Abstract
Invasive pulmonary aspergillosis (IPA) is typically considered a disease of immunocompromised patients, but, recently, many cases have been reported in patients without typical risk factors. The aim of our study is to develop a risk predictive model for IPA through machine learning techniques (decision trees) in patients with influenza. We conducted a retrospective observational study analyzing data regarding patients diagnosed with influenza hospitalized at the University Hospital “Umberto I” of Rome during the 2018-2019 season. We collected five IPA cases out of 77 influenza patients. Although the small sample size is a limit, the most vulnerable patients among the influenza-infected population seem to be those with evidence of lymphocytopenia and those that received corticosteroid therapy.
1. Introduction
The association between invasive pulmonary aspergillosis (IPA) and influenza has begun to attract particular attention over the last 20 years. It represents a worldwide phenomenon, reported in at least 16 countries from across Europe, Asia, and the United States of America [1]. Since IPA often has severe consequences, with a mortality rate between 50 and 90%, especially if the diagnosis is delayed [2,3], it is essential to identify patients at higher risk so that they can be monitored closely, and prophylaxis can be considered. Therefore, we designed a retrospective observational study on hospitalized patients diagnosed with influenza during the 2018-2019 winter season with the aim of developing a risk predictive model for IPA through machine learning techniques (decision trees) [4,5].
2. Results
During the 2018-2019 flu season, 104 episodes of influenza among patients hospitalized at the “Umberto I” hospital were recorded. Twenty-seven patients presented the exclusion criteria; therefore, 77 patients were included in the study (Table 1) and five (6.5%) out of these developed IPA superinfections. In one case, an autopsy was performed with a histopathological diagnosis; in three cases, the mycological criterion was the positivity of GM on BAL (in all three cases the culture for Aspergillus was negative), in one case GM on serum. In all cases, a worsening of respiratory exchanges was observed, and two patients also presented a refractory fever. Thirty-four episodes were classified as mild influenza (44%) and 43 as severe flu (56%). Univariate analysis of preexisting and hospitalization-related risk factors for IPA are presented in Table 2 and Table 3. All cases of IPA were patients with severe flu (Table 4).

Table 1.
Population characteristics.

Table 2.
Preexisting risk factors associated with IPA in influenza patients.

Table 3.
Hospitalization-related risk factors associated with IPA in influenza patients.

Table 4.
Clinical impact of Aspergillus superinfection in patients with influenza.
We have subsequently developed a risk predictive model of IPA through a machine learning technique: decision-making trees, with the aim of providing clinicians with a tool capable of accurately identifying IPA high-risk patients, in order to monitor them closely and evaluate antifungal prophylaxis.
The predictive model has highlighted two variables as decisive in risk assessment: lymphocytes count ≤340/µL and methylprednisolone administration >0.65 mg/kg/day, as shown in Figure 1 (see Section 4.3 Statistical analysis for more details). A 10-fold cross-validation [4] test revealed 87.5% as the average sensitivity and 96.4% as the average specificity.
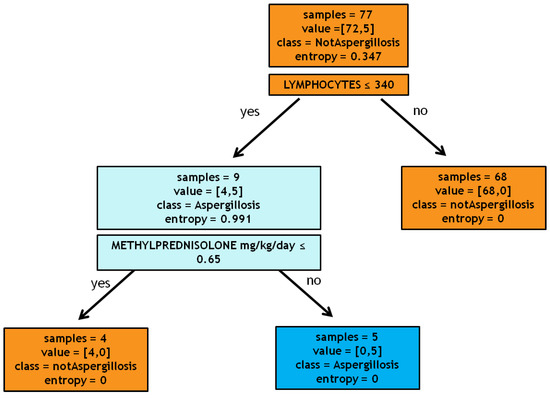
Figure 1.
Decision tree.
