Abstract
Ocellatins are a family of antimicrobial peptides found exclusively in the Leptodactylus genus. To date, 10 species have been studied and more than 23 peptides described. Here we report the sequences of five new peptides from the skin of the frog Leptodactylus latrans (Anura: Leptodactylidae) determined by cDNA cloning of the complete prepro-peptide structures. The mature peptides were characterized with in silico tools and compared with those previously described. With 21 amino acid residues, this new set of peptides not previously described in the Leptodactylus genus share between 100 and 76.2% similarity to ocellatin antimicrobial peptides. These novel peptides are cationic and their three-dimensional (3D) structure holds the highly conserved residues G1, D4, K7, and K11 and a high theoretical amphipathic α-helix content. Furthermore, in silico analyses of these new peptides predicted antimicrobial activity. This study is framed in the context of previous work published about ocellatins, and therefore, provides a review of this intriguing family of peptides.
1. Introduction
Antimicrobial peptides (AMPs) are short polypeptides found in the species of six kingdoms and they play a crucial role in innate immune systems. In amphibians, skin secretions have at least two main functions: host-shielding antimicrobial activity and a toxic effect against predators [1,2]; therefore, research into amphibian AMPs is focused mainly on the design of new drugs to treat human diseases. There is increasing concern about the antibiotic resistance of several pathogenic microorganisms, and thus, an urgent need to tackle this problem [3,4]. Despite drawbacks [5,6], AMPs are a novel class of molecules with a broad spectrum of activity and low capacity to induce bacterial resistance [7]. In combination with advanced chemoinformatics tools, they offer the possibility to address obstacles by peptide design approaches [8,9].
Despite advances in molecular and cell biology, in silico design, and automatization such as high-throughput screening, natural products are still one of the most successful sources of drug leads. The impact of natural products on drug discovery is not limited to the peptide domain but extends to all types of drugs [10]. Although recent years have witnessed revived interest of the pharmaceutical industry in natural products, many analysts attributed the crisis that this industry experienced at the end of the last century to the closure of most internal Natural Product departments, which had been in place since the 1980s.
In the field of peptides and natural products there are several interesting examples to highlight as Ziconotide (Prialt®), a potent analgesic discovered from the venom of a marine cone snail of the Conus magus species, and Romidepsin (Istodax®), first isolated from the bacterium Chromobacterium violaceum found in a soil sample in Japan that has a potent anticancer activity against some non-Hodgkin lymphomas.
As a naturally occurring antibiotic peptide, we can mention Daptomycin, isolated from the actinobacterium Streptomyces roseosporus in the soil and used in hospitals for the treatment of serious infections caused by multi-drug resistant bacteria and Colistin (or Polymyxin E), both cyclic peptides. Colistin was first isolated in Japan during the fermentation of Bacillus polymyxa var, and it is also used for infections that are difficult to treat. A good example of a peptide arising from frog skin is Pexiganan, a synthetic analog of magainin 2 derived from Xenopus laevis skin that reached phase III clinical trials as a topical antimicrobial agent for diabetic foot ulcers. The history of this peptide reflects the long and risk-laden process of drug discovery. Magainin 2 was identified in 1987. In 1999, the Food and Drug Administration (FDA) rejected the application of Magainin Pharmaceuticals, but another company, Dipexium Pharmaceuticals, ran the drug until phase III clinical trials but in 2016 failed to achieve FDA authorization again. This is not a problem unique to peptides as drugs but rather a dominant issue in the pharmaceutical industry, where approximately 1 out of every 10,000 or more compounds identified and analyzed reach the market [11]. However, peptides as potential drugs are considered to be “friendlier” than small molecules in this drug discovery journey due to the ease of analog synthesis and lack of toxicity, which, therefore, facilitates the progress of these biomolecules through the different phases and makes them a viable alternative to small molecules.
In the field of AMPs, more than 3240 natural AMPs enrich databases [12]. With computer-assisted design and bioinformatics analysis strategies, researchers can anticipate and optimize peptide properties and launch medicinal chemistry programs. Such programs involve data set preparation, peptide representation, model construction, design, and optimization of the active AMP.
AMPs are highly diverse. They can be categorized into different families on the basis of their structure and physical-chemical properties. Magainins, caerins, dermaseptins, brevinins, esculentins, and ocellatins are some of the types that have been described in amphibians [13,14]. The first ocellatin peptides were reported in the species Leptodactylus ocellatus (now L. latrans) [15,16], to which they owe their name. Members of the ocellatin family were described only in the skin secretion of frog species belonging to Leptodactylus. Grouping 78 species, Leptodactylus is a widely distributed genus throughout the south of North America, South America, and the West Indies [17]. Ocellatins have been identified by direct de novo sequencing using Edman′s mass spectrometry and microsequencing techniques, although molecular biology techniques to isolate mRNA and clone the full cDNA of complete prepro-peptides have also been performed [18].
Using molecular biology techniques, the present study aimed to identify novel complete prepro-peptide structures of AMPs in the skin of a specimen of L. latrans collected in Chaco, Argentina. The newly identified mature peptides were studied in relation to the ocellatin peptide family, comparing their physical-chemical characteristics and analyzing their sequences and theoretical structures to predict their activity against pathogen strains.
2. Results and Discussion
2.1. Identification of Peptide-Encoding cDNA Sequences
To identify the sequences of new peptides from the skin of the frog L. latrans, total RNA was isolated, followed by amplification and sequencing of specific cDNAs of the prepro-peptide precursors. After cloning the cDNA structures, 83 colonies were selected and grown separately in liquid media for subsequent plasmid purification. Size evaluation of the insert fragments was performed for selection and posterior sequencing (Figures S1 and S2). The nucleic acid and the deduced amino acid sequences of cDNA encoding new ocellatins are detailed in the supporting information (Figure S3). The results, as shown in Figure 1, indicate the identification of five de novo sequences of complete prepro-peptides with a typical AMP tripartite structure, namely, a 21-residue long highly conserved signal peptide ending with the characteristic Cys residue; variable acidic domain (19–22 residues), which present the typical Lys-Arg prohormone processing signal at their carboxyl terminus; and the mature peptide. All mature peptides presented a Gly residue at the C-terminal end of their sequences, which involved the amidation of the peptide at that end [19].
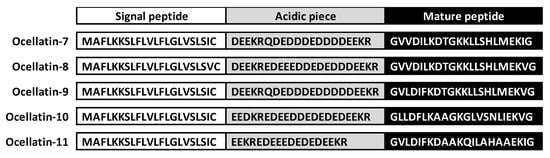
Figure 1.
Deduced amino acid sequences of the ocellatin precursors of Leptodactylus latrans. Prepro-regions (signal peptide and acidic region boxed white and gray respectively) and a variable domain (boxed black) that correspond to mature peptide are indicated. Cys residue indicates the end of the signal peptide and the Lys–Arg processing site shows the end of the acidic region.
2.2. Structural Comparison and Nomenclature
Comparison of mature peptide sequences with the peptides deposited in the APD and DRAMP databases confirmed the identification of a set of five previously unknown peptides with high degrees of similarity between them and to AMPs of the ocellatin family [20,21]. Since L. latrans was defined originally as L. ocellatus [22], from which the name of ocellatin-1 to -6 was derived, we did not incorporate letters and the newly identified peptides were named ocellatin-7 to -11, following the proposed nomenclature [23,24,25].
A comparison of the primary structures of the novel peptides showed highly conserved residues. Six amino acid residues (Gly1, Asp4, Lys7, Lys11, Glu19, and Lys20) are invariant, and 11 residues present conservative substitutions (Figure S4). Pairwise alignment between the identified peptides using DRAMP revealed between 52.4 and 100% similarity (Table S2), the highest values being between ocellatin-7 and ocellatin-9 and ocellatin-8, which show 90% and 100% of similarity (90.5% and 95.2% identity) to each other, respectively.
