Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Isolates
2.2. Confirmation of Bacterial Identities
2.3. Multilocus Sequence Typing (MLST)
2.4. Biofilm Formation
2.5. Quantification of Biofilm Mass
2.6. Detection of Biofilm-Associated Genes
2.7. Determination of Minimal Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.8. Determination of Minimal Biofilm Inhibitory Concentration (MBIC) and Minimum Biofilm Eradication Concentration (MBEC) Assay
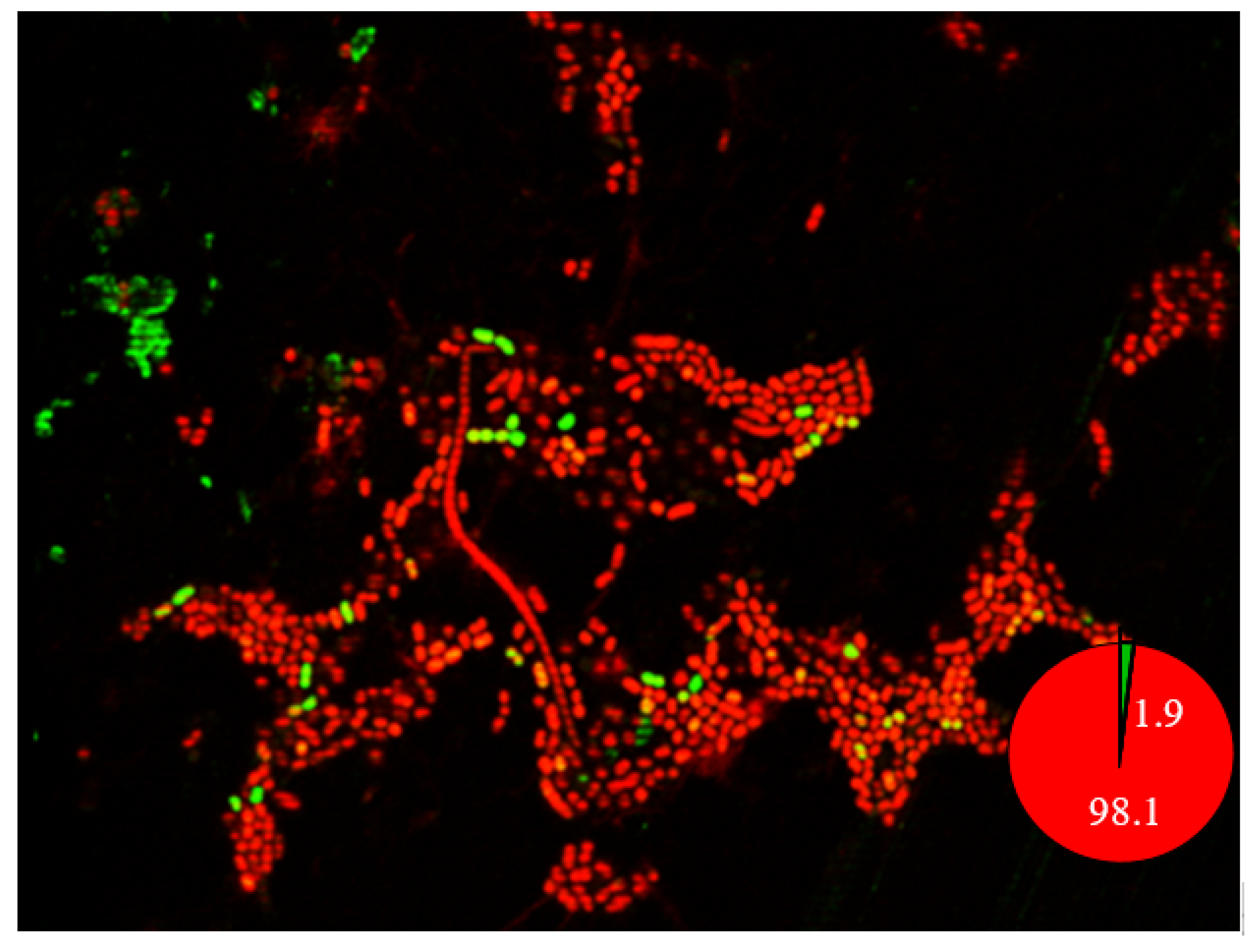
2.9. Confocal Laser Scanning Microscopy Imaging of Biofilm
3. Reversibility of Antibiotic Resistance of the Biofilm Cells
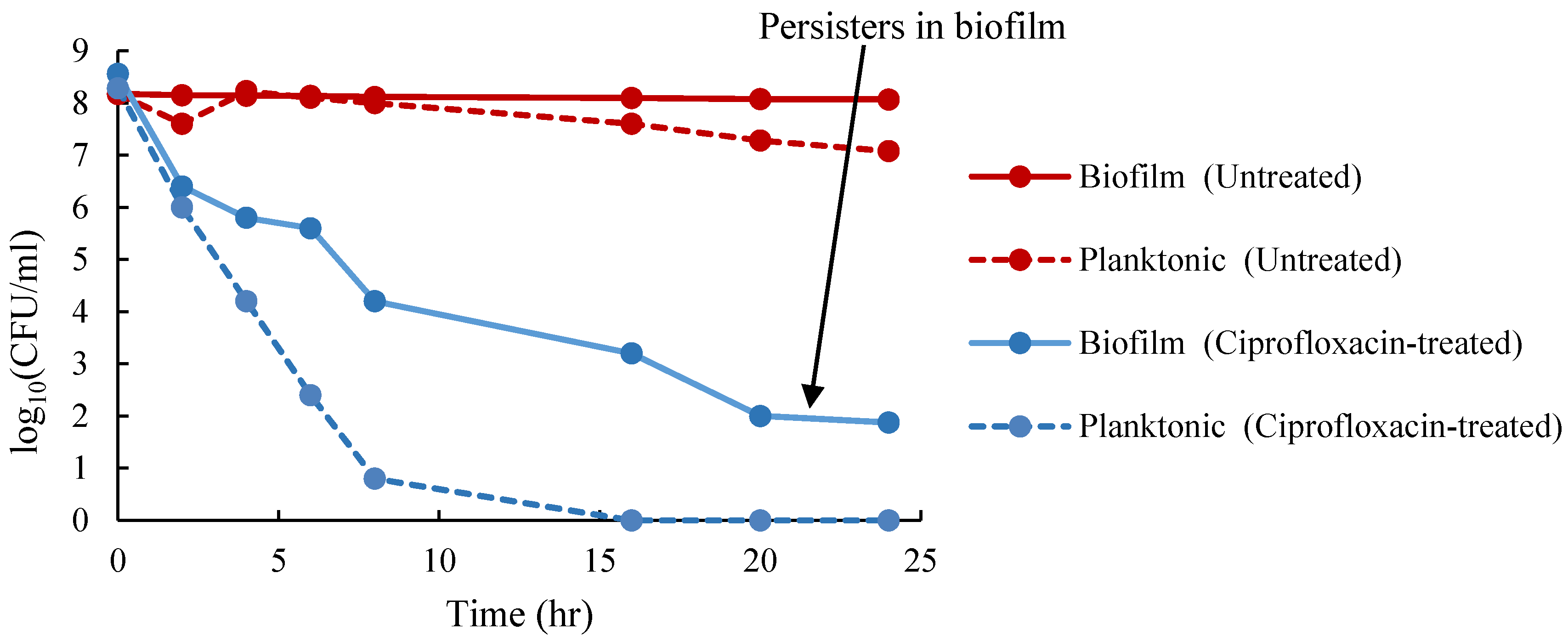
3.1. Enumeration of Persister Cells from Planktonic and Biofilm Populations
3.2. Statistical Analysis
4. Results
4.1. Antibiotic Susceptibility Profiles of the A. baumannii Strains
4.2. Biofilm-Forming Capacities of the A. baumannii Isolates
4.3. Distribution of Biofilm-Associated Virulence Genes in the Clinical A. baumannii Strains
4.4. Comparison of MIC and MBIC
4.5. Comparison of MBC and MBEC
4.6. Reversibility of Antibiotic Susceptibility in Planktonic Cells Regrown from Biofilm
4.7. Isolation of Persister Cells in Planktonic and Biofilm Cells
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fournier, P.E.; Richet, H.; Weinstein, R.A. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Priority Pathogens List for R&D of New Antibiotics. Available online: https://www.who.int/en/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-forwhich-new-antibiotics-are-urgently-needed (accessed on 10 August 2020).
- Piperaki, E.T.; Tzouvelekis, L.; Miriagou, V.; Daikos, G. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Wu, T.C.; Chao, D.V.K.; Hung, I.F.N.; Liu, L.; Lung, D.C. Reducing Bacterial Resistance with IMPACT, 5th ed.; Centre for Health Protection: Hong Kong, China, 2017.
