Characterization and Antimicrobial Activity of the Teleost Chemokine CXCL20b
Abstract
1. Introduction
2. Results
2.1. Producing of Recombinant Grass Carp CXCL20b
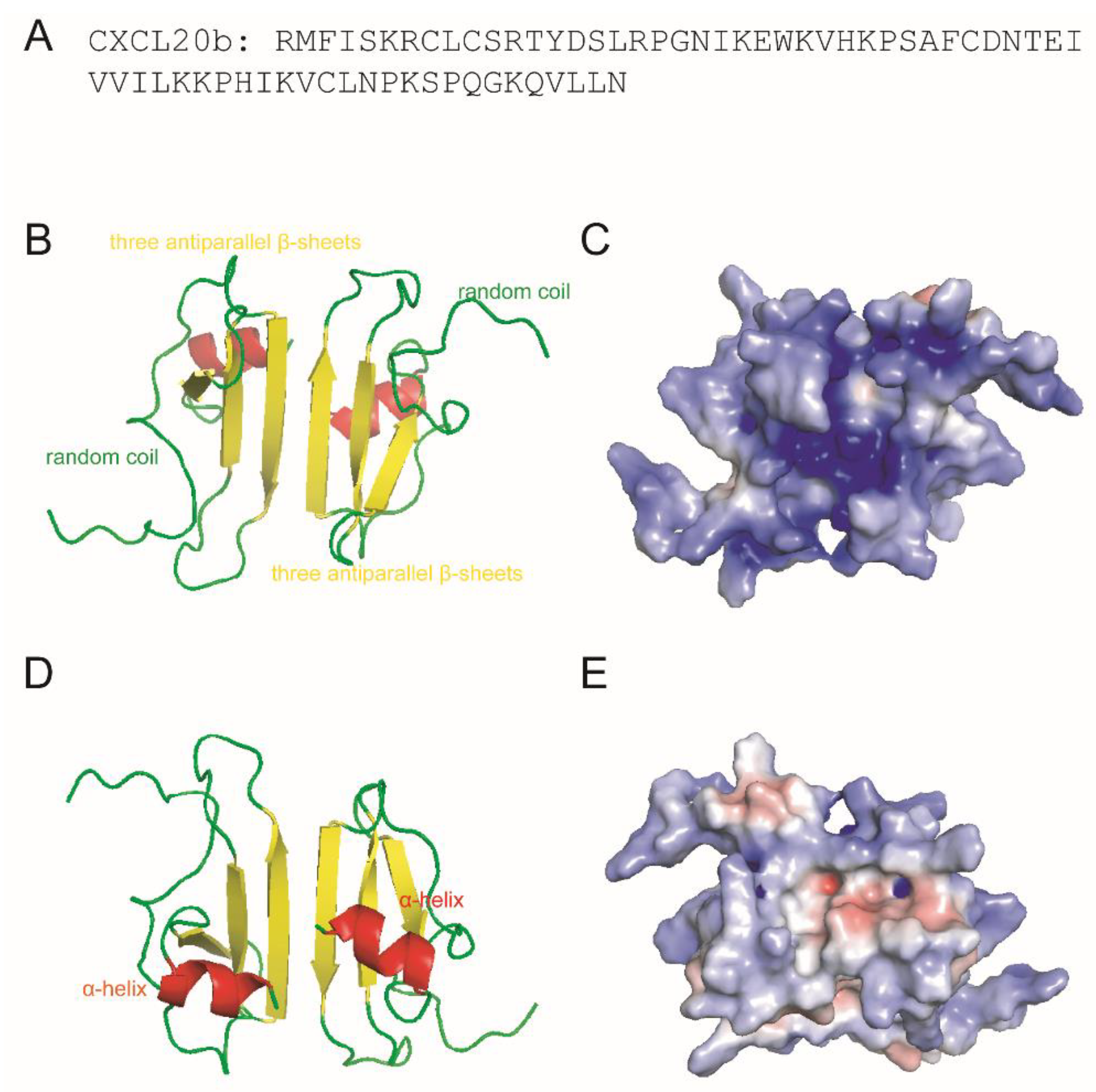
2.2. 3D Modeling and Net Charges Distribution Analysis of CXCL20b
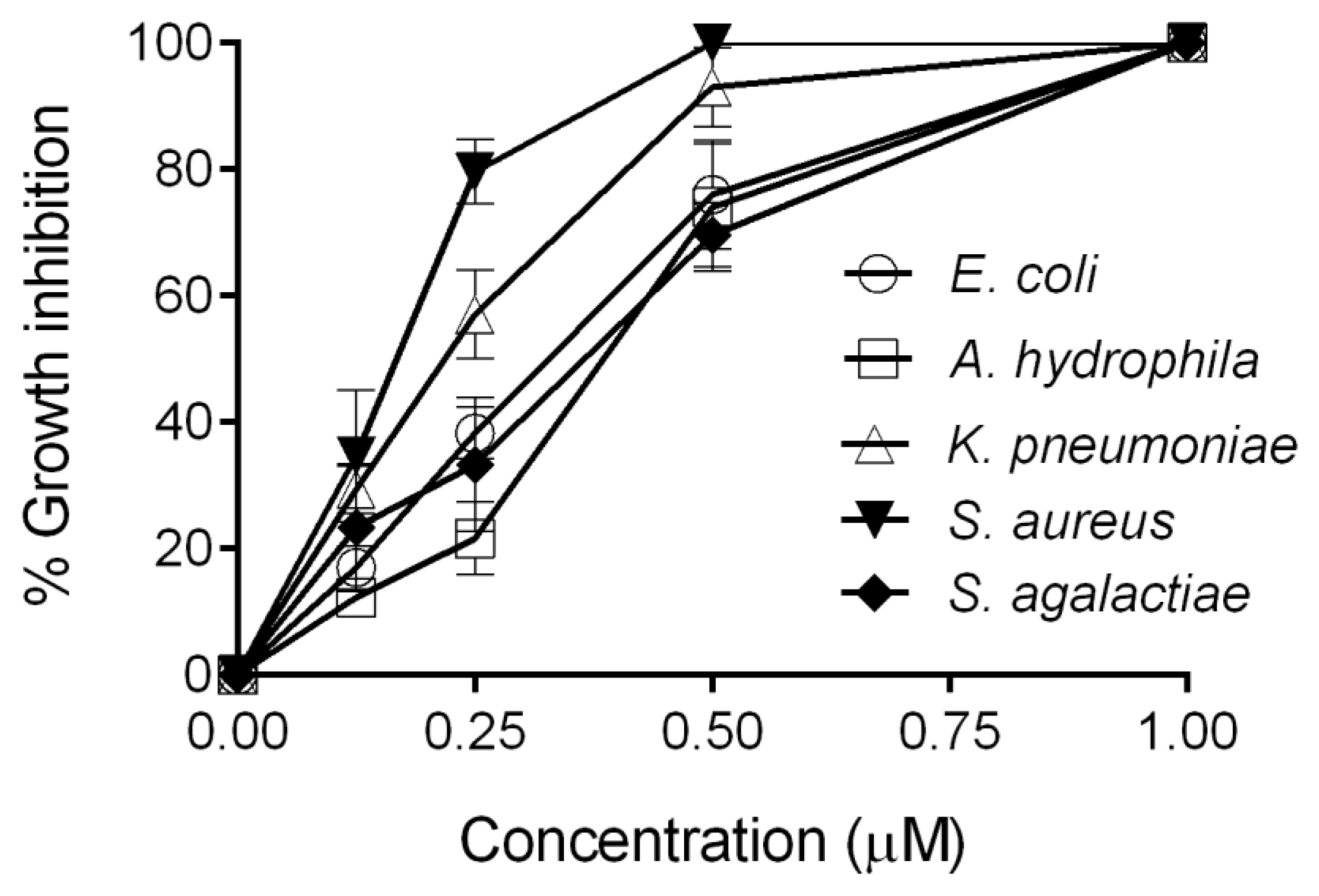
2.3. Antimicrobial Activity Assay of CXCL20b
2.4. DiOC2(3) Assay
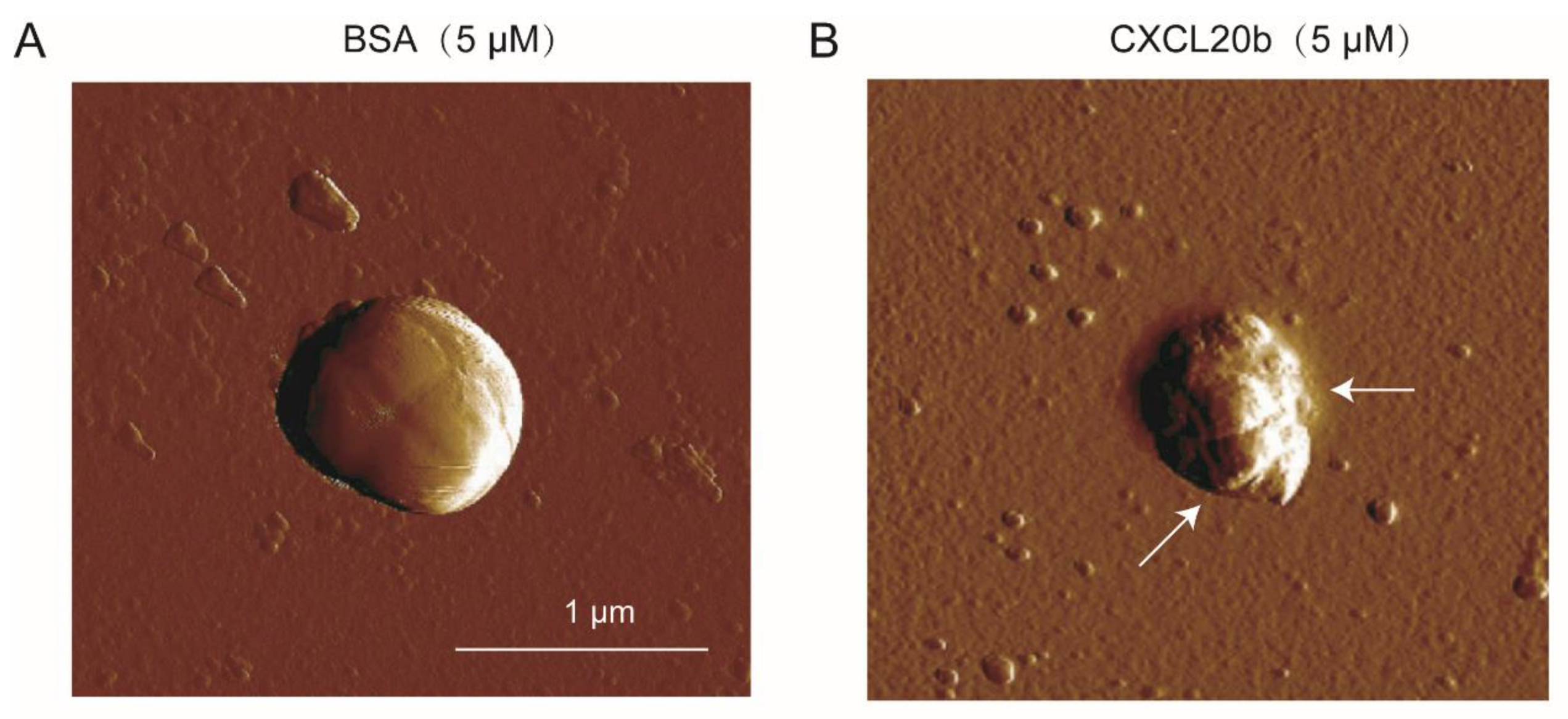
2.5. Atomic Force Microscopy (AFM) Assay
2.6. Structure Antimicrobial Assay of Grass Carp CXCL20b
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Recombinant Grass Carp CXCL20b Protein
4.2. Microorganisms and Peptides
4.3. Antimicrobial Assay
4.4. Fluorescence Microscopy
4.5. Atomic Force Microscopy (AFM)
4.6. Grass Carp CXCL 20b Modeling
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DiOC2(3) | 3,3′-Diethyloxacarbocyanine Iodide |
| AFM | Atomic Force Microscopy |
| BSA | Bovine Serum Albumin |
| MBC | Minimum Bactericidal concentration |
| PDB | Protein Data Bank |
| CCCP | Carbonyl cyanide 3-chlorophenylhydrazone |
| AMP | Antimicrobial peptide |
| NMR | Nuclear magnetic resonance |
References
- Harder, J.; Glaser, R.; Schroder, J.M. Human antimicrobial proteins effectors of innate immunity. J Endotoxin Res 2007, 13, 317–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Human Antimicrobial Peptides and Proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, D.; Landuyt, B.; Luyten, W.; Schoofs, L. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell. Immunol. 2012, 280, 22–35. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W.Y. alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Kavanagh, K.; Dowd, S. Histatins: Antimicrobial peptides with therapeutic potential. J. Pharm. Pharmacol. 2004, 56, 285–289. [Google Scholar] [CrossRef]