As shown in Table 1, ocellatin-11 presents 95.2% of similarity to ocellatin-2. Ocellatin-9, -8, and -7 show 76.2% similarity to ocellatin-5. Ocellatin-10 is 86.4% similar to ocellatin-6 and interestingly presents 90.47% identity and 95.5% of similarity to the peptide analog P3-Ll-2085, a hybrid peptide combination of two fragments [26].

Table 1.
Theoretical molecular weight (Mw), total net charge, identity, and similarity percentage of newly identified ocellatin peptides.
2.3. The Ocellatin Family
The information described in this section was obtained after an exhaustive search of the Antimicrobial Peptide Database (APD3), the Database of Antimicrobial Activity and Structure of Peptides (DBAASP), DRAMP, and BLAST and an in-depth analysis of the literature. In all cases, the isolation and identification method, specific associated activity, structural analysis, and study of the mechanism of action were considered (Table S1).
To date, a set of 28 ocellatin peptide sequences, including the five reported in the present study, have been described in 10 species of the genus Leptodactylus (Table 2). Ocellatin-1 to -3 [15], ocellatin-4 [16], ocellatin-5 and -6 [27], and ocellatin-7 to -11 (this work) were identified in the skin secretion of L. latrans (formerly L. ocellatus). Ocellatin-F (fallaxin) was identified in L. fallax [28]; ocellatin-P (pentadactylin) in L. pentadactylus [29]; ocellatin-L1 (laticeptin) and ocellatin-L2 in L. laticeps [30]; ocellatin-S (syphaxin) in L. syphax [31]; ocellatin-V1 to -V3 in L. validus [32]; ocellatin-K1 in L. knudseni [33]; ocellatin-PT1 to -PT8 in L. pustullatus [18]; and ocellatin-LB1 and -LB2 in L. labyrinticus [34]. In our opinion, these last two peptides should have been classified as ocellatin-F (1–22) and ocellatin-F (1–23) as their sequences are fragments of ocellatin-F (see below). It is interesting to note that ocellatin-F and ocellatin-K1 were identified in more than one species of the Leptodactylus genus. Ocellatin-K1 was present not only in L. knudseni but also in L. vastus [35], whereas ocellatin-F was present in three species, namely L. fallax, L. pentadactylus, and L. labyrinticus [29,34].

Table 2.
Amino acid sequence alignment of the 28 ocellatin peptides identified in the skin of Leptodactylus frog species.
As observed in the literature, numerous ocellatin fragments were also identified by de novo sequencing techniques in the skin secretions of leptodactylid frogs (Table S3). Ocellatin-S (1–16), (1–22), (1–23), (1–24), and (16–25) were present in the skin secretion of L. syphax; ocellatin-K1 (1–16), and (1–21) in L. vastus; and P2-Ll-1298 in L. latrans [26]. Ocellatin-F (1–22) was present in the skin secretion of L. fallax [28] and ocellatin-F (1–23), named Des-Lys24-Leu25-ocellatin-F1, was present in the skin secretion of L. labyrinticus. Two ocellatin-F fragments, ocellatin-LB1 and -LB2, were also identified in L. labyrinticus [34]. In our opinion, these peptides should be described as ocellatin-F (1–22) and (1–23), since their sequences do not present originality with respect to the peptides already described, and even ocellatin-F was present in the secretion of that species. The fragment ocellatin-L1 (1–22) was found in L. laticeps. Five fragments were identified in skin secretion of specimens of L. latrans (ocellatin-1 (1–16), ocellatin-2 (1–15), ocellatin-3 (1–15), ocellatin-5 (1–14), and ocellatin-6 (1–18)) [27]. Furthermore, Nascimento et al. described ocellatin-5 [36]. Given its length (17 amino acid residues) and the presence of ocellatin-5 (1-16), we propose that this ocellatin-5 is indeed a fragment of a distinct undescribed ocellatin. Considering that Leite Jr. et al. [27] have been described a different 21-residue sequence as ocellatin-5, we propose to call the first one ocellatin-5* until definitive identification of the corresponding ocellatin. The ocellatin-5* sequence is almost identical to the N-terminal region of ocellatin-10, except for one residue (T15S). Table S3 summarizes the experimental data obtained to date for the antimicrobial activity of ocellatin fragments.
Interestingly, the skin secretions of leptodactylid frogs also contain peptides with remarkably different sequences, such as LASP (GLWDDLKAAAKKVVSSLASAAIEKL-NH2), which have been described as Leptodactylus aggression-stimulating peptides in male frogs of L. fallax [37]. Gly-rich peptides have also been described: plasticin-L1 (GLVNGLLSSVLGGGQGGGGLLGGIL-OH) in L. laticeps [38], and leptoglycin (GLLGGLLGPLLGGGGGGGGGLL-OH) in L. pentadactylus [39]. Curiously, anionic peptide P1-Ll-15577 (DEMKLDGFNMHLE-NH2) was also reported in L. latrans [26]. These types of peptide are unusual in frog secretions, although several have been identified in other species [40,41].
Numerous synthetic peptide analogs have also been designed from ocellatins to improve activity. The peptide KLLKFVTKVGKAIFKALIKAI-OH is an ocellatin-4 analog. It has an increase in the positive net charge from +1 to +6, which improves its activity against plant pathogens [42]. P3-Ll-2085 [26], a hybrid peptide combination of two fragments (the N-terminal region of ocellatin-5* and the fragment P2-Ll-1298, identified in the secretion of L. latrans) showed a minimal inhibitory concentration (MIC) of 15 µM. Furthermore, analogs of ocellatin-F and ocellatin-4 were also designed for structure-activity relationship studies (SAR) [43].
2.4. Amino Acid Frequency and Particular Motif Occurrence
The peptides of the ocellatin family described hitherto contain 21, 25, or 32 residues, are cationic, and present conserved positions. However, they usually present a low cationic character, varying between net charges +3 and +1, compared with other peptides isolated from the skin of amphibians, which have double or triple loads, such as esculentin-2P from Rana pipiens with a net charge of +6, hymenochirin-1Pa from Pseudhymenochirus merlini with +7, or cathelicidin-RC1 and -RC2 from Rana catesbeiana with +9 and +8, respectively [44,45,46]. The positive character depends not only on the balance between positive and negative charges but also on the posttranslational amidation of the C-terminus, all ocellatins being amidated at the C-terminus, with the exception of the longest ones.
Only one residue, namely Asp4, is invariant for all the ocellatins described. However, several positions are highly conserved in the 28 ocellatins (Table 2). The distribution of acidic and basic amino acids plays a central role in the approach to the bacterial membrane. Analysis of the ocellatin sequences identified preferred locations for the basic and acidic residues. Sites 7, 11, 20, 24, and 29 are practically exclusive for the Lys residue and site 16 for a highly conserved His (Figure 2). As expected, SAR studies showed that Lys seems to have a greater effect than His because replacing the positively charged Lys in sites 7, 11, 20, and 24 with Ala results in a loss of activity compared to the original peptide, while the substitution of the His16 residue with Ala16 has no observable effect on antibacterial or hemolytic activity [43].
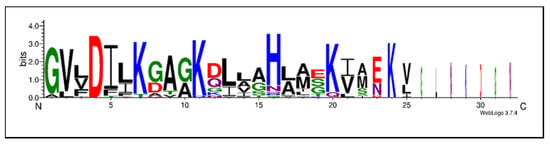
Figure 2.
Sequence motifs common to the 28 ocellatin family peptides described to date. The amino acids are stacked on top of each other in increasing order of frequency and plotted. The height of each letter is proportional to the observed frequency of the corresponding amino acid. The overall height of each stack is proportional to the sequence conservation at that position. Amino acid color code: green = polar (G, S, T, Y, C, Q, N), blue = basic (K, R, H), red = acidic (D, E), and black = hydrophobic (A, V, L, I, P, W, F, M).
