Abstract
Humidimycin (MDN-0010) is a ribosomally synthesized and post-translationally modified peptide (RiPP) belonging to class I lasso peptides, and is structurally related to siamycins, which have been shown to have strong antimicrobial activities against Gram-positive bacteria and to possess anti-HIV activity. Humidimycin was isolated from the strain Streptomyces humidus CA-100629, and was shown to synergize the activity of the fungal cell wall inhibitor caspofungin. In this work, the biosynthetic gene cluster of humidimycin was identified by genome mining of S. humidus CA-100629, cloned by Gibson assembly, and heterologously expressed.
1. Introduction
Natural products are non-essential small molecules produced by plants, microbes, and invertebrates. More than 70% of the natural product scaffolds used to produce clinically relevant anti-infective molecules are produced by Actinobacteria [1,2,3]. Advances in next-generation sequencing (NGS) technologies and genome mining approaches have reinvigorated the search for novel bioactive natural products [4,5]. The continuous development of bioinformatics tools and databases for the automated scanning and annotation of secondary metabolite gene clusters, such as antiSMASH (antibiotics and secondary metabolites analysis shell) [6], MIBiG (minimum information about a biosynthetic gene cluster) [7], or PRISM (prediction informatics for secondary metabolomes) [8], is enabling the discovery of new biosynthetic gene clusters (BGCs), including those encoding ribosomally synthesized and post-translationally modified peptides (RiPPs), which have been shown to be more widespread than first estimated [9,10].
Among RiPPs, lasso peptides are cyclic peptides, containing an N-terminal macrolactam ring comprised of seven to nine amino acid residues and a linear C-terminal peptide tail threaded through the ring. Lasso peptides are classified into four classes, depending on the number of disulfide bonds present (Figure 1A) [11]: Class I lasso peptides, only produced by strains of Streptomyces, have two disulfide bridges in the molecule; class II lasso peptides have no disulfide bonds; and class III have only one disulfide bond. Tietz et al. [12] described the new class IV to include the topology of lasso peptide LP2006, which contains two Cys forming a disulfide bridge on the tail and not linking the tail to the macrocycle, as occurs in class I and III lasso peptides.
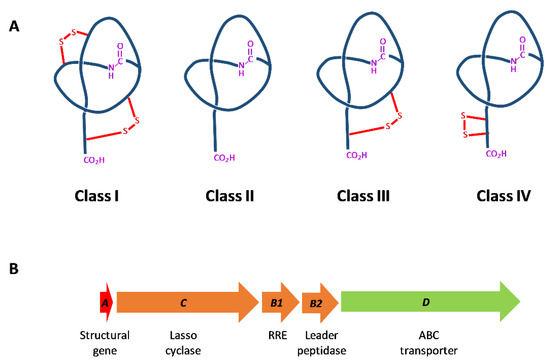
Figure 1.
Classification of lasso peptides according to their different topologies (A) and general overview of lasso peptide biosynthetic genes (B).
In recent years, genome mining approaches have considerably increased the number of identified lasso peptides and other RiPPs BGCs, and contributed to a better understanding of their biosynthesis [9,13]. In addition to antiSMAH and PRISM, platforms like BAGEL (bacteriocin genome mining tool) [14], RiPPER (RiPP precursor peptide enhanced recognition) [15], RiPPMiner (ribosomally synthesized post-translationally modified peptides miner) [16], and RODEO (rapid ORF description & evaluation online) [17] can efficiently mine genomic sequence data to search for lasso peptides.
Lasso peptides are synthesized from a precursor peptide, named A protein, which is processed by a leader peptidase (the B protein), homologous to a transglutaminase, and by a lasso cyclase (C-protein), homologous to an asparagine synthase, which is involved in the formation of the 7-9 residue macrolactam ring [12] (Figure 1B). Some lasso peptide gene clusters present B1 gene encoding for the N-terminal domain of the B protein and B2 gene encoding for the C-terminal domain. The B1 protein, also described as E-protein [12], contains the RiPP recognition element (RRE) [18], which recognizes the leader peptide and directs enzymatic modification [19]. Then, the B2 protein cleaves the leader peptide. The structural basis of leader peptide recognition by these so-called “split-B” proteins have been characterized in depth in Actinobacteria and Firmicutes [20].
Other genes present in many lasso peptide gene clusters encode for an ABC-transporter, presumably involved in the secretion and the self-immunity of the lasso peptide [21]. In other cases, a gene encoding for an isopeptidase is present instead of the ABC transporter. It has been proposed that clusters containing ABC-transporters might produce antimicrobial lasso peptides, while those containing isopeptidases might produce lasso peptides with another function [21]. Lasso peptide gene clusters are often flanked by regulatory proteins, but these have been rarely studied [13].