3. Discussion
The innovative aspect of our study is the development of a methodology based on a machine learning technique, the decision tree, for building a risk predictive model for IPA in patients with influenza. Since IPA is a disease with many possible complications and often poor prognosis, a risk predictive score would be a tool of crucial importance. This predictive model, although built on a very small dataset, highlighted two variables as determinants in the assessment of risk: lymphocytopenia at diagnosis of influenza with a lymphocyte count ≤340/µL and administration of methylprednisolone during hospitalization at a dosage of more than 0.65 mg/kg/day. These results, although preliminary, seem to be of particular interest. Various studies have shown the role of lymphocytes in invasive Aspergillus infections, especially the Th1 and Th17 subpopulations [6,7], and furthermore, lymphocytopenia was recognized among the independent risk factors for pulmonary superinfections [8]. Another study proposes virus-induced lymphocytopenia among the risk factors associated with IPA in patients with influenza [9]. The degree of lymphocytopenia is an expression of an increased apoptotic rate in the context of an abnormal anti-inflammatory response with immunoparalysis and anergy of the immune system, a condition that typically occurs in severe influenza infection, as well as increased recruitment at extensively inflamed lung tissue. The role of corticosteroids as a risk factor for invasive fungal infections has been known for many years [10]. They alter the immune system by affecting different pathways [11]. In addition to immune dysfunction, which certainly plays a key role in the pathogenesis of invasive aspergillosis, it has been hypothesized that glucocorticoids have a direct effect on Aspergillus species, inducing their growth [12]. Our study has a number of limitations: (1) the small sample number does not allow us to build a score but only to identify the factors related to an increased risk of superinfection; (2) the retrospective nature makes the study less precise and more susceptible to error; (3) in this study, the decision tree is highly based on the “use of corticosteroids”. However, several previous studies reported that a number of the patients developing IPA did not use corticosteroids: in this case, the decision tree could have significant limitations. One of the innovative aspects of our study is to propose a methodology for the prediction of the risk through machine learning techniques, whose applications remain almost unexplored in this field.
4. Materials and Methods
4.1. Study Design, Inclusion and Exclusion Criteria, and Definitions
We conducted a retrospective observational study analyzing data regarding patients diagnosed with influenza hospitalized at the University Hospital “Umberto I” of Rome during the 2018-2019 season. The first case dates back to the 3rd of January and the last to the 23rd of April 2019. Exclusion criteria were history of chronic or invasive aspergillosis or immunosuppression according to EORTC/MSG criteria [13]: a recent history of neutropenia (<500 neutrophils/mm3) for >10 days, receipt of an allogeneic stem cell transplant, prolonged use of corticosteroids at a mean minimum dose of 0.3 mg/kg/die of prednisone equivalent for >3 weeks, treatment with other recognized T-cell immunosuppressants, and inherited severe immunodeficiency. Influenza virus infection was diagnosed if viral RNA on respiratory samples resulted as positive. A real-time PCR technique (CEPHEID kit) was used with primers for the RNA of influenza A, B, and H1N1 subtypes. This analysis was requested by the ward physicians in patients with suspected influenza infection. To define a case of IPA, the criteria of the modified AspICU algorithm were adopted [14]. One or more of the following mycological criteria had to be present: histopathology or direct microscopic evidence of dichotomous septate hyphae with a positive culture for Aspergillus from tissue; a positive Aspergillus culture from a bronchoalveolar lavage (BAL); a galactomannan optical index on BAL of ≥1; a galactomannan optical index on serum of ≥0.5; associated with at least one of the following clinical criteria: fever refractory to at least 3 days of appropriate antibiotic therapy; recrudescent fever after a period of defervescence of at least 48 h while still on antibiotics and without other apparent cause; dyspnea; hemoptysis; pleural friction, rub or chest pain; worsening respiratory insufficiency in spite of appropriate antibiotic therapy and ventilatory support; associated with the presence of the radiographic criteria: any pulmonary infiltrate detected at RX or CT.
4.2. Population Analysis
Demographic, clinical, and anamnestic patients’ data were collected from the patient’s medical records, from the access to first aid until discharge or death, all discharged patients were contacted three months after the dismission in order to confirm the absence of new hospital admission. In particular, age, sex, weight, ethnicity, smoking, alcoholic habits, comorbidities, and the Charlson Comorbidity Index were recorded. For each patient, at the diagnosis of influenza, type of influenza virus, community or nosocomial-acquisition, the value of white blood cells, neutrophils, lymphocytes and CRP (C reactive protein), SOFA score (sequential organ failure assessment), radiographic findings, need for oxygen supplementation (via Venturi mask, non-invasive mechanical ventilation or invasive mechanical ventilation), were recorded as well as the eventual need for ECMO support (extracorporeal membrane oxygenation) and hemodialysis during hospitalization. We divided patients diagnosed with influenza into two groups: patients with influenza who developed IPA, and patients who did not undergo this superinfection, we therefore developed a risk predictive model for IPA through machine learning techniques (decision trees), to try to identify IPA high-risk patients. We also assessed the risk factors and the clinical impact associated with IPA. In a first analysis, we compared patients with IPA with patients without. We defined “severe flu” as all episodes of influenza that required oxygen therapy for at least 48 h. The study protocol was approved by the Hospital Ethics Commission and, considering the retrospective nature of the study, the request for informed consent to patients was omitted.