Acidic amino acids also have preferred sites. In general, ocellatins have no more than two or three acidic amino acids, except for ocellatin-PT6. Site 4 of ocellatins is exclusively for the residue Asp. However, the second or third acidic residue is located at sites 8, 12, 19, or 23, positions 8 or 12 being excluding sites. At sites 19 and 23, the residue Glu is present instead of Asp, site 19 being preferred in short ocellatins. In the case of longer ocellatins (32 residues), described to date only in L. pustullatus, an acidic Asp residue is present at the C-terminal end at position 30. However, these particularly long ocellatins do not have acidic amino acids at site 8 or 12.
It is important to note that ocellatins maintain a low cationic character through a combination of positive and negative residues since they conserve both the basic amino acids and the number of acid residues in specific positions. Acidic and basic residues play a crucial role in the electrostatic attraction between charged peptides and negatively charged bacterial membranes. Therefore, higher positively charged peptides will have a greater capacity to interfere with the membranes of microorganisms. SAR studies with ocellatins demonstrated that the replacement of acid residues by Lys or by the electrically neutral Ala leads to an increase in antibacterial activity [36,43]. However, to date, no ocellatin without acidic residues has been described, and Asp4 is the only residue exclusively preserved in all ocellatins.
The five new ocellatins identified herein belong to the group of shortest ocellatins. They present 21 residues and a +2 net charge for ocellatin-7, -8, -9, and -10 and +1 for ocellatin-11. All of these compounds have Asp4, Lys7, Lys11, Glu19, and Lys20, whereas Asp8 and His16 are present in four of the five peptides. Translation of the nucleotide sequences of the prepro-peptides revealed the presence of a Gly residue at their C-terminal end. This residue represents a signal for C-terminus amidation during the posttranslational maduration process of the peptides. C-terminus amidation is common in natural AMPs, where a difference of approximately 1u is observed between the theoretical and experimental molecular masses according to the amino acid sequence [18,47]. All ocellatins are amidated at the C-terminus. Only the longer ocellatins present an acidic C-terminus (ocellatin-PT6, -PT7 and -PT8). C-terminus amidation is essential for the biological activity of many neuropeptides and hormones [48]. Several studies have proposed that C-terminus amidation confers enhanced antimicrobial activity compared with analogs with an acidic C-terminus, although this effect is not universal [49,50].
Hydrophobic amino acids also play a significant role in antimicrobial activity performance. Most natural peptides have a 40–60% content of hydrophobic residues, which are generally located in a specific sector on a helical wheel projection, thereby giving an amphipathic character to the molecule [51]. Of note, all ocellatins have hydrophobic residues at fixed positions among the charged residues, these being conservative substitutions. This arrangement results in the motif GXXDXXK for most ocellatin peptides, where X could be hydrophobic amino acids such as Val, Leu, Ile, and Phe, with a preference for Val in position X2 and Ile in position X6. These aliphatic amino acids are also typical in other positions such as X13, X14, X17, X21, and X25 (Figure 2 and Figure 3). These arrangements seem to be related to the phylogeny between Leptodactylus species, where ocellatin peptides identified in L. syphax, L. laticeps, L. vastus, L. knudseni, and L. fallax are phylogenetically close, differing only in conservative residues. However, certain positions present radical substitutions or replacements. Site 8 varies between small amino acids, such as Gly in L. vastus, and the electrically charged Asp in L. latrans and L. pustullatus. Position 12 varies between simple residues such as Gly or amino acids with acidic (Asp), basic (Lys), or polar uncharged (Gln, Asn) side chains. While in position 19, Glu is present for most of the ocellatins isolated from L. latrans, in L. vastus this position is occupied by Ser and in L. pustullatus by Gly. Some hydrophobic residues play a crucial role in the activity of ocellatins. In this regard, the replacement of Leu with Ala results in decreased activity compared with the original peptide [43].
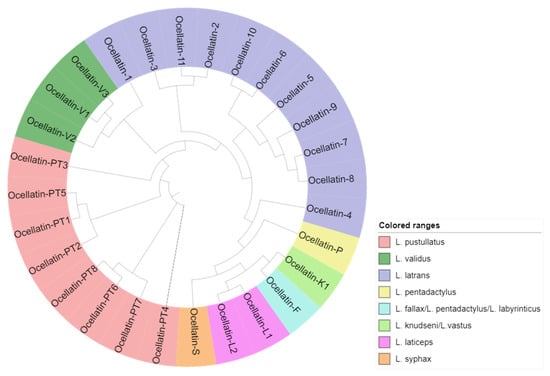
Figure 3.
Cladogram of all ocellatin peptides identified from the Leptodactylus genus to display similarity, and the associated anuran species in which they were identified (following the color code).
2.5. Phylogenetic Relationships
Analysis of the alignment of the ocellatin peptides (Table 2) and its associated cladogram (Figure 3) reveals that although the pressure to conserve the complete amino acid sequences of the peptides has been weak, with only one invariable residue (Asp4), there is a structure relationship indicative of a common evolutionary origin. De Sá et al. analyzed more than 80% of the recognized species of Leptodactylus and, based on a combined analysis of molecular and non-molecular data, suggested that evolution in this genus took the form of a pectinate tree with four main groups. A basal position group called L. fuscus (36 species), followed by the L. pentadactylus species group (17 species), which is the sister group to L. latrans (10 species), and the L. melanonotus (17 species) group [52].
Among the 10 species of Leptodactylus from which ocellatin peptides were identified, there are species of the four proposed groups. Two belong to the L. fuscus group (L. syphax and L. laticeps), five to the L. pentadactylus group (L. vastus, L. fallax, L. labyrintycus, L. knudseni, and L. pentadactylus), one to the L. latrans group (L. latrans), and two to the L. melanonotus group (L. pustullatus and L. validus).
It is noteworthy that the peptides from L. vastus, L. fallax, L. labyrinticus, L. knudseni, and L. pentadactylus are grouped, thereby validating the common origin perceived in the phylogram, and the same was observed for L. pustullatus and L. validus. All the new peptides identified from L. latrans in this work are segregated, together with four of the six ocellatins previously described in specimens of the same species. Ocellatin-10 and -6 form one cluster; ocellatin-9, -7, -8, and -5 form another sister cluster, and ocellatin-11, -2, and -3 are in a third cluster.
Some authors [13,53] have postulated that skin peptide profiling can be employed in combination with other sources of information, such as molecular and morphological characters, to discriminate hybrids, species, and clades at higher taxonomic levels. However, a systematic analysis to evaluate this approach has not been conducted to date. Furthermore, since peptide profiles can fluctuate depending on environmental factors [54], an extensive sampling and deeper understanding of the variability and evolution of these substances are required before considering their use in systematic [2].
2.6. Physical-Chemical Properties, 3D Structure, and Activity Prediction
To predict the activity of the novel ocellatins, we carried out a comparative study of the physicochemical parameters of the peptides identified with those already described.
The activities of AMPs and their ability to disrupt the plasma membrane of eukaryotic cells are dependent upon complex interactions between charge, hydrophobicity, conformation (helix stability), and amphiphathicity [55,56].
2.6.1. Physical-Chemical Properties
All the ocellatins described to date are cationic peptides and they present a high α-helix content prediction, and therefore, high values of pI, between 9.66 and 10.7, except ocellatin-2, which is the only member of this family with a net charge of 0 (Zero). Three-dimensional (3D) structure analysis using NMR and CD experiments has been performed for only a few ocellatins. NMR of ocellatin-F1 showed well defined helical segments along the polypeptide 3D structures. In the presence of micelles, ocellatin-F1 is bent and the helix curvature is consistent with the internalization of the hydrophobic residues into the membrane interior [57]. Additionally, although in aqueous media most of tested peptides present spectra consistent with random coil conformations, CD techniques demonstrated an enhancement of the helical contents with high percentages of α- helix in the presence of trifluoroethanol (TFE), dodecylphosphocholine (DPC), sodium dodecyl sulfate (SDS), or vesicles with dipalmitoyl phosphatidylglycerol (DPPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and POPC:1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPG) [3,26,34]. These observations indicate that the hydrophobic environment and the negative charge density of the membranes induce the adoption of such a structure of the peptide, and therefore, subsequent interaction. Ocellatins-LB1, -LB2, and -F1 from the skin of L. labyrinticus acquire higher helical contents, reaching approximately 90% in the presence of phospholipid vesicles, when compared to the peptides in the presence of TFE-H2O or aqueous micellar solutions [34]. The predicted α-helix content for the rest of the ocellatins is between 53–86%, except ocellatin-6 (Figure 4 and Figure S5C).