Humidimycin is a class I lasso peptide that was previously discovered by our research team as a new synergist of the of the fungal cell wall biosynthesis inhibitor caspofungin (CAS) [22]. The dramatic increase in the number of cases of life-threatening fungal infections and the lack of effective drugs make the development of novel approaches to fight against these infectious diseases necessary. The combination of humidimycin and CAS strongly increases the CAS efficacy against Aspergillus fumigatus and Candida albicans. The analysis of transcriptomes and selected A. fumigatus mutants has suggested that humidimycin affects the high osmolarity glycerol (HOG) response pathway and hits the CAS salvage pathway in human pathogenic fungi, a target that can be used to potentiate CAS activity [22]. Since humidimycin has no antifungal activity on its own and lacks in vitro cytotoxicity, this molecule could be an excellent example to propose the development of new combined treatments of invasive fungal infections [22].
Humidimycin was isolated from liquid culture broths of the actinomycete Streptomyces humidus CA-100629, and its structural elucidation showed that it is closely related to siamycins, which have anti-HIV activity and strong antimicrobial activities against Gram-positive bacteria [23,24,25,26]. The structure of humidimycin, a 21-amino-acid peptide that is cyclized from the side chain of Asp9 to the N-terminus of Cysl, has the topology of class I lasso peptides. Two disulfide bonds containing linkages Cys1 ➔ Cys13 and Cys7 ➔ Cys19 form the tricyclic structure [22]. Recently, Tan et al. [27] have found that siamycin I binds and inhibits the action of lipid II, the essential precursor molecule involved in peptidoglycan formation. Given the few differences in the amino acid sequences of class I lasso peptides, it might be also expected that other molecules of this group, including humidimycin, also inhibit peptidoglycan biosynthesis [27].
The BGCs of additional class I siamycin-like molecules (aborycin, specialicin, and MS-271) have been recently identified by genome mining, and some of them have been heterologously expressed [28,29,30].
In this work, we present the identification, cloning, and heterologous expression of the humidimycin BGC (hum) after genome mining of the producer strain S. humidus CA-100629. We also demonstrate that the C-terminal tryptophan of humidimycin is epimerized in both wild-type and heterologous strains.
2. Results and Discussion
2.1. Identification and Analysis of Humidimycin Biosynthetic Gene Cluster
The humidimycin BGC (hum) was located by mining the genome of strain CA-100629 (data not shown). The core sequence of humidimycin (CLGIGSCDDFAGCGYAIVCFW) was used to query the genome sequence in a tblastn and allowed us to find an open reading frame (ORF) (humA) that unambiguously corresponds to the structural gene. Among the described siamycin-like peptides, humidimycin is the only one having an aspartic residue in position 8 (Figure 2). From the nucleotide sequence of humA, we were able to deduce the amino acid sequence of the leader peptide of humidimycin (MSAIYEPPALQEIGDFDELTK). As shown in Figure 2, the leader peptide is very similar to the leader peptides of MS-271, aborycin, siamycin II, and specialicin, and contains the conserved YxxPxL motif and a threonine residue that are required for leader peptide recognition by the RRE in Actinobacteria [11,12].
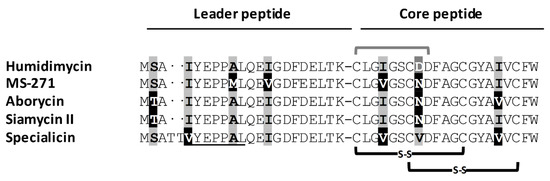
Figure 2.
Comparison of the leader and core sequences of the siamycin-like pre-propeptides. The disulfide bonds are indicated in black, and the amide bond between the N-terminal Cys and the side chain of the aspartic residue at position 9 is indicated in grey. Underlined residues correspond to the YxxPxL motif recognized by the RiPP recognition element (RRE)-containing protein. Differences in the sequences are highlighted.
The analysis of the ORFs present in a region of ~15 Kb up- and downstream the structural gene indicated that this cluster (hum) has a gene organization similar to those of other siamycin-like peptides (Figure 3). In fact, the proteins encoded in the hum BGC share a high degree of similarity with those described in other class I lasso peptides, which has permitted the proposal of a function for each gene based on these homologies [29,30] (Table 1). The hum cluster encodes a precursor peptide (HumA) that is processed by a split B protein HumB1 containing an RRE that joins the pre-propeptide and prefolds it via ATP-driven hydrolysis. Then, HumB2 cleaves the leader peptide to generate a 21-amino-acid linear peptide. Next, the lasso peptide cyclase HumC forms a macrolactam ring between the amino group of N-terminal Cys1 and the carboxyl group of Asp9, and the oxidoreductases HumE and HumF establish disulfide bonds between Cys1 and Cys13 and between Cys7 and Cys19, respectively. Finally, proteins HumD1/D2/D3/D4 should be involved in the export the peptide. The cluster also has one histidine kinase (HumG) and two DNA-binding response regulators (HumR1/R2) that are most probably involved in the regulation of gene expression.