- Niu, T.; Guo, L.; Luo, Q.; Zhou, K.; Yu, W.; Chen, Y.; Huang, C.; Xiao, Y. Wza gene knockout decreases Acinetobacter baumannii virulence and affects wzy-dependent capsular polysaccharide synthesis. Virulence 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.C.M.; Leung, P.H.M.; Lai, R.W. Emergence of carbapenem-resistant Acinetobacter baumannii ST195 harboring blaOxa-23 isolated from bacteremia in Hong Kong. Microb. Drug Resist. 2019, 25, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Cheng, V.C.; Wong, S.; Chen, J.H.; So, S.Y.; Wong, S.C.; Ho, P.L.; Yuen, K.Y. Control of multidrug-resistant Acinetobacter baumannii in Hong Kong: Role of environmental surveillance in communal areas after a hospital outbreak. Am. J. Infect. Control 2018, 46, 60–66. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Yang, C.H.; Su, P.W.; Moi, S.H.; Chuang, L.Y. Biofilm formation in Acinetobacter baumannii: Genotype-phenotype correlation. Molecules 2019, 24, 1849. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef]
- Loehfelm, T.W.; Luke, N.R.; Campagnari, A.A. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 2008, 190, 1036–1044. [Google Scholar] [CrossRef]
- Tomaras, A.P.; Dorsey, C.W.; Edelmann, R.E.; Actis, L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiology 2003, 149, 3473–3484. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lu, F.; Yuan, F.; Jiang, D.; Zhao, P.; Zhu, J.; Cheng, H.; Cao, J.; Lu, G. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the adeFGH efflux pump. Antimicrob. Agents Chemother. 2015, 59, 4817–4825. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Visca, P.; Seifert, H.; Towner, K.J. Acinetobacter infection–an emerging threat to human health. IUBMB Life 2011, 63, 1048–1054. [Google Scholar] [CrossRef]
- Chen, T.L.; Lee, Y.T.; Kuo, S.C.; Yang, S.P.; Fung, C.P.; Lee, S.D. Rapid identification of Acinetobacter baumannii, Acinetobacter nosocomialis and Acinetobacter pittii with a multiplex PCR assay. J. Med. Microbiol. 2014, 63, 1154–1159. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.; Stremick, C.; Read, R.; Morck, D.; Buret, A. The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A Modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Manchanda, V.; Sanchaita, S.; Singh, N. Multidrug resistant Acinetobacter. J. Glob. Infect. Dis. 2010, 2, 291. [Google Scholar] [CrossRef]
- Moskowitz, S.M.; Foster, J.M.; Emerson, J.; Burns, J.L. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 2004, 42, 1915–1922. [Google Scholar] [CrossRef]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, D.R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, I.S.; Kayumov, A.R. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.N. Isolation of persister cells from biofilm and planktonic populations of Pseudomonas aeruginosa. Bio-Protocol 2015, 5, e1590. [Google Scholar] [CrossRef]
- Anghel, I.; Chifiriuc, M.C.; Anghel, G.A. Pathogenic features and therapeutical implications of biofilm development ability in microbial strains isolated from rhinologic chronic infections. Farmacia 2011, 59, 770–783. [Google Scholar]