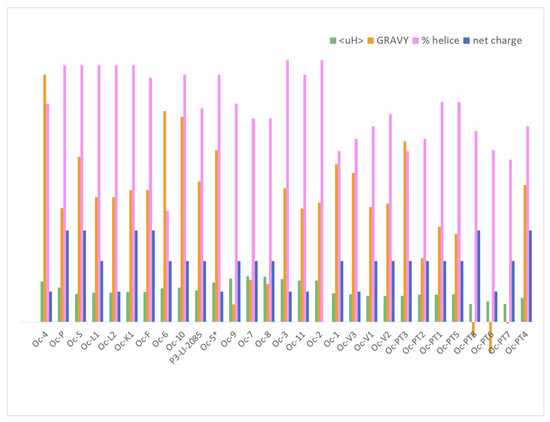
Figure 4.
Physical-chemical properties of ocellatins. Net charge, hydrophobicity (GRAVY), amphipathicity (µH), and percentage of α-helix content.
To interact with negative density charged bacterial membranes not only must the peptide have a positive net charge but also a high hydrophobic content, at least 50%, and an amphipathic arrangement. The GRAVY value is a useful index to evaluate hydrophobicity. It is calculated by adding the hydropathy values [58] of each amino acid residue and dividing by the number of residues in the sequence. A higher positive score indicates increased hydrophobicity. Figure 4 shows the extensive variability of hydrophobicity among ocellatins, which GRAVY values range between −0.103 and 0.81, the aggregation value also being variable (Figure S5B,C). The ocellaltin-10 described by our group is among the most hydrophobic ocellatins (together with ocellatin-4, -5, and -6), while the novel ocellatin-7, -8, and -9 are the least hydrophobic. Ocellatin-PT6, -PT7, and -PT8 are the only hydrophilic peptides in the ocellatin family (Figure 4).
2.6.2. D Theoretical Structures
The 3D theoretical structures of ocellatins predicted by the PEP-FOLD server vary between two types: a single α-helix (15 ocellatins, Figure S6) or an α-helix linked by a kink (13 ocellatins, Figure S7). An analysis of the sequences revealed that the amino acid residue at position 10 and the surrounding amino acids are decisive in defining the 3D structure adopted (see supporting information, 3D structure section). Ocellatin-7, -8, -9, and -11 are predicted as a single α-helix, while ocellatin-10 shows a structure with two helices joined by a coil (Figure 5).
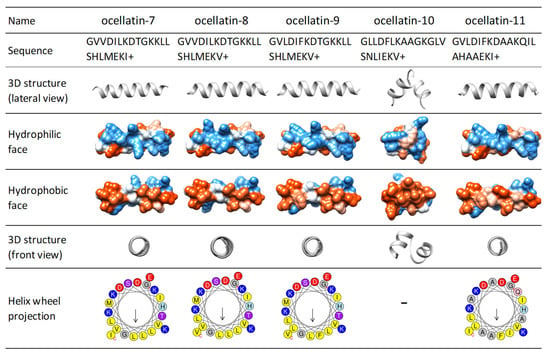
Figure 5.
Three-dimensional (3D) structure prediction, hydrophobic, and hydrophilic faces and Schiffer and Edmundson helical wheel projection diagrams of the peptides identified from the skin secretion of Leptodactylus latrans. Diagrams show α-helix motifs with amphiphilic peptide structures having a hydrophobic (red region) and hydrophilic region (blue region). Amino acid color code for the wheel projections: yellow = non-polar/hydrophobic (Leu, Val), gray = Gly, blue = basic (Lys, Arg), purple = polar without charge (Thr), pink = polar without charge (Asn), and green = Pro.
The arrangement plays a significant role in the balance between charged and hydrophobic amino acids. It should be amphipathic, with a hydrophilic side and an opposite hydrophobic side in the projection of the helical wheel. The amphipathic helix improves activity and selectivity, although some studies have observed that an imperfect amphipathicity provides certain benefits [59]. The hydrophobic moment is a measure of the amphipathicity of a helix. Each amino acid side chain is assigned a positive or negative value on the basis of its hydrophobicity. These values are used to weight vectors for each residue as they are displayed around the helix [60,61]. Ocellatin peptides show variable hydrophobic moments (between 0.6–1.49) (Kyte-Doolittle scale), with values that indicate amphipathic helical structures. The Schiffer and Edmundson wheel projection diagram allows visualization of the arrangement of amino acids in α-helix type structures. Figure S8 shows the wheel projections of all ocellatins that have a single-α-helix theoretical 3D. Among the single-α-helix structured ocellatins, although the newly identified ocellatin-9, -7, -8, and -11 are the least hydrophobic, they present the highest hydrophobic moments (Figure 5 and Figure S4A,B).
2.6.3. Activity Prediction
Table 3 shows the compilation of the experimental results of the antimicrobial activities of the ocellatin family peptides tested thus far against two characteristic Gram-negative and Gram-positive strains.

Table 3.
Experimental results of antimicrobial activities of ocellatin family peptides.
Comparison of the physical-chemical parameters of all members of the ocellatin family (Figure 4) with their experimental activity (Table 3) shows that antimicrobial activity does not depend on a specific combination of these parameters but is rather more complex. The antimicrobial activity of a peptide cannot be predicted solely on the basis of physical-chemical parameters as a different combination may result in peptides with similar activity and vice versa. As shown elsewhere, the MICs of peptides with the same charge, <H>, and µH differ by 10 orders of magnitude [43]. These parameters are useful when comparing peptides derived from the same peptide.
Ocellatins show variable antimicrobial activity against E. coli, with MICs ranging from 14 to 320 µM. Additionally, innovative activities were tested for ocellatin family members as combining the antiviral activity with alkaloid, antioxidant activity, or neuromodulatory therapeutic applications. Ocellatin-F1, cooperatively with bufotenine, presented an antiviral effect on the inhibition of rabies virus infection in BHK-21 cells [62]. Additionally, ocellatin-K1 fragments increased superoxide dismutase activity and glutathione concentration, as well as preventing the formation of reactive oxygen species (ROD) induced by lipopolysaccharides [35]. Moreover, within the ocellatin family, peptides with synergistic properties with antibiotics and antibiofilm activity have been identified against the multidrug-resistant strain Pseudomonas aeruginosa [3]. An ocellatin-4 analog has also been proposed for the control of bacterial pathogens affecting agriculture [42].
We focused on the physical-chemical properties and available experimental data on the activity of ocellatins-2, -5, and -6 since they are the ocellatins that present the highest percentages of similarity to the newly identified peptides. Ocellatin-2 is 99% similar to ocellatin-11, ocellatin-5 is 76% similar to ocellatin-7, -8, and -9, and ocellatin-6 is 86.4% similar to ocellatin-10 (Table 1). We also considered the synthetic peptide P3-Ll-2085 since it is 90.5% identical and 95.2% similar to ocellatin-10, differing in only one amino acid (I18L) (preserved substitution) (Table 1).