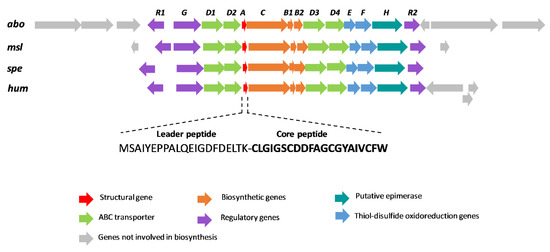
Figure 3.
Schematic representation of the biosynthetic gene cluster (BGC) of siamycin-like lasso peptides (abo: aborycin BGC from Streptomyces sp. SCSIO ZS0098; msl: MS-271 BGC from Streptomyces sp. M-271; spe: specialicin BGC from S. specialis JCM 16611T; hum: humidimycin BGC from S. humidus CA-100629). The sequences of the leader and core peptides of humidimycin are shown.

Table 1.
Proposed functions and homologies of the proteins encoded in the hum cluster.
Feng et al. [30] demonstrated the involvement of the homologous regulators MslR1, MslG, and MslR2 in the production of MS-271. They independently cloned mslR1, mslG, and mslR2 genes under the control of an ermEP* promoter and determined that MslR2 and MslG significantly increased product yield, while MslR1 decreased the production of MS-271. Given the high degree of homology of hum and msl regulators (Table 1), the production of humidimycin might follow the same regulation.
The cluster also harbors a gene encoding HumH, a protein homologous to MslH and SpeH, which have been proposed to epimerize Trp21 in MS-271 and specialicin, respectively [29,30]. Since spe, msl, and hum clusters lack an obvious epimerase, and the H protein is the only one of unknown function, is likely to be responsible for the epimerization of the C-terminal Trp of humidimycin [31]. The deletion of the mslH gene completely abolished MS-271 production [30], a fact that demonstrated that MslH is required for MS-271 biosynthesis, as most probably is HumH. Recently, Ogasawara et al. [32] described that PgsA, which has a 52% identity to MslH, catalyzes the epimerization of L-Glu residues to D-Glu in the growing poly-γ-glutamic acid (PGA) chain, a polymer that is present in the mucilage of many bacteria and which contains D- and L-Glu connected via an amide bond. The authors propose that MslH would act in a similar manner to PgsA in MS-271 biosynthesis, although the detailed reaction mechanism is still unclear.
2.2. Cloning and Heterologous Expression of Humidimycin Biosynthetic Gene Cluster
To prove that the hum cluster is responsible for humidimycin production, and to check if the C-terminal Trp of humidimycin is epimerized, the 14,987 bp fragment containing the hum genes was cloned in pCAP01 by Gibson assembly, yielding the plasmid pHUM. To that end, four overlapping fragments of 4.590 Kb, 4.452 Kb, 3.070 Kb, and 3.063 Kb were amplified by PCR. A XhoI-linearized vector pCAP01 (9 Kb) was also amplified. The five fragments were combined for Gibson assembly, and the reaction was transformed into E. coli NEB 10-beta competent cells. Clones were checked by restriction analysis, and one of the clones harboring pHUM was selected to purify the vector.
Then, pHUM was used to transform CmR E. coli ET12567. Since pHUM contains the kanamycin-resistant marker, we could not directly transform non-methylating CmR KmR E. coli ET12567/pUB307. Thus, we performed three independent, triparental, intergeneric conjugations, using E. coli ET12567/pHUM and ET12567/pUB307 as donor strains and the spores of each of the heterologous hosts (S. coelicolor M1152, M1154 and S. albus J1074) as recipient strains. For the negative controls, a triparental conjugation was also made, using E. coli ET12567/pCAP01 and ET12567/pUB307 as donor strains with the same recipient strains. Transconjugants were checked by PCR with primers HumidA_fw and HumidA_rv to confirm the integration of the hum BGC into the chromosomes of the heterologous hosts.
Five positive transconjugants from each heterologous host, together with the negative controls, were grown on liquid RAM2, the original humidimycin-producing medium, over 7 days at 28 °C, and then acetone extracts from the cultures were obtained. After removing of the solvent, the residue was resuspended in 20% dimethyl sulfoxide (DMSO)/water and analyzed by LC-HRESI-TOF.