- Becknell, B.; Spencer, J.D. A review of ribonuclease 7’s structure, regulation, and contributions to host defense. Int. J. Mol. Sci. 2016, 17, 423. [Google Scholar] [CrossRef]
- Burian, M.; Schittek, B. The secrets of dermcidin action. Int. J. Med. Microbiol. 2015, 305, 283–286. [Google Scholar] [CrossRef]
- Yang, D.; Chen, Q.; Hoover, D.M.; Staley, P.; Tucker, K.D.; Lubkowski, J.; Oppenheim, J.J. Many chemokines including CCL20/MIP-3 alpha display antimicrobial activity. J. Leukoc. Biol. 2003, 74, 448–455. [Google Scholar] [CrossRef]
- Gallo, R.L.; Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012, 12, 503–516. [Google Scholar] [CrossRef]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Edderkaoui, B. Potential role of chemokines in fracture repair. Front. Endocrinol. 2017, 8, 39. [Google Scholar] [CrossRef]
- Nomiyama, H.; Hieshima, K.; Osada, N.; Kato-Unoki, Y.; Otsuka-Ono, K.; Takegawa, S.; Izawa, T.; Yoshizawa, A.; Kikuchi, Y.; Tanase, S.; et al. Extensive expansion and diversification of the chemokine gene family in zebrafish: Identification of a novel chemokine subfamily CX. BMC Genom. 2008, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.Y.; Yeaman, M.R. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 337–360. [Google Scholar] [CrossRef] [PubMed]
- Pomponi, S.A. The bioprocess-technological potential of the sea. J. Biotechnol. 1999, 70, 5–13. [Google Scholar] [CrossRef]
- Bird, S.; Tafalla, C. Teleost chemokines and their receptors. Biology 2015, 4, 756–784. [Google Scholar]
- Zlotnik, A.; Yoshie, O.; Nomiyama, H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006, 7, 243. [Google Scholar] [CrossRef]