The MICs of ocellatin-5 against E. coli and S. aureus are 64 µg/mL (28 µM) and 128 µg/mL (56 µM), respectively [27] (Table 3). An in silico tool indicates that ocellatin-5 has a 0.81 chance of being an antimicrobial peptide. Although ocellatin-7, -8, and -9 show similarities of 71–76% to ocellatin-5, the prediction programs indicate that the probability of them being AMPs is less than 0.4 (Table S1). The main difference between these ocellatins is their hydrophobicity and 3D structure. Ocellatin-7, -8, and -9 show much lower hydrophobicity (GRAVY 0.14-0.06) than ocellatin-5 (GRAVY 0.56). They present a hydrophobic residue percentage of 42%, while ocellatin-5 has 52%. They also show differences in the % of α-helix, with lower percentages for ocellatin-7, -8, and -9, between 66 and 71%, whereas ocellatin-5 presents >80% (Table S1). Ocellatin-7, -8, and -9 show very high similarity (90.5–100%), and the residues in which they differ correspond to conservative substitutions. We expect them to behave in a similar fashion and not to exceed the antimicrobial capacity of ocellatin-5.
Ocellatin-11 shows high similarity to ocellatin-2, differing in only one amino acid (K20Q). In preliminary antimicrobial assays, ocellatin-2 inhibited the growth of E. coli but at millimolar level (probably influenced by its zero-net charge). Both peptides present very similar physical-chemical characteristics (hydrophobic moment value, (µH), hydrophobicity (GRAVY), percentage of α-helix, and percentage of aggregation (Table S1 and Figure 4). Therefore, considering that ocellatin-11 presents a net charge of +1, it is conceivable that it could outperform ocellatin-2 in terms of activity against these strains. The influence of the charge in the mechanism of action of AMP was widely studied [63,64]. SARS studies demonstrate that charge played a greater role in translocation efficacy of the peptides than hydrophobicity, with a higher net positive charge leading to a higher level of translocation into bacterial or C. albicans membranes [65]. The prediction of the penetration depth of the DBAASP tool for ocellatin-11 also supports this observation. The value of penetration depth is even slightly higher than for ocellatin-2 (23 vs. 22 for ocellatin-10 vs. ocellatin-2, Table S1). Therefore, this additional change could improve the interaction of the peptide with the membrane.
However, ocellatin-10 has a better projection since its sequence differs from P3-Ll-2085 in only one conservative substitution (I18L). P3-Ll-2085 shows activity of 15 µM against both E. coli and S. aureus (Table 3). No significant differences in the values of most of the physical-chemical parameters of the two peptides were observed (Figure 4 and Table S1). Both peptides present the same net charge (+2), µH, and GRAVY. However, there is a difference in the percentage of aggregation, with ocellatin-10 presenting an intermediate value between ocellatin-6 (32%) and P3-Ll-2085 (7%). Given these observations, we do not consider this parameter relevant for activity projection. Although ocellatin-10 differs from ocellatin-6 in the percentage of α-helix (81 vs. 36%), it has a similar value to P3-Ll-2085 (70%). Circular dichroism studies of P3-Ll-2085 in different environments [H2O, TFE-H20 (50% v/v)], and in the presence of DPPG or DPPC vesicles confirmed the adoption of a helical conformation in TFE and DPPG with over 70% of α-helix content. The theoretical 3D structure prediction indicates that ocellatin-6, ocellatin-10, and P3-Lla-2085 have very similar arrangements, with two helices joined by a slit (Figure S7). Therefore, the similarities between the identified peptides and those already described lead us to expect that ocellatin-10 will have a MIC of around 15 µM.
3. Materials and Methods
3.1. Amphibian Collection
One adult specimen of Leptodactylus latrans was collected from a peri-urban site in Resistencia, Chaco, Argentina 27°23′50.9″ S 58°55′33.5″ W (−27.397460, −58.925960), (Transit Guide No. 5144, and authorization of the Subsecretary of Environment and Biosafety of the province of Chaco). The specimen was deposited in the herpetological collection, voucher numbers CNP 4208, Colección Herpetológica-Anfibios of the Instituto de Diversidad y Evolución Austral (IDEAus)—CONICET. The authors state that all animal manipulation was carried out following the ARRIVE guidelines [66].
3.2. cDNA Cloning
The dorsal skin of a L. latrans specimen (previously anesthetized with lidocaine hydrochloride) was isolated, frozen in liquid nitrogen, and pulverized in a mortar using the Trizol (Invitrogen) reagent to extract total RNA. Agarose gel was used for quality analysis of RNA. Reverse transcription and synthesis of first-strand cDNA were performed using M-MLV Reverse Transcriptase (Promega) and the primer 008 (5′- GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTT-3). After synthesis of the first strand, the 3′RACE procedure was performed using a primer that anneals to a conserved region as a forward primer, and 002 (5′-ATGGCTTTCCTGAAGAAATCTCTTTTCCTTGTACTATTCCTTGG-3′) and 009 (5′- GACCACGCGTATCGATGTCGAC-3′) as the reverse primer. The cycling parameters were as follows: one cycle of 94 °C/120 s; four cycles of 94 °C/60 s, 53 °C/60 s, and 72 °C/60 s; 35 cycles of 94 °C/30 s, 53 °C/30 s, and 72 °C/60 s; and one cycle of 72 °C/420 s. The PCR products were purified using ADN PuriPrep-GP Kit (Highway) and linked to the pCR4-TOPO (TOPOTA, LifeTechnologies) for sequencing (Invitrogen). Escherichia coli Top 10 competent cells were prepared and transformed using the Inoue method described by Sambrook and Russell [67]. Following the selection and growth of bacterial colonies, the resulting plasmids were purified using the protocol Preparation of plasmid DNA by alkaline lysis with SDS: Minipreparation [67].
3.3. Prepro-Peptide cDNA Amplification and Selection
The inserts linked in the plasmids were amplified using the forward and reverse M13 primers (Invitrogen), which annealed to the M13 reverse and forward priming sites present in the plasmid pCRTM4-TOPO® by PCR according to the parameters specified: one cycle of 94 °C/120 s; 32 cycles of 94 °C/30 s, 53 °C/30 s, and 72 °C/60 s; and one cycle of 72 °C/420 s. Size evaluation of the insert fragments was performed by comparing band sizes in 2% agarose gels, (1X TAE buffer) and fragments with a molecular weight greater than 500 bp were selected for purification with the DNA PuriPrep-GP Kit (Highway). The quality of the amplified fragments was tested with agarose gels.
3.4. Insert Sequencing and Prepro-Peptide Identification
Selected inserts were sequenced by the BigDye terminator reaction (Applied Biosystems), using an ABI 3130 DNA analyzer (Applied Biosystems) and universal M13 forward and reverse primers. Sequences were studied using the Lasergene sequence analysis software (DNASTAR, Inc., Madison, WI, USA). EMBOOS Transseq was used to translate nucleic acid sequences to the corresponding peptide sequences.
3.5. Sequence Analysis
The deduced peptide sequences were compared with those of the peptides deposited in the Antimicrobial Peptide Database (APD3) [12], Database of Antimicrobial Activity and Structure of Peptides (DBAASP) [68], DRAMP [21], and BLAST [69]. Several software packages were used for the characterization of the physical-chemical properties and the prediction of the activity of the sequences. The PEP-FOLD program [70] was used to predict the 3D structure of the peptide, UCSF Chimera was used for the renderings [71], and the HeliQuest program was used to evaluate amphipathic character [72]. The computer software DBAASP v2.702 [68] and the software ProtParam [73] were used to determine the physical and chemical parameters of the peptide sequences. The ClustalX tool of the MEGA7 software [74] was used to align the primary sequences of the peptides described for the genus Leptodactylus. The online program WebLogo [75] was used for the graphical representation of amino acid sequence alignment. The software packages DBAASP v2.702 and Collection of Anti-Microbial Peptides (CAMPR3) [76] allowed the prediction of antimicrobial activity, and the bioinformatics program Hemolytic Peptide Identification Server (HemoPI) [77] the prediction of hemolytic character.