All the transconjugants showed a peak at 4.8 minutes, which was absent in the negative controls (Figure 4A) and which coincided with the retention time of elution of the humidimycin standard. The perfect correlation between the UV spectrum, exact mass, and isotopic distribution between the humidimycin standard (Figure 4B, top) and the component isolated from the transconjugant S. coelicolor M1154/pHUM (Figure 4B, bottom) unequivocally confirmed that the latter corresponded to humidimycin. S. albus J1074 and the engineered strains S. coelicolor M1152 and S. coelicolor M1154 have been largely used as heterologous hosts to produce of a broad range of compound classes with improved yields [33]. In most cases, S. albus J1074 provides better yields than the S. coelicolor strains, a fact that could be due to the presence of an additional active vector integration attB site in its chromosome [34]. However, in the case of pHUM transconjugants, the production of humidimycin was higher in the transconjugant S. coelicolor M1154-pHUM (Figure 4A).
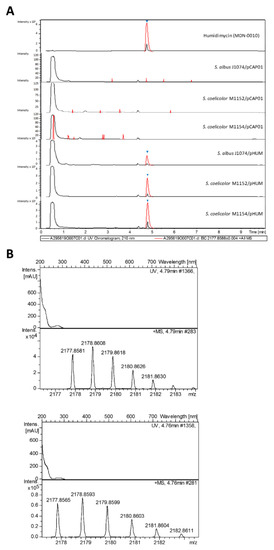
Figure 4.
UV profiles at 210 nm (black) and extracted ion chromatograms (red) (A) at 2177.8588 ± 0.004 (C98H133N22O27S4+) of humidimycin, the negative controls S. albus J1074/pCAP01, S. coelicolor M1152/pCAP01, and S. coelicolor M1154/pCAP01, and the transconjugants S. albus J1074/pHUM, S. coelicolor M1152/pHUM, and S. coelicolor M1154/pHUM. ▼: humidimycin. (B) Experimental UV and mass spectra of M + H+ humidimycin adduct (C98H133N22O27S4+) from S. coelicolor M1154/pHUM (top), and from the original producer strain CA-100629 (bottom).
2.3. Marfey’s Analysis of Humidimycin C-terminal Tryptophan
When the structure of humidimycin was determined [22], all the amino acids were assumed to have an L-configuration analogous to what had been described for siamycin II [35]. However, considering the similarity of the biosynthesis with that of MS-271 [30] and specialicin [29], which have been described as having a D-tryptophan, it was reasonable to think that humidimycin, and by extension the rest of siamycins, could also contain a D-Trp. We determined the absolute configuration of the C-terminal Trp using Marfey’s method, and confirmed the D-stereochemistry of the C-terminal Trp of humidimycin isolated from both the original producer strain and the transconjugant S. coelicolor M1154/pHUM (Figure 5). As the hum cluster lacks obvious epimerase genes to catalyze the stereochemical inversion of Trp21, and (as stated above) HumH is homologous to SpeH and MslH, we propose this protein to also be responsible for the epimerization.
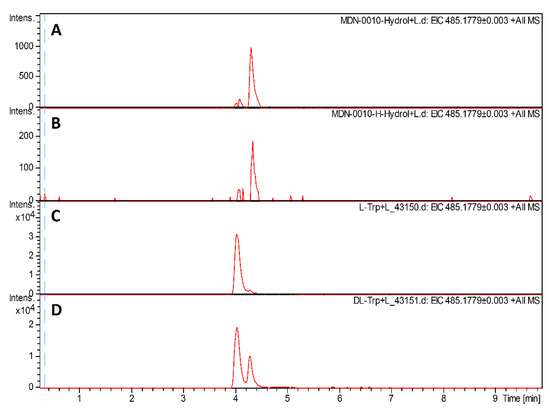
Figure 5.
Marfey’s chiral analysis of the C-terminal tryptophan (Trp) using 1-fluoro-2,4-dinitrophenyl-5-L-valine-amide (L-FDVA). The LC-MS profile was monitored at 485.1779 m/z (M + H+ of FDVA derivatives of Trp). (A) Humidimycin from strain S. humidus CA-100629, (B) humidimycin from S. coelicolor M1154/pHUM cultures, (C) L-Trp standard. and (D) L-Trp + D-Trp standard.
2.4. Genome Mining of Additional Humidimycin-like Biosynthetic Gene Clusters
A tblastn search of the humidimycin core peptide against both nucleotide and Streptomyces whole genome sequences (WGS) databases from NCBI has shown that no other humidimycin gene clusters are contained in these genomes, and that siamycin I/MS-271 and siamycin IIII/RP 71955/aborycin are the siamycin-like lasso peptides most frequently detected (Figure 6). We also found an undescribed BGC for siamycin II in the genome of the strain Streptomyces sp. F-7 (GenBank FKJH01000010.1) that shares a high degree of homology with the rest of siamycin-like clusters (Figure 7B).

