaijaa, A.; Knappe, D.; Hoffmann, R.; Al-Balas, Q. Antimicrobial and antibiofilm activity of UP-5, an ultrashort antimicrobial peptide designed using only arginine and biphenylalanine. Pharmaceuticals 2018, 11, 3. [Google Scholar] [CrossRef]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Téllez, G.A.; Castaño, J.C. Péptidos antimicrobianos. Infectio 2010, 14, 55–67. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Lai, P.K.; Kaznessis, Y.N. Free energy calculations of microcin J25 variants binding to the FhuA receptor. J. Chem. theory Comput. 2017, 13, 3413–3423. [Google Scholar] [CrossRef]
- Farahmandian, N.; Sifipour, M.; Sefid, F. Article Structure Engineering of Fhua as a Vaccine. IIOAB J. 2016, 7, 352–358. [Google Scholar]
- Flórez-Castillo, J.M.; Perullini, M.; Jobbágy, M.; Jesús Cano Calle, H. Enhancing Antibacterial Activity Against Escherichia coli K-12 of Peptide Ib-AMP4 with Synthetic Analogues. Int. J. Pept. Res. Ther. 2014, 20, 365–369. [Google Scholar] [CrossRef]
- Balhara, V.; Schmidt, R.; Gorr, S.U.; Dewolf, C. Membrane selectivity and biophysical studies of the antimicrobial peptide GL13K. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2193–2203. [Google Scholar] [CrossRef]
- Ashby, M.; Petkova, A.; Hilpert, K. Cationic antimicrobial peptides as potential new therapeutic agents in neonates and children: A review. Curr. Opin. Infect. Dis. 2014, 27, 258–267. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid A Modification Systems in Gram-Negative Bacteria. NIH Public Access 2007, 76, 295–329. [Google Scholar] [CrossRef]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1184–1202. [Google Scholar] [CrossRef]
- Farfán-García, A.E.; Zhang, C.; Imdad, A.; Arias-Guerrero, M.Y.; Sánchez-Alvarez, N.T.; Shah, R.; Iqbal, J.; Tamborski, M.E.; Gómez-Duarte, O.G. Case-Control Pilot Study on Acute Diarrheal Disease in a Geographically Defined Pediatric Population in a Middle Income Country. Int. J. Pediatr. 2017. [Google Scholar] [CrossRef]
- CLSI Performance Standards for Antimicrobial Susceptibility Testing. Wayne PA Clin. Lab. Stand. Inst. 2019, 39, 1–285.
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, 288–293. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta FlexPepDock web server—High resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef]
- Schrödinger, L. The PyMOL Molecular Graphics System, Version 1.8; Schrodinger LLC: New York, NY, USA, 2015. [Google Scholar]
- Alford, R.F.; Leaver-Fay, A.; Jeliazkov, J.R.; O’Meara, M.J.; DiMaio, F.P.; Park, H.; Shapovalov, M.V.; Renfrew, P.D.; Mulligan, V.K.; Kappel, K.; et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J. Chem. Theory Comput. 2017, 13, 3031–3048. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

| Name Strain | MIC * (µM) | MBC * (µM) | |
|---|---|---|---|
| Ib-M6 | Streptomycin | ||
| EAEC_1 | 1.5 | 6.25 | 25 |
| EAEC_2 | 0.7 | 6.25 | 1.5 |
| ETEC_1 | 3.1 | 6.25 | 12.5 |
| ETEC_2 | 12.5 | 6.25 | > 100 |