4. Conclusions
The ocellatin peptides characterized to date belong exclusively to leptodactylid frogs. A review of the members of the ocellatin family shows that they have a length of 21, 25, or 32 amino acids, are cationic with net charges between +1 and +3, present a hydrophobic residue content between 42% and 57%, have a variable combination of physical-chemical properties, and have a high helix content, as indicated by 3D structure predictions.
The amino acid frequency analysis and particular motif occurrence demonstrates that certain amino acid residues in ocellatins are highly conserved or have conservative substitutions. Polar charged residues have a significant role in the electrostatic attraction between charged peptides and negatively charged bacterial membranes. SAR studies with ocellatins proved the effect of replacing charged residues by the electrically neutral Ala. The antibacterial activity of ocellatin-F proved to be dependent on the overall charge. The substitution of Asp4 or Asp12 for Ala resulted in increased antibacterial activity and the replacement of Lys with Ala resulted in decreased activity. Therefore, higher positively charged peptides will show greater capacity to disturb microorganism membranes. However, interestingly, it would seem that, for reasons unknown, ocellatins maintain a low cationic character as they preserve both the cationic amino acids and also the number of acidic residues in specific positions.
The present study was designed to identify novel peptides from the skin of the Argentinian frog L. latrans. Using molecular biology techniques to identify the complete prepro-peptide precursors, here, we describe five new members of the ocellatin family, namely ocellatin-7 to -11. The methodology used allowed us to obtain the entire sequence of mature peptide, which is relevant because ocellatin fragments were found to be always less active than ocellatins. This is the case of ocellatin-F (1–22) and ocellatin-L1 (1–22) fragments, which did not inhibit E. coli, while the mature peptides inhibited the strain at a concentration of 40 and 50 µM, respectively. Therefore, concerning the bioprospection of new molecules as antimicrobial alternatives, the identification of the entire prepro-peptide may be paramount.
The primary structure of the mature peptides identified indicates that they are orthologs of active peptides of the ocellatin family previously isolated from other species of South American frogs of the genus Leptodactylus. Ocellatin-7, -8, and -9 were found to be the members of the family with the lowest percentage of hydrophobic residues but with the highest amphipathicity µH values. Through comparison with peptides from the same family, we estimate that ocellatin-10 will be the most active of the five novel peptides, followed by ocellatin-7, -8, -9, and -11.
These results increase the data available on the ocellatin peptide family. Future work will address the synthesis, purification, and testing of the activity of the new peptides described herein to corroborate the predictions. Further work with species of the Leptodactylus genus will be required to contribute to phylogeny relationship studies among species of this particular genus. Finally, we envisage that a general ocellatin structure could be used as a template for the preparation of libraries of peptides with improved antimicrobial activities.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/11/751/s1. Figure S1. Size evaluation of the amplified insert fragments in 2% agarose gels.; Figure S2. Fragments selected for sequencing.; Figure S3. Nucleic acid and deduced amino acid sequence of cDNA encoded ocellatin-7, ocellatin-8, ocellatin-9, ocellatin-10, ocellatin-11 from the skin of Leptodactylus latrans.; Figure S4. Amino acid sequence alignment of the five novel peptides identified in the skin of Leptodactylus latrans; Figure S5. Hydrophobic moment (µH), Hydrophobicity (GRAVY), percentage of α-helix, and percentage of aggregation of the ocellatins described to date; Figure S6. Ocellatins with α-helix 3D theoretical structure; Figure S7. Ocellatins with 3D theoretical structure of two α-helix linked by a kink; Figure S8. Schiffer and Edmundson wheel projection diagrams of ocellatins with complete α-helix 3D structure prediction.; Table S1: Ocellatins and other peptides described to date from the skin secretion of Leptodactylid frogs. In vivo and in silico parameters; Table S2. Similarity percentages of the identified ocellatins by pairwise alignment; Table S3. Fragments of ocellatin peptides identified in the skin secretion of frog of the Leptodactylus genus; Supporting information: 3D structures.
Author Contributions
A.P.C.B., S.A., N.L.C., and N.G.B. performed the experiments. M.M.M. conceived and supervised the experimental work. M.M.M. and N.G.B. provided reagents and materials. M.M.M. and S.A. performed the in silico search and analyzed the data. M.M.M. planned and wrote the original draft preparation; S.A. and A.P.C.B. prepared the figures and tables; N.L.C. formatted the references; F.A. critically revised the manuscript and contributed extensively to its improvement. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2019 N° 2278). This work was framed within the P-UE CONICET 2016 N° 22920160100044.
Acknowledgments
S.A. and N.L.C. acknowledge doctoral grants awarded by CONICET, and A.P.C.B. the initiation grant awarded by the Universidad Nacional del Nordeste. M.M.M. and N.G.B. are researchers of CONICET.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raaymakers, C.; Verbrugghe, E.; Hernot, S.; Hellebuyck, T.; Betti, C.; Peleman, C.; Claeys, M.; Bert, W.; Caveliers, V.; Ballet, S.; et al. Antimicrobial peptides in frog poisons constitute a molecular toxin delivery system against predators. Nat. Commun. 2017, 8, 1495. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Marani, M.M.; Soldi, R.A.; Mendonça, J.N.; Faivovich, J.; Cabrera, G.M.; Lopes, N.P. Cleavage of Peptides from Amphibian Skin Revealed by Combining Analysis of Gland Secretion and in Situ MALDI Imaging Mass Spectrometry. ACS Omega 2018, 3, 5426–5434. [Google Scholar] [CrossRef]
- Bessa, L.J.; Eaton, P.; Dematei, A.; Placido, A.; Vale, N.; Gomes, P.; Delerue-Matos, C.; Leite, J.R.S.A.; Gameiro, P. Synergistic and antibiofilm properties of ocellatin peptides against multidrug-resistant Pseudomonas aeruginosa. Future Microbiol. 2018, 13, 151–163. [Google Scholar] [CrossRef]
- Mobarki, N.S.; Almerabi, B.A.; Hattan, A. Antibiotic resistance crisis. Int. J. Med. Dev. Ctries 2019, 3, 561–564. [Google Scholar] [CrossRef]
- Seo, M.-D.; Won, H.-S.; Kim, J.-H.; Mishig-Ochir, T.; Lee, B.-J. Antimicrobial Peptides for Therapeutic Applications: A Review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, e24122. [Google Scholar] [CrossRef]
- Baindara, P.; Ghosh, A.K.; Mandal, S.M. Coevolution of resistance against antimicrobial peptides. Microb. Drug Resist. 2020, 26, 880–899. [Google Scholar] [CrossRef]
- Ramesh, S.; Govender, T.; Kruger, H.G.; de la Torre, B.G.; Albericio, F. Short AntiMicrobial peptides (SAMP) as a class of extraordinary promising therapeutic agentes. J. Pept. Sci. 2016, 22, 438–451. [Google Scholar] [CrossRef]
- Fjell, C.; Hiss, J.; Hancock, R.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Pennington, M.W.; Czerwinski, A.; Norton, R. Peptide therapeutics from venom: Current status and potential. Bioorgan. Med. Chem. 2018, 26, 2738–2758. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Pukala, T.L.; Bowie, J.H.; Maselli, V.M.; Musgrave, I.F.; Tyler, M.J. Host-defence peptides from the glandular secretions of amphibians: Structure and activity. Nat. Prod. Rep. 2006, 23, 368–393. [Google Scholar] [CrossRef]
- Wang, G. Bioinformatic Analysis of 1000 Amphibian Antimicrobial Peptides Uncovers Multiple Length-Dependent Correlations for Peptide Design and Prediction. Antibiotics 2020, 9, 491. [Google Scholar] [CrossRef]
- Nascimento, A.C.C.; Zanotta, L.C.; Kyaw, C.M.; Schwartz, E.N.F.; Schwartz, C.A.; Sebben, A.; Sousa, M.V.; Fontes, W.; Castro, M.S. Ocellatins: New Antimicrobial Peptides from the Skin Secretion of the South American Frog Leptodactylus ocellatus (Anura: Leptodactylidae). Protein J. 2004, 23, 501–508. [Google Scholar] [CrossRef]
- Nascimento, A.; Chapeaurouge, A.; Perales, J.; Sebben, A.; Sousa, M.; Fontes, W.; Castro, M. Purification, characterization and homology analysis of ocellatin 4, a cytolytic peptide from the skin secretion of the frog Leptodactylys ocellatus. Toxicon 2007, 50, 1095–1104. [Google Scholar] [CrossRef]
- American Museum of Natural History. Amphibian Species of the World: An Online Reference; Version 6.1; Frost, D.R., Ed.; American Museum of Natural History: New York, NY, USA, 2020; Available online: https://amphibiansoftheworld.amnh.org/index.php (accessed on 13 September 2020).