- Fu, Q.; Zeng, Q.; Li, Y.; Yang, Y.; Li, C.; Liu, S.; Zhou, T.; Li, N.; Yao, J.; Jiang, C.; et al. The chemokinome superfamily in channel catfish: I. CXC subfamily and their involvement in disease defense and hypoxia responses. Fish Shellfish Immunol. 2017, 60, 380–390. [Google Scholar] [CrossRef]
- Liao, Z.; Wan, Q.; Xiao, X.; Ji, J.; Su, J. A systematic investigation on the composition, evolution and expression characteristics of chemokine superfamily in grass carp Ctenopharyngodon idella. Dev. Comp. Immunol. 2018, 82, 72–82. [Google Scholar] [CrossRef]
- Huising, M.O.; Stolte, E.; Flik, G.; Savelkoul, H.F.J.; Verburg-van Kemenade, B.M.L. CXC chemokines and leukocyte chemotaxis in common carp (Cyprinus carpio L.). Dev. Comp. Immunol. 2003, 27, 875–888. [Google Scholar] [CrossRef]
- Huising, M.O.; van der Meulen, T.; Flik, G.; Verburg-van Kemenade, B.M. Three novel carp CXC chemokines are expressed early in ontogeny and at nonimmune sites. Eur. J. Biochem. 2004, 271, 4094–4106. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, Q.; Wang, T.; Collet, B.; Corripio-Miyar, Y.; Bird, S.; Xie, P.; Nie, P.; Secombes, C.J.; Zou, J. Phylogenetic analysis of vertebrate CXC chemokines reveals novel lineage specific groups in teleost fish. Dev. Comp. Immunol. 2013, 41, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-J.; Chen, Q.; Rong, Y.-J.; Chen, F.; Chen, J. CXCR3.1 and CXCR3.2 differentially contribute to macrophage polarization in teleost fish. J. Immunol. 2017, 198, 4692–4706. [Google Scholar] [CrossRef]
- Zhou, S.; Mu, Y.; Ao, J.; Chen, X. Molecular characterization and functional activity of CXCL8_L3 in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2018, 75, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Zuniga, S.; Morales, R.A.; Munoz-Sanchez, S.; Munoz-Montecinos, C.; Parada, M.; Tapia, K.; Rubilar, C.; Allende, M.L.; Pena, O.A. CXCL12a/CXCR4b