- Babapour, E.; Haddadi, A.; Mirnejad, R.; Angaji, S.A.; Amirmozafari, N. Biofilm formation in clinical isolates of nosocomial Acinetobacter baumannii and its relationship with multidrug resistance. Asian Pac. J. Trop. Biomed. 2016, 6, 528–533. [Google Scholar] [CrossRef]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Baño, J.; Marti, S.; Soto, S.; Fernández-Cuenca, F.; Cisneros, J.M.; Pachón, J.; Pascual, A.; Martínez-Martínez, L.; McQueary, C.; Actis, L. Biofilm formation in Acinetobacter baumannii: Associated features and clinical implications. Clin. Microbiol. Infect. 2008, 14, 276–278. [Google Scholar] [CrossRef]
- Wang, Y.C.; Huang, T.W.; Yang, Y.S.; Kuo, S.C.; Chen, C.T.; Liu, C.P.; Liu, Y.M.; Chen, T.L.; Chang, F.Y.; Wu, S.H. Biofilm formation is not associated with worse outcome in Acinetobacter baumannii bacteraemic pneumonia. Sci. Rep. 2018, 8, 7289. [Google Scholar] [CrossRef]
- Perez, L.R.R. Acinetobacter baumannii displays inverse relationship between meropenem resistance and biofilm production. J. Chemother. 2015, 27, 13–16. [Google Scholar] [CrossRef]
- Brossard, K.A.; Campagnari, A.A. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 2012, 80, 228–233. [Google Scholar] [CrossRef]
- Bagge, N.; Hentzer, M.; Andersen, J.B.; Ciofu, O.; Givskov, M.; Høiby, N. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents. Chemother. 2004, 48, 1168–1174. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Lebeaux, D.; DeFrancesco, A.S.; Valomon, A.; Soubigou, G.; Coppée, J.Y.; Ghigo, J.M.; Beloin, C. Starvation, together with the sos response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013, 9, e1003144. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]

| Classification | OD Range | ||
|---|---|---|---|
| Non-biofilm producer | OD | ≤ODc | |
| Weak biofilm producer | ODc | <OD | ≤2 × ODc |
| Moderate biofilm producer | 2 × ODc | <OD | ≤4 × ODc |
| Strong biofilm producer | OD | >4 × ODc | |
| Antimicrobials | MIC Breakpoints (µg/mL) | Susceptible (%) | Intermediate (%) | Resistant (%) | ||
|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||||
| Colistin | ≤2 | - | ≥4 | 79.8 | - | 20.2 |
| Imipenem | ≤2 | 4 | ≥8 | 48.1 | - | 51.9 |
| Meropenem | ≤2 | 4 | ≥8 | 48.1 | - | 51.9 |
| Cefotaxime | ≤8 | 16–32 | ≥64 | 29.8 | 13.5 | 56.7 |
| Ceftazidime | ≤8 | 16 | ≥32 | 39.4 | - | 60.6 |
| Ciprofloxacin | ≤1 | 2 | ≥4 | 48.1 | - | 51.9 |
| Levofloxacin | ≤1 | 2 | ≥4 | 48.1 | - | 51.9 |
| Gentamycin | ≤2 | 4 | ≥8 | 52.9 | - | 47.1 |
| Tetracycline | ≤4 | 8 | ≥16 | 48.1 | - | 51.9 |
| Ampicillin-sulbactam | ≤4 | 8 | ≥16 | 4.8 | 3.8 | 91.3 |
| Biofilm-Producing Ability | % of Isolates | % of Isolates with Different Antibiotic Susceptibility Profiles | ||
|---|---|---|---|---|
| Non-MDR 46.2% (48/104) | XDR 30.8% (32/104) | PDR 23.1% (24/104) | ||
| Biofilm-forming | >59.6% (62/104) | >66.1% (41/62) | >17.7% (11/62) | >16.1% (10/62) |
| Strong | 25% (26/104) | 92.3% (24/26) | 7.7% (2/26) | 0% (0/26) |
| Moderate | 14.4% (15/104) | 66.7% (10/15) | 26.7% (4/15) | 6.7% (1/15) |
| Weak | 20.2% (21/104) | 33.3% (7/21) | 23.8% (5/21) | 42.9% (9/21) |
| Non-biofilm-forming | 40.4% (42/104) | 16.7% (7/42) | 50% (21/42) | 33.3% (14/42) |
| Biofilm-Associated Gene | Biofilm-Forming No. (%) | Non-Biofilm-Forming No. (%) | p-Value |
|---|---|---|---|
| bap | 0.005 | ||
| Positive | 10 (100) | 0 (0) | |
| Negative | 52 (55.3) | 42 (44.7) | |
| csuE | 0.02 | ||
| Positive | 49 (55.1) | 40 (44.9) | |
| Negative | 13 (86.7) | 2 (13.3) | |
| adeFGH | 0.07 | ||
| Positive | 57 (57.6) | 42 (42.4) | |
| Negative | 5 (100.0) | 0 (0) | |
| ompA | 0.193 | ||
| Positive | 53 (57) | 40 (43) | |
| Negative | 9 (81.8) | 2 (18.2) | |