Figure 6.
Humidimycin homologs found by BLAST against the nucleotide and Streptomyces WGS databases from NCBI. Letters in bold indicate amino acid differences.
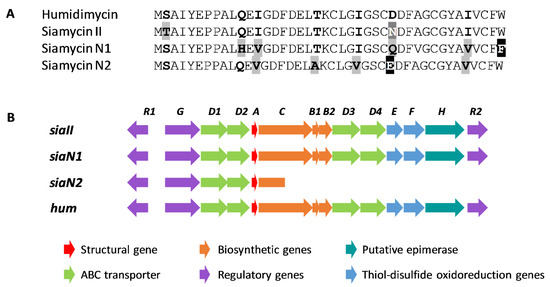
Figure 7.
Comparison of the pre-propeptide sequences (A) and the biosynthetic gene cluster (BGC) (B) of humidimycin, siamycin II, and the new siamycins (N1 and N2) identified by genome mining.
This search also allowed us to find two new siamycin-like peptides (Figure 6 and Figure 7A); one, named for clarity as siamycin N1, in the genomes of Streptomyces sp. CB02056 (GenBank LIPD01000011.1) and Streptomyces xanthocidicus MMS17-GH009 (GenBank QVIG01000001.1), and another, named siamycin N2, in the genome of Streptomyces sp. NL15-2K (GenBank BHXA01000012.1). In the case of siamycin N2, the pathway lacks the genes necessary to synthesize a lasso peptide, and contains a truncated C protein. However, in the case of siamycin N1, a complete and typical siamycin cluster was found in both genomes (Figure 7B). It is interesting to note in the case of siamycin N1 that the residue in position 21 is a phenylalanine and not a tryptophan, as has been described for all the siamycin-like peptides (Figure 7A). Considering that the siamycin N1 BGC also harbors a HumH homolog, it would be of great interest to study whether this gene is also able to epimerize Phe21. The study of the biological activities of siamycin N1 would pave the way to the detailed molecular characterization of this group of molecules.
3. Materials and Methods
3.1. Bacterial Strains and Plasmids
Strain Streptomyces humidus CA-100629 was isolated from a soil collected in Almería (Spain) and deposited in MEDINA culture collection [22]. NEB 10-beta competent E. coli (New England BioLabs, Ipswich, MA, United States), E. coli ET12567 (LGC Standards, Manchester, NH, United States), and E. coli ET12567/pUB307 [36] (kindly provided by José Antonio Salas) were employed throughout the cloning and conjugation processes. Vector pCAP01, a S. cerevisiae/E. coli/actinobacteria shuttle vector allowing site-specific φC31 integration of the cloned gene cluster into the chromosomes of heterologous Actinobacteria hosts [37], was a gift from Bradley Moore (Addgene plasmid #59981; http://n2t.net/addgene:59981; RRID: Addgene_59981). Streptomyces coelicolor M1152 and M1154 heterologous hosts [38] were kindly gifted by Mervyn Bibb. The Streptomyces albus J1074 heterologous host [39] was generously provided by José Antonio Salas.
3.2. Growth and Culture Conditions
Streptomyces humidus CA-100629 was cultured on ATCC-2 liquid medium (soluble starch 20 g/L, glucose 10 g/L, NZ Amine Type E 5 g/L, meat extract 3 g/L, peptone 5 g/L, yeast extract 5 g/L, calcium carbonate 1 g/L; pH 7) and grown in an orbital shaker at 28 °C, 220 rpm, and 70% relative humidity. E. coli strains were routinely cultured in LB broth (Miller) (Merck KGaA, Darmstadt, Germany) (37 °C, 250 rpm) and Difco LB agar Lennox (Fisher Scientific, Madrid, Spain) (37 °C, static). Intergeneric conjugations were carried out on MA (MOPS 21 g/L, glucose 5 g/L, yeast extract 0.5 g/L, beef extract 0.5 g/L, casamino acids 1 g/L, agar 25 g/L; pH 7). Transconjugants were grown on MA. Antibiotics were supplemented when required for selection of transformants at the following final concentrations: kanamycin (50 μg/mL), nalidixic acid (25 μg/mL), and choramphenicol (25 μg/mL). For heterologous expression experiments, RAM2 medium was used (corn meal 4 g/L, glucose 10 g/L, maltose 15 g/L, pharmamedia 7.5 g/L, primary yeast 5 g/L; pH 7), and the recombinant strains were incubated for 7 days on an orbital shaker at 28 °C, 220 rpm, and 70% relative humidity.