- Marani, M.M.; Dourado, F.S.; Quelemes, P.V.; Araujo, A.R.; Perfeito, M.L.G.; Barbosa, E.A.; Veras, L.M.C.; Coelho, A.L.R.; Andrade, E.B.; Eaton, P.; et al. Characterization and Biological Activities of Ocellatin Peptides from the Skin Secretion of the Frog Leptodactylus pustulatus. J. Nat. Prod. 2015, 78, 1495–1504. [Google Scholar] [CrossRef]
- Vanhoye, D.; Bruston, F.; Nicolas, P.; Amiche, M. Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur. J. Biochem. 2003, 270, 2068–2081. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Zasloff, M. A database view of naturally occurring antimicrobial peptides: Nomenclature, classification and amino acid sequence analysis. In Antimicrobial Peptides: Discovery, Design, and Novel Therapeutic Strategies; Wang, G., Ed.; CAB International: Cambridge, MA, USA, 2010; pp. 1–21. [Google Scholar]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef]
- Lavilla, E.O.; Langone, J.A.; Caramaschi, U.; Heyer, W.R.; De Sá, R.O. The identification of Rana ocellata Linnaeus, 1758. Nomenclatural impact on the species currently known as Leptodactylus ocellatus (Leptodactylidae) and Osteopilus brunneus (Gosse, 18514) (Hylidae). Zootaxa 2010, 2346, 1–16. [Google Scholar]
- Amiche, M.; Ladram, A.; Nicolas, P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides 2008, 29, 2074–2082. [Google Scholar] [CrossRef]
- Conlon, J.M. A proposed nomenclature for antimicrobial peptides from frogs of the genus Leptodactylus. Peptides 2008, 29, 1631–1632. [Google Scholar] [CrossRef]
- Conlon, J.M. Reflections on a systematic nomenclature for antimicrobial peptides from the skin of frogs of the family Ranidae. Peptides 2008, 29, 1815–1819. [Google Scholar] [CrossRef]
- Siano, A.; Humpola, M.V.; de Oliveira, E.; Albericio, F.; Simonetta, A.C.; Lajmanovich, R.; Tonarelli, G.G. Leptodactylus latrans Amphibian Skin Secretions as a Novel Source for the Isolation of Antibacterial Peptides. Molecules 2018, 23, 2943. [Google Scholar] [CrossRef]
- Leite, J.M.A., Jr.; Silva, L.P.; Silva-Leite, R.R.; Ferrari, A.S.; Noronha, S.E.; Silva, H.R.; Bloch, C., Jr.; Leite, J.R.S.A. Leptodactylus Ocellatus (Amphibia): Mechanism of defense in the skin and molecular phylogenetic relationships. J. Exp. Zool. A Ecol. Genet. Physiol. 2010, 313, 1–8. [Google Scholar] [CrossRef]
- Rollins-Smith, L.A.; King, J.D.; Nielsen, P.F.; Sonnevend, A.; Conlon, J.M. An antimicrobial peptide from the skin secretions of the mountain chicken frog Leptodactylus fallax (Anura:Leptodactylidae). Regul. Pept. 2005, 124, 173–178. [Google Scholar] [CrossRef]
- King, J.D.; Al-Ghaferi, N.; Abraham, B.; Sonnevend, A.; Leprince, J.; Nielsen, P.F.; Conlon, J.M. Pentadactylin: An antimicrobial peptide from the skin secretions of the South American bullfrog Leptodactylus pentadactylus. Comp. Biochem. Physiol. C 2005, 141, 393–397. [Google Scholar] [CrossRef]
- Conlon, J.M.; Al-Ghaferi, N.; Abraham, B.; Sonnevend, A.; King, J.D.; Nielsen, P.F. Purification and properties of laticeptin, an antimicrobial peptide from skin secretions of the South American frog Leptodactylus laticeps. Protein Pept. Lett. 2006, 13, 411–415. [Google Scholar] [CrossRef]
- Dourado, F.S.; Leite, J.R.S.A.; Silvab, L.P.; Melo, J.A.T.; Bloch, C., Jr.; Schwartz, E.F. Antimicrobial peptide from the skin secretion of the frog Leptodactylus syphax. Toxicon 2007, 50, 572–580. [Google Scholar] [CrossRef]
- King, J.D.; Leprince, J.; Vaudry, H.; Coquet, L.; Jouenne, T.; Conlon, J.M. Purification and characterization of antimicrobial peptides from the Caribbean frog, Leptodactylus validus (Anura: Leptodactylidae). Peptides 2008, 29, 1287–1292. [Google Scholar] [CrossRef]
- Cardozo-Filho, J.L.; Soares, A.A.; Bloch, C., Jr.; Silva, L.P.; Stabeli, R.G.; Calderon, L.A. Identification of peptides from Amazonian Leptodactylus knudseni skin secretion by MALDI TOF/TOF; UniProtKB/Swiss-Prot. P86711 (OCE1_LEPKN). 2010. Available online: http://www.uniprot.org/uniprot/P86711 (accessed on 1 July 2020).
- Gusmão, K.A.G.; dos Santos, D.M.; Santos, V.M.; Cortes, M.E. Ocellatin peptides from the skin secretion of the South American frog Leptodactylus labyrinthicus (Leptodactylidae): Characterization, antimicrobial activities and membrane interactions. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 4. [Google Scholar] [CrossRef]
- Sousa, N.A.; Oliveira, G.A.L.; de Oliveira, A.P.; Lopes, A.L.F.; Iles, B.; Nogueira, K.M.; Araújo, T.S.L.; Souza, L.K.M.; Araújo, A.R.; Ramos-Jesus, J.; et al. Novel Ocellatin Peptides Mitigate LPS-induced ROS Formation and NF-kB Activation in Microglia and Hippocampal Neurons. Sci. Rep. 2020, 10, 2696. [Google Scholar] [CrossRef]
- Nascimento, A.C.C. Cytolytic Peptides and Proteases from the Skin Secretion of the Frog Leptodactylus Ocellatus; University of Brasilia: Brasilia, Brazil, 2007; Available online: https://www.uniprot.org/uniprot/P85443 (accessed on 1 July 2020).