acts to retain neutrophils in caudal hematopoietic tissue and to antagonize recruitment to an injury site in the zebrafish larva. Immunogenetics 2017, 69, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zhu, S.; Shen, J.; Han, X.; Gao, J.; Wu, M.; Yu, Y.; Lu, H.; Han, W. Expression and purification of recombinant human Mig in Escherichia coli and its comparison with murine Mig. Protein Expr. Purif. 2012, 82, 205–211. [Google Scholar] [CrossRef]
- Yang, F.; Pan, Y.; Chen, Y.; Tan, S.; Jin, M.; Wu, Z.; Huang, J. Expression and purification of Canis interferon alpha in Escherichia coli using different tags. Protein Expr. Purif. 2015, 115, 76–82. [Google Scholar] [CrossRef]
- Hanzawa, H.; Haruyama, H.; Watanabe, K.; Tsurufuji, S. The 3-dimensional structure of rat cytokine cinc/gro in-solution by homonuclear 3D-nmr. Febs Lett. 1994, 354, 207–212. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.W.; Wong, G.C.L. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr. Opin. Solid State Mater. Sci. 2013, 17, 151–163. [Google Scholar] [CrossRef]
- Meller, S.; di Domizio, J.; Voo, K.S.; Friedrich, H.C.; Chamilos, G.; Ganguly, D.; Conrad, C.; Gregorio, J.; le Roy, D.; Roger, T.; et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat. Immunol. 2015, 16, 970–979. [Google Scholar] [CrossRef]
- Baindara, P.; Singh, N.; Ranjan, M.; Nallabelli, N.; Chaudhry, V.; Pathania, G.L.; Sharma, N.; Kumar, A.; Patil, P.B.; Korpole, S. Laterosporulin10: A novel defensin like Class IId bacteriocin from Brevibacillus sp strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology 2016, 162, 1286–1299. [Google Scholar] [CrossRef]