3.3. General Molecular Biology Techniques
Restriction endonucleases and Q5 High-Fidelity polymerase were purchased from New England Biolabs (Ipswich, MA, United States). Calf intestinal alkaline phosphatase (CIAP) was purchased from Invitrogen (Waltham, MA, United States). Gibson Assembly® Ultra Kit was purchased from VWR (Chester, PA, United States). All the primers employed were purchased at Sigma-Aldrich (St. Louis, MO, United States) and are described in Table 2. A QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) was employed for the purification of plasmid DNA, and the IllustraTM GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare, Boston, MA, United States) was used for the purification of DNA amplicons from agarose gels and enzymatic reactions.

Table 2.
Primers used in this study.
3.4. Location and Analysis of Humidimycin Biosynthetic Gene Cluster
The sequence of the humidimycin BGC has been deposited at GenBank under the accession number MN956991. To identify the structural gene and the rest of ORFs present in the cluster, and to assign functions, the tBlastn and blastp algorithms were used (https://blast.ncbi.nlm.nih.gov), as well as antiSMASH [6].
3.5. Gibson Assembly Cloning of Humidimycin Biosynthetic Gene Cluster
Cloning of humidimycin BGC was performed by Gibson assembly. Four overlapping fragments between 3 and 4.5 Kb were PCR amplified using the genomic DNA of strain CA-100629 and the primers listed in Table 2. Also, vector pCAP01 was linearized with XhoI, and then used as the template in a PCR with primers pCAPHumGibF and pCAPHumGibR (Table 2), which have 5’ overhang sequences that would allow for Gibson assembly with the ends of the hum BGC. The PCR was digested with DpnI to eliminate the template DNA.
All the PCRs were purified after agarose gel electrophoresis using the IllustraTM GFXTM PCR DNA and Gel Band Purification Kit, and employed for a Gibson assembly reaction according to the manufacturer’s protocol. E. coli NEB 10-beta was transformed with the reaction, and kanamycin was used as a selection marker to screen for cells carrying the target plasmid.
Plasmid DNA was isolated from the transformants, and the correct cloning of the hum BGC was checked by restriction enzyme digestion and PCR amplification with primers HumidA_fw and HumidA_rv (Table 2).
3.6. Intergeneric Conjugation
Plasmid pHUM was introduced into Streptomyces hosts by triparental conjugation, as previously described [40]. In summary, purified pHUM was used to electroporate the methylation-deficient strain E. coli ET12567. Cells from E. coli ET12567/pUB307 and E. coli ET12567/pHUM were collected at an O.D. of 0.4-0.6, washed twice with LB to remove the antibiotics, resuspended in 100 µL of LB, and mixed with 50 µl of freshly activated spores of S. coelicolor M1152, M1154, and S. albus J1074. The mixtures were plated on MA and overlaid after about 16 h with nalidixic acid (25 μg/mL) and kanamycin (50 μg/mL). After few days of incubation, five colonies from each Streptomyces heterologous host were picked and streaked on MA plates containing nalidixic acid (25 μg/mL) and kanamycin (50 μg/mL). The insertion of pHUM into the Streptomyces hosts chromosomes was checked by PCR, employing the genomic DNA of the transconjugants and the primers HumidA_fw and HumidA_rv (Table 2).
3.7. Heterologous Expression of Humidimycin Biosynthetic Gene Cluster and LC-ESI-TOF Analysis
Seed cultures on ATCC-2 of the recombinant strains S. coelicolor M1152/pHUM, M1154/pHUM and S. albus J1074/pHUM, together with the corresponding negative controls harboring empty pCAP01 vector, were used to inoculate EPA vials containing 10 mL of RAM2 fermentation medium. The vials were incubated at 28 °C for 7 days at 220 rpm and 70% humidity, and then the broths were subjected to extraction with acetone. The organic solvent was evaporated to dryness and the extract resuspended to a final ratio of 20% DMSO/water. The microbial extracts were filtered and analyzed employing a Bruker maXis QTOF high resolution mass spectrometer coupled to a HPLC system, as previously described [41].
3.8. Marfey’s Analysis of Tryptophan
Two samples of humidimycin from strain CA-100629 (0.1 mg) and from S. coelicolor M1154/pHUM were treated with 6 N HCl containing a 5% of thioglicolic acid in a sealed vial at 110 °C for 24 h. The solutions were concentrated to dryness in vacuo. The hydrolysates were reconstituted in H2O (100 μL) and treated with a solution of 1-fluoro-2,4-dinitrophenyl-5-L-valine-amide (L-FDVA, 300 μL, 1% in acetone) and a 1 M solution of NaHCO3 (50 μL) in a sealed vial at 40 °C for 1 h. The reaction mixtures were neutralized with 1 N HCl (50 μL), and an aliquot (10 μL) was diluted with CH3CN (40 μL). The resulting solutions were analyzed by LC-MS employing a Zorbax SB-C8 column (2.1 × 30 mm, 3.5 μm) and a gradient elution profile of 10% B (90% CH3CN, 10% H2O, 0.01% TFA, 1.3 mM ammonium formiate)/90% A (10% CH3CN, 90% H2O, 0.01% TFA, 1.3 mM ammonium formiate) to 100% B over 6 min at a flow rate of 0.3 mL/min.