| abaI | 0.033 | ||
| Positive | 47 (54.7) | 39 (45.3) | |
| Negative | 15 (83.3) | 3 (16.7) |
| Colistin | Ciprofloxacin | Imipenem | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MLST | MIC for Planktonic Cells (µg/mL) | MBIC for Biofilm Cells (µg/mL) | Fold Change | MIC for planktonic Cells (µg/mL) | MBIC for Biofilm Cells (µg/mL) | Fold Change | MIC for Planktonic Cells (µg/mL) | MBIC for Biofilm Cells (µg/mL) | Fold Change |
| ST1894 | 0.5 | 16 | 32 | 1 | 64 | 64 | 0.125 | 256 | 2048 |
| ST1990 | 0.5 | 8 | 16 | 0.5 | 2 | 4 | 0.125 | 2 | 16 |
| ST1417 | 0.25 | 8 | 32 | 0.5 | 2 | 4 | 0.25 | 4 | 16 |
| ST1992 | 2 | 4 | 2 | 0.5 | 4 | 8 | 0.25 | 32 | 128 |
| ST373 | 1 | 32 | 32 | 0.5 | 16 | 32 | 0.25 | 32 | 128 |
| ST1862 | 1 | 4 | 4 | 1 | 8 | 8 | 0.015 | 8 | 533.3 |
| ST1964 | 0.5 | 16 | 32 | 1 | 64 | 64 | 4 | 16 | 4 |
| ST1855 | 0.5 | 2 | 4 | 0.5 | 16 | 32 | 0.015 | 8 | 533.3 |
| ST1861 * | 0.5 | 16 | 32 | 1 | 64 | 64 | 0.5 | 32 | 64 |
| Colistin | Ciprofloxacin | Imipenem | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MLST | MBC for Planktonic Cells (µg/mL) | MBEC for Biofilm Cells (µg/mL) | Fold Change | MBC for Planktonic Cells (µg/mL) | MBEC for Biofilm Cells (µg/mL) | Fold Change | MBC for Planktonic Cells (µg/mL) | MBEC for Biofilm Cells (µg/mL) | Fold Change |
| ST1894 | 4 | 256 | 64 | 8 | 1024 | 128 | 4 | 1024 | 256 |
| ST1990 | 0.5 | 32 | 64 | 1 | 1024 | 1024 | 1 | 64 | 64 |
| ST1417 | 1 | 32 | 32 | 2 | 64 | 32 | 0.5 | 64 | 128 |
| ST1992 | 2 | 8 | 4 | 1 | 1024 | 1024 | 1 | 1024 | 1024 |
| ST373 | 4 | 128 | 32 | 32 | 1024 | 32 | 4 | 32 | 8 |
| ST1862 | 2 | 128 | 64 | 2 | 512 | 256 | 1 | 1024 | 1024 |
| ST1964 | 4 | 256 | 64 | 2 | 32 | 16 | 4 | 512 | 128 |
| ST1855 | 0.5 | 32 | 64 | 1 | 1024 | 1024 | 2 | 1024 | 512 |
| ST1861 * | 4 | 16 | 4 | 8 | 1024 | 128 | 1 | 128 | 128 |
| Strain | Biofilm Forming | Colistin | Ciprofloxacin | Imipenem | Reason for Reduced Susceptibility in Biofilm | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBIC | MIC of Reverted Planktonic Cells | MIC | MBIC | MIC of Reverted Planktonic Cells | MIC | MBIC | MIC of Reverted Planktonic Cells | |||
| ST1894 Non-MDR | Strong | 0.5 | 16 | 0.5 | 1 | 64 | 1 | 0.125 | 256 | 0.125 | Tolerance |
| ST373 Non-MDR | Strong | 1 | 32 | 1 | 0.5 | 16 | 16 | 0.25 | 32 | 0.25 | Tolerance or resistant mutant for ciprofloxacin |
| ST195 XDR | Weak | 1 | 16 | 16 | 4 | 64 | 64 | 8 | 32 | 32 | Resistant mutant |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shenkutie, A.M.; Yao, M.Z.; Siu, G.K.-h.; Wong, B.K.C.; Leung, P.H.-m. Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates. Antibiotics 2020, 9, 817. https://doi.org/10.3390/antibiotics9110817
Shenkutie AM, Yao MZ, Siu GK-h, Wong BKC, Leung PH-m. Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates. Antibiotics. 2020; 9(11):817. https://doi.org/10.3390/antibiotics9110817
Chicago/Turabian StyleShenkutie, Abebe Mekuria, Mian Zhi Yao, Gilman Kit-hang Siu, Barry Kin Chung Wong, and Polly Hang-mei Leung. 2020. "Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates" Antibiotics 9, no. 11: 817. https://doi.org/10.3390/antibiotics9110817
APA StyleShenkutie, A. M., Yao, M. Z., Siu, G. K.-h., Wong, B. K. C., & Leung, P. H.-m. (2020). Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter baumannii Isolates. Antibiotics, 9(11), 817. https://doi.org/10.3390/antibiotics9110817