- Dai, C.; Basilico, P.; Cremona, T.P.; Collins, P.; Moser, B.; Benarafa, C.; Wolf, M. CXCL14 displays antimicrobial activity against respiratory tract bacteria and contributes to clearance of Streptococcus pneumoniae pulmonary Infection. J. Immunol. 2015, 194, 5980–5989. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Lee, M.W.; Wolf, A.J.; Limon, J.J.; Becker, C.A.; Ding, M.; Murali, R.; Lee, E.Y.; Liu, G.Y.; Wong, G.C.L.; et al. Direct antimicrobial activity of IFN-beta. J. Immunol. 2017, 198, 4036–4045. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wilson, E. The antimicrobial activity of CCL28 is dependent on C-terminal positively-charged amino acids. Eur. J. Immunol. 2010, 40, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Sathyamoorthi, A.; Bhatt, P.; Ravichandran, G.; Kumaresan, V.; Arasu, M.V.; Al-Dhabi, N.A.; Arockiaraj, J. Gene expression and in silico analysis of snakehead murrel interleukin 8 and antimicrobial activity of C-terminal derived peptide WS12. Vet. Immunol. Immunopathol. 2017, 190, 1–9. [Google Scholar] [CrossRef]
- Santana, P.A.; Salinas, N.; Alvarez, C.A.; Mercado, L.A.; Guzman, F. Alpha-helical domain from IL-8 of salmonids: Mechanism of action and identification of a novel antimicrobial function. Biochem. Biophys. Res. Commun. 2018, 498, 803–809. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Wu, Z.; Nuding, S.; Groscurth, S.; Marcinowski, M.; Beisner, J.; Buchner, J.; Schaller, M.; Stange, E.F.; Wehkamp, J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature 2011, 469, 419. [Google Scholar] [CrossRef]