4. Conclusions
In this work, we have cloned and heterologously expressed in three different hosts the biosynthetic gene cluster of humidimycin (MDN-0010) from Streptomyces humidus CA-100629. The cluster is highly homologous to those of other siamycin-like peptides (aborycin, specialicin, MS-271), and contains genes encoding the precursor peptide (HumA), the enzymes involved in post-translational modifications (HumB1, HumB2, HumC, HumE, HumF), transporters (HumD1-D4), regulators (HumR1-R2, HumG), and a protein (HumH) that have been postulated to catalyze the epimerization of the tryptophan in position 21. We have in fact confirmed that the stereochemistry of Trp21 is D-, a modification that could increase the stability of the protein.
A search of novel siamycins in the NCBI microbial genomes database showed that new siamycin-like peptides are yet to be discovered. The few differences in the amino acid sequences of class I lasso peptides suggest that, as has been demonstrated with siamycin I and humidimycin, these molecules could also inhibit peptidoglycan biosynthesis and/or potentiate the activity of the antifugal compound caspofungin. The increasing number of sequenced genomes will help to better characterize of this group of molecules.
Author Contributions
M.S.-H., J.M. conceived, designed, and performed the experiments, as well as the data analysis. M.S.-H., J.M., and O.G. contributed to/wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We thank Ignacio González, Mar Estévez, José Rubén Tormo, and Paqui Muñoz (Fundación MEDINA) for their technical support. We thank Bradley Moore for providing plasmid pCAP01. We thank José Antonio Salas and the University of Oviedo for kindly providing strains Streptomyces albus J1074 and Escherichia coli ET12567/pUB307. We also thank Mervyn Bibb and the John Innes Centre for generously providing strains Streptomyces coelicolor M1152 and Streptomyces coelicolor M1154.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gomez-Escribano, J.P.; Bibb, M.J. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: From genome mining to manipulation of biosynthetic pathways. J. Ind. Microbiol. Biot. 2014, 41, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Genilloud, O. Natural products discovery and potential for new antibiotics. Curr. Opin. Microbiol. 2019, 51, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Alt, S.; Bibb, M.J. Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar. Drugs 2016, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Mining actinomycetes for novel antibiotics in the omics era: Are we ready to exploit this new paradigm? Antibiotics 2018, 7, 85. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef]
- Poorinmohammad, N.; Bagheban-Shemirani, R.; Hamedi, J. Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria. Antonie Leeuwenhoek 2019, 112, 1477–1499. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Tan, S.; Moore, G.; Nodwell, J. Put a Bow on It: Knotted Antibiotics Take Center Stage. Antibiotics 2019, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Link, A.J. Genome mining for lasso peptides: Past, present, and future. J. Ind. Microbiol. Biot. 2019, 46, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Santos-Aberturas, J.; Chandra, G.; Frattaruolo, L.; Lacret, R.; Pham, T.H.; Vior, N.M.; Eyles, T.H.; Truman, A.W. Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool. Nucleic Acids Res. 2019, 47, 4624–4637. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef]
- Schwalen, C.J.; Mitchell, D. Discovery of Antibiotic Peptides from Novelty-Prioritized Natural Product Genome Mining. FASEB J. 2017, 31 (Suppl. 1), 939.8. [Google Scholar] [CrossRef]
- Burkhart, B.J.; Hudson, G.A.; Dunbar, K.L.; Mitchell, D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 2015, 11, 564–570. [Google Scholar] [CrossRef]
- Zhu, S.; Fage, C.D.; Hegemann, J.D.; Mielcarek, A.; Yan, D.; Linne, U.; Marahiel, M.A. The B1 protein guides the biosynthesis of a lasso peptide. Sci. Rep. 2016, 6, 35604. [Google Scholar] [CrossRef]