- King, J.D.; Rollins-Smith, L.A.; Nielsen, P.F.; John, A.; Conlon, J.M. Characterization of a peptide from skin secretions of male specimens of the frog, Leptodactylus fallax that stimulates aggression in male frogs. Peptides 2005, 26, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Abdel-Wahab, Y.H.; Flatt, P.R.; Leprince, J.; Vaudry, H.; Jouenne, T.; Condamine, E. A glycine-leucine-rich peptide structurally related to the plasticins from skin secretions of the frog Leptodactylus laticeps (Leptodactylidae). Peptides 2009, 30, 888–892. [Google Scholar] [CrossRef]
- Sousa, J.C.; Berto, R.F.; Gois, E.A.; Fontenele-Cardi, N.C.; Honório, J.E., Jr.; Konno, K.; Richardson, M.; Rocha, M.F.; Camargo, A.A.; Pimenta, D.C.; et al. Leptoglycin: A new glycine/leucine-rich antimicrobial peptide isolated from the skin secretion of the South American frog Leptodactylus pentadactylus (Leptodactylidae). Toxicon 2009, 54, 23–32. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.R.; Phoenix, D.A. Anionic antimicrobial peptides from eukaryotic organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef]
- Marani, M.M.; Pérez, L.O.; Rodrigues de Araujo, A.; Plácido, A.; Sousa, C.F.; Veras Quelemes, P.; Oliveira, M.; Gomes-Alves, A.G.; Pueta, M.; Gameiro, P.; et al. Thaulin-1: The first antimicrobial peptide isolated from the skin of a Patagonian frog Pleurodema thaul (Anura: Leptodactylidae: Leiuperinae) with activity against Escherichia coli. Gene 2017, 605, 70–80. [Google Scholar] [CrossRef]
- Kishi, I.R.N.; Stach-Machado, D.; Singulani, J.d.L.; dos Santos, C.T.; Fusco-Almeida, A.M.; Cilli, E.M.; Freitas-Astúa, J.F.; Picchi, S.C.; Machado, M.A. Evaluation of cytotoxicity features of antimicrobial peptides with potential to control bacterial diseases of citrus. PLoS ONE 2018, 13, e0203451. [Google Scholar]
- Nielsen, S.L.; Frimodt-Møller, N.; Kragelund, B.B.; Hansen, P.R. Structure–activity study of the antibacterial peptide fallaxin. Protein Sci. 2007, 16, 1969–1976. [Google Scholar] [CrossRef]
- Goraya, J.; Wang, Y.; Li, Z.; O’Flaherty, M.; Knoop, F.C.; Platz, J.E.; Conlon, J.M. Peptides with antimicrobial activity from four different families isolated from the skins of the North American frogs Rana luteiventris, Rana berlandieri and Rana pipiens. Eur. J. Biochem. 2000, 267, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Prajeep, M.; Mechkarska, M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; King, J.D. Characterization of the host-defense peptides from skin secretions of Merlin’s clawed frog Pseudhymenochirus merlini: Insights into phylogenetic relationships among the Pipidae. Comp. Biochem. Physiol. Part. D 2013, 8, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Ling, G.; Gao, J.; Zhang, S.; Xie, Z.; Wei, L.; Yu, H.; Wang, Y. Cathelicidins from the bullfrog Rana catesbeiana provides novel template for peptide antibiotic design. PLoS ONE 2014, 9, e93216. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Ma, C.; Zhou, M.; Xi, X.; Li, L.; Wu, D.; Wang, L.; Lin, C.; Lopez, J.C.; Chen, T.; et al. Phylloseptin-PBa—A novel broad-spectrum antimicrobial peptide from the skin secretion of the peruvian purple-sided leaf frog (Phyllomedusa Baltea) which exhibits cancer cell cytotoxicity. Toxins 2015, 7, 5182–5193. [Google Scholar] [CrossRef]
- Chufán, E.E.; De, M.; Eipper, B.A.; Mains, R.E.; Amzel, L.M. Amidation of bioactive peptides: The structure of the lyase domain of the amidating enzyme. Structure 2009, 17, 965–973. [Google Scholar] [CrossRef]
- Pan, Y.; Wan, J.; Roginski, H.; Lee, A.; Shiell, B.; Michalski, W.P.; Coventry, M.J. Comparison of the effects of acylation and amidation on the antimicrobial and antiviral properties of lactoferrin. Lett. Appl. Microbiol. 2007, 44, 229–234. [Google Scholar] [CrossRef]
- Dennison, S.R.; Harris, F.; Bhatt, T.; Singh, J.; Phoenix, D.A. The effect of C-terminal amidation on the efficacy and selectivity of antimicrobial and anticancer peptides. Mol. Cell. Biochem. 2009, 332, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic a helical antimicrobial peptides A systematic study of the effects of structural and physical properties on biological activity. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef]
- De Sá, R.; Grant, T.; Camargo, A.; Heyer, W.R.; Ponssa, M.L.; Stanley, E. Systematics of the neotropical genus Leptodactylus Fitzinger, 1826 (Anura: Leptodactylidae): Phylogeny, the relevance of non-molecular evidence, and Species accounts. S. Am. J. Herpetol. 2014, 9, 1–128. [Google Scholar]
- Jackway, R.J.; Pukala, T.L.; Donnellan, S.C.; Sherman, P.J.; Tyler, M.J.; Bowie, J.H. Skin peptide and cDNA profiling of Australian anurans: Genus and species identification and evolutionary trends. Peptides 2011, 32, 161–172. [Google Scholar] [CrossRef]
- Wabnitz, P.A.; Bowie, J.H.; Tyler, M.J.; Wallace, J.C.; Smith, B.P. Differences in the skin peptides of the male and female Australian tree frog Litoria splendida. Eur. J. Biochem. 2000, 267, 269–275. [Google Scholar] [CrossRef]
- Conlon, J.M.; Mechkarska, M.; Leprince, J. Peptidomic analysis in the discovery of therapeutically valuable peptides in amphibian skin secretions. Expert Rev. Proteom. 2019, 16, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Unifying themes in host defense effector polypeptides. Nat. Rev. Microbiol. 2007, 5, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Augusto, M.T.; Felicio, M.R.; Hollmann, A.; Franco, O.L.; Goncalves, S.; Nuno, C.S. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Juretić, D.; Simunić, J. Design of α-helical antimicrobial peptides with a high selectivity index. Expert Opin. Drug Discov. 2019, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 1982, 299, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Libério, M.S.; Joanitti, G.A.; Azevedo, R.B.; Cilli, E.M.; Zanotta, L.C.; Nascimento, A.C.; Sousa, M.V.; Pires Júnior, O.R.; Fontes, W.; Castro, M.S. Anti-proliferative and cytotoxic activity of pentadactylin isolated from Leptodactylus labyrinthicus on melanoma cells. Amino Acids 2011, 44, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Cunha Neto, R.D.; Vigerelli, H.; Jared, C.; Antoniazzi, M.M.; Chaves, L.B.; Rodrigues da Silva, A.C.; Lopes de Melo, R.; Sciani, J.M.; Pimenta, D.C. Synergic effects between ocellatin-F1 and bufotenine on the inhibition of BHK-21cellular infection by the rabies virus. J. Venom. Anim. Toxins 2015, 21. [Google Scholar] [CrossRef]
- Gagnon, M.C.; Strandberg, E.; Grau-Campistany, A.; Wadhwani, P.; Reichert, J.; Bürck, J.; Rabanal, F.; Auger, M.; Paquin, J.F.; Ulrich, A.S. Influence of the length and charge on the activity of α-helical amphipathic antimicrobial peptides. Biochemistry 2017, 56, 1680–1695. [Google Scholar] [CrossRef]
- Shagaghi, N.; Palombo, E.A.; Clayton, A.H.A.; Bhave, M. Antimicrobial pptides: Biochemical determinants of activity and biophysical techniques od elucidating their functionality. World J. Microb. Biot. 2018, 34. [Google Scholar] [CrossRef]
- Gong, Z.; Doolin, M.T.; Adhikari, S.; Stroka, K.M.; Karlsson, A.J. Role of charge and hydrophobicity in translocation of cell-penetrating peptides into Candida albicans cells. AIChe J. 2019, e16768. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, R. Molecular Cloning. A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring, NY, USA, 2001; Volume 1. [Google Scholar]
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E.; Grigolava, M.; Chubinidze, M.; Gogoladze, G.; Vishnepolsky, B.; et al. DBAASP v.2: An Enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. [Google Scholar] [CrossRef]
- Protein BLAST. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 15 April 2020).
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theor. Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Higgins, D.; Thompson, J.; Gibson, T.; Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).