| EPEC_1 | 3.1 | 6.25 | 6.25 |
| EPEC_2 | 1.5 | 100 | 3.1 |
| DAEC_1 | 6.25 | 200 | 12.5 |
| DAEC_2 | 3.1 | 50 | 6.25 |
| EIEC | 25 | 200 | 50 |
| STEC_1 | 12.5 | 50 | 25 |
| STEC_2 | 6.25 | 50 | 6.25 |
| Non-pathogenic_1 | 1.5 | 200 | 3.1 |
| Non-pathogenic_2 | 1.5 | 3.1 | 25 |
| Non-pathogenic_3 | 3.1 | 400 | 3.1 |
| Complex | I_sc (REU) |
|---|---|
| FhuA+M6_A | −8.928 |
| FhuA+M6_B | −7.407 |
| FhuA+M6_C | −8.334 |
| FhuA+M6_D | −7.099 |
| FhuA+M6_E | −10.305 |
| Complex | I_sc (REU) | Hydrogen Bonds | Length (Å) |
|---|---|---|---|
| FhuA+M6_A_1 | −42.545 | ALA411-GLY3 | 1.858 |
| FhuA+M6_A_2 | −41.622 | GLY548-ARG14 | 2.216 |
| FhuA+M6_A_3 | −41.466 | GLY548-ARG14 | 2.277 |
| FhuA+M6_B_1 | −33.700 | TYR307-GLY 8 | 1.849 |
| FhuA+M6_B_2 | −33.421 | TYR 307-GLY 8 | 2.616 |
| FhuA+M6_B_3 | −33.071 | TYR 307-GLY 8 | 2.363 |
| FhuA+M6_D_1 | −39.595 | PRO415-GLU1 | 1.843 |
| Complex | I_sc (REU) |
|---|---|
| LPS–FhuA+M6_A | −6.038 |
| LPS–FhuA+M6_B | −3.975 |
| LPS–FhuA+M6_C | −3.772 |
| LPS–FhuA+M6_D | −4.021 |
| LPS–FhuA+M6_E | −4.833 |
| Complex * | I_sc (REU) | Hydrogen Bonds ** | Length (Å) |
|---|---|---|---|
| LPS+M6_B_1 | −40.625 | FTT718-ARG5 | 1.976 |
| FTT718-ARG5 | 2.227 | ||
| FTT718-ARG5 | 1.989 | ||
| FTT719-GLU1 | 1.759 | ||
| FTT721-GLU1 | 1.800 | ||
| FTT721-TRP2 | 1.985 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flórez-Castillo, J.M.; Rondón-Villareal, P.; Ropero-Vega, J.L.; Mendoza-Espinel, S.Y.; Moreno-Amézquita, J.A.; Méndez-Jaimes, K.D.; Farfán-García, A.E.; Gómez-Rangel, S.Y.; Gómez-Duarte, O.G. Ib-M6 Antimicrobial Peptide: Antibacterial Activity against Clinical Isolates of Escherichia coli and Molecular Docking. Antibiotics 2020, 9, 79. https://doi.org/10.3390/antibiotics9020079
Flórez-Castillo JM, Rondón-Villareal P, Ropero-Vega JL, Mendoza-Espinel SY, Moreno-Amézquita JA, Méndez-Jaimes KD, Farfán-García AE, Gómez-Rangel SY, Gómez-Duarte OG. Ib-M6 Antimicrobial Peptide: Antibacterial Activity against Clinical Isolates of Escherichia coli and Molecular Docking. Antibiotics. 2020; 9(2):79. https://doi.org/10.3390/antibiotics9020079
Chicago/Turabian StyleFlórez-Castillo, J. M., P. Rondón-Villareal, J. L. Ropero-Vega, S. Y. Mendoza-Espinel, J. A. Moreno-Amézquita, K. D. Méndez-Jaimes, A. E. Farfán-García, S. Y. Gómez-Rangel, and Oscar Gilberto Gómez-Duarte. 2020. "Ib-M6 Antimicrobial Peptide: Antibacterial Activity against Clinical Isolates of Escherichia coli and Molecular Docking" Antibiotics 9, no. 2: 79. https://doi.org/10.3390/antibiotics9020079
APA StyleFlórez-Castillo, J. M., Rondón-Villareal, P., Ropero-Vega, J. L., Mendoza-Espinel, S. Y., Moreno-Amézquita, J. A., Méndez-Jaimes, K. D., Farfán-García, A. E., Gómez-Rangel, S. Y., & Gómez-Duarte, O. G. (2020). Ib-M6 Antimicrobial Peptide: Antibacterial Activity against Clinical Isolates of Escherichia coli and Molecular Docking. Antibiotics, 9(2), 79. https://doi.org/10.3390/antibiotics9020079