- Sumida, T.; Dubiley, S.; Wilcox, B.; Severinov, K.; Tagami, S. Structural basis of leader peptide recognition in lasso peptide biosynthesis pathway. ACS Chem. Biol. 2019, 14, 1619–1627. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015, 48, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Valiante, V.; Monteiro, M.C.; Martín, J.; Altwasser, R.; El Aouad, N.; González, I.; Kniemeyer, O.; Mellado, E.; Palomo, S.; de Pedro, N.; et al. Hitting the caspofungin salvage pathway of human-pathogenic fungi with the novel lasso peptide humidimycin (MDN-0010). Antimicrob. Agents Chemother. 2015, 59, 5145–5153. [Google Scholar] [CrossRef] [PubMed]
- Helynck, G.; Dubertret, C.; Mayaux, J.F.; Leboul, J. Isolation of RP 71955, a new anti-HIV-1 peptide secondary metabolite. J. Antibiot. (Tokyo) 1993, 46, 1756–1757. [Google Scholar] [CrossRef] [PubMed]
- Potterat, O.; Stephan, H.; Metzger, J.W.; Gnau, V.; Zähner, H.; Jung, G. Aborycin–A Tricyclic 21-Peptide Antibiotic Isolated from Streptomyces griseoflavus. Liebigs Annalen Chemie 1994, 1994, 741–743. [Google Scholar] [CrossRef]
- Tsunakawa, M.; Hu, S.L.; Hoshino, Y.; Detlefson, D.J.; Hill, S.E.; Furumai, T.; White, R.J.; Nishio, M.; Kawano, K.; Yamamoto, S.; et al. Siamycins I and II, new anti-HIV peptides: I. Fermentation, isolation, biological activity and initial characterization. J. Antibiot. (Tokyo) 1995, 48, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.F.; Samanta, H.; Bechtold, C.M.; Deminie, C.A.; Patick, A.K.; Alam, M.; Riccardi, K.; Rose, R.E.; White, R.J.; Colonno, R.J. Characterization of siamycin I, a human immunodeficiency virus fusion inhibitor. Antimicrob. Agents Chemother. 1996, 40, 133–1387. [Google Scholar] [CrossRef]
- Tan, S.; Ludwig, K.C.; Müller, A.; Schneider, T.; Nodwell, J.R. The lasso peptide siamycin-I targets lipid II at the Gram-positive cell surface. ACS Chem. Biol. 2019, 14, 966–974. [Google Scholar] [CrossRef]
- Shao, M.; Ma, J.; Li, Q.; Ju, J. Identification of the Anti-Infective Aborycin Biosynthetic Gene Cluster from Deep-Sea-Derived Streptomyces sp. SCSIO ZS0098 Enables Production in a Heterologous Host. Mar. Drugs 2019, 17, 127. [Google Scholar] [CrossRef]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Harada, S.; Kodani, S. Isolation and structure determination of a new lasso peptide specialicin based on genome mining. Bioorg. Med. Chem. 2018, 26, 6050–6055. [Google Scholar] [CrossRef]
- Feng, Z.; Ogasawara, Y.; Nomura, S.; Dairi, T. Biosynthetic Gene Cluster of ad-Tryptophan-Containing Lasso Peptide, MS-271. ChemBioChem 2018, 19, 2045–2048. [Google Scholar] [CrossRef]
- Ogasawara, Y. New enzymes for peptide biosynthesis in microorganisms. Biosci. Biotechnol. Biochem. 2019, 83, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, Y.; Shigematsu, M.; Sato, S.; Kato, H.; Dairi, T. Involvement of Peptide Epimerization in Poly-γ-glutamic Acid Biosynthesis. Org. Lett. 2019, 21, 3972–3975. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Luzhetskyy, A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, B.; Luzhetskyy, A. Unusual site-specific DNA integration into the highly active pseudo-attB of the Streptomyces albus J1074 genome. Appl. Microbiol. Biotechnol. 2014, 98, 5095–5104. [Google Scholar] [CrossRef]
- Constantine, K.L.; Friedrichs, M.S.; Detlefsen, D.; Nishio, M.; Tsunakawa, M.; Furumai, T.; Ohkuma, H.; Oki, T.; Hill, S.; Bruccoleri, R.E.; et al. High-resolution solution structure of siamycin II: Novel amphipathic character of a 21-residue peptide that inhibits HIV fusion. J. Biomol. NMR 1995, 5, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Flett, F.; Mersinias, V.; Smith, C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Nat. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef]
- Chater, K.F.; Wilde, L.C. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J. Gen. Microbiol. 1980, 116, 323–334. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000; Volume 291. [Google Scholar]
- Martin, J.; Crespo, G.; González-Menéndez, V.; Pérez-Moreno, G.; Sánchez-Carrasco, P.; Pérez-Victoria, I.; Ruiz-Pérez, L.M.; González-Pacanowska, D.; Vicente, F.; Genilloud, O.; et al. MDN-0104, an antiplasmodial betaine lipid from Heterospora chenopodii. J. Nat. Prod. 2014, 77, 2118–2123. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).