- Munoz-Atienza, E.; Aquilino, C.; Syahputra, K.; Al-Jubury, A.; Araujo, C.; Skov, J.; Kania, P.W.; Hernandez, P.E.; Buchmann, K.; Cintas, L.M.; et al. CK11, a teleost chemokine with a potent antimicrobial activity. J. Immunol. 2019, 202, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hu, Y.; Zhou, J.; Hu, S.; Xiao, X.; Liu, X.; Su, J.; Yuan, G. Chitosan reduces the protective effects of IFN-gamma 2 on grass carp (Ctenopharyngodon idella) against Flavobacterium columnare infection due to excessive inflammation. Fish Shellfish Immunol. 2019, 95, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wu, H.; Cai, P.; Fein, J.B.; Chen, W. Atomic force microscopy measurements of bacterial adhesion and biofilm formation onto clay-sized particles. Sci. Rep. 2015, 5, 16857. [Google Scholar] [CrossRef] [PubMed]



| Bacteria Strain | MBC90 of CXCL20b (μM) | G+/G− |
|---|---|---|
| Staphylococcus aureus (ATCC 25923) | 0.25 | G+ |
| Streptococcus agalactiae (ATCC 13813) | 0.25 | G+ |
| Escherichia coli (ATCC 25922) | 0.5 | G− |
| Aeromonas hydrophila (ATCC 7966) | 0.5 | G− |
| Klebsiella pneumoniae (K13) | 0.5 | G− |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Zhang, Y.; Liao, Z.; Su, J. Characterization and Antimicrobial Activity of the Teleost Chemokine CXCL20b. Antibiotics 2020, 9, 78. https://doi.org/10.3390/antibiotics9020078
Xiao X, Zhang Y, Liao Z, Su J. Characterization and Antimicrobial Activity of the Teleost Chemokine CXCL20b. Antibiotics. 2020; 9(2):78. https://doi.org/10.3390/antibiotics9020078
Chicago/Turabian StyleXiao, Xun, Yanqi Zhang, Zhiwei Liao, and Jianguo Su. 2020. "Characterization and Antimicrobial Activity of the Teleost Chemokine CXCL20b" Antibiotics 9, no. 2: 78. https://doi.org/10.3390/antibiotics9020078
APA StyleXiao, X., Zhang, Y., Liao, Z., & Su, J. (2020). Characterization and Antimicrobial Activity of the Teleost Chemokine CXCL20b. Antibiotics, 9(2), 78. https://doi.org/10.3390/antibiotics9020078