4.3. Statistical Analysis
We developed a decision tree-based predicting model [4,5]. The algorithm has been coded in Python 3.5. As splitting criteria, we used entropy. In order to assess the quality of the model, we performed a 10-fold cross-validation test, considering the sensitivity and specificity as performance measures.
Precisely, the algorithm processes the database containing all patients with influenza through a progressive sequence of tests. These tests, with a positive or negative outcome for the categorical variable and greater or smaller than a given threshold for the continuous variables, were performed by the algorithm on all the instances in the database. Subsequently, we asked the algorithm to provide the variables that split the samples into classes (IPA and non-IPA). For our specific cohort, as the first test, the algorithm selected the variable lymphocytes ≤340/µL; this variable divides the root (i.e., the set of all patients) into two leaves: a pure leaf, patients with lymphocytes more than 340/µL, consisting of 68 all non-IPA patients, and an impure leaf, patients with lymphocytes less than 340/µL, consisting of 4 non-IPA patients and 5 IPA patients. Then, we asked the algorithm to choose a new test, namely the variable that most divided IPA from non-IPA patients among this population of 9 subjects. The algorithm identified the administration of methylprednisolone at a dosage greater than 0.65 mg/kg/day. Eventually, we obtained two pure leaves: one containing the 4 patients taking less than 0.65 mg/kg/day, none of which were affected by IPA, and one containing 5 patients taking more than 0.65 mg/kg/day, all with the diagnosis of IPA. The new cases of influenza will be processed with the built model to predict the probability of developing aspergillosis as follows: the patient will be tested according to the sequence of questions identified by the tree and eventually lie in a leaf that will determine the class (IPA or non-IPA). Other statistical analyses were performed using Microsoft Excel (Office 2019) or Statistical Program for the Social Sciences (SPSS) version 20. To assess the normal distribution, the coefficients of kurtosis, skewness, and histogram plots were used. The data, unless otherwise stated, were reported as median with interquartile range (IQR: 25th and 75th percentile) for continuous variables and as simple frequencies (n) and proportions (or percentages) for dichotomous variables. Group comparisons in univariate analysis were made using the Mann–Whitney or Kruskal–Wallis tests for continuous variables. The Fisher’s exact or Chi-square tests were used as appropriate to test group differences of proportions. Lastly, parameters that were statistically significant in a univariate way were used in the logistic regression analysis to estimate adjusted odds ratios (ORs) and 95% confidence intervals (95% CI) for the risk factors associated with the development of IPA, including the confounding factors (age, gender, etc.). A p-value of less than 0.05 was considered statistically significant.
5. Conclusions
From our data, we confirm the risk of developing IPA during influenza. The onset of this complication severely conditions the prognosis and results in high mortality rates. The most vulnerable patients among the influenza-infected population are those that present the following risk factors: smoking habit, diagnosis of COPD, evidence of lymphocytopenia, and administration of corticosteroids. A methodology for the prediction of IPA risk that uses a technique of machine learning, decision trees, seems to provide interesting results but would need to be corroborated and possibly validated in larger patient cohorts.
Author Contributions
Conceptualization, V.B.; methodology, E.C.; software, L.C. (Luigi Celani) and G.S.; formal analysis, C.B.; investigation, L.S., G.P.I., L.C. (Livia Conte), and C.P.; data curation, L.C. (Luigi Celani) and G.S; writing—original draft preparation, G.C.; writing—review and editing, M.V.; visualization, supervision and project administration, G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Verweij, P.E.; Brüggemann, R.J.M.; Wauters, J.; Rijnders, B.J.A.; Chiller, T.; van de Veerdonk, F.L. Influenza coinfection: Be(a)ware of invasive aspergillosis. Clin. Infect. Dis. 2020, 70, 349–350. [Google Scholar] [CrossRef] [PubMed]
- Meersseman, W.; Vandecasteele, S.J.; Wilmer, A.; Verbeken, E.; Peetermans, W.E.; Van Wijngaerden, E. Invasive aspergillosis in critically ill patients without malignancy. Am. J. Respir. Crit. Care Med. 2004, 170, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Cornillet, A.; Camus, C.; Nimubona, S.; Gandemer, V.; Tattevin, P.; Belleguic, C.; Chevrier, S.; Meunier, C.; Lebert, C.; Aupée, M.; et al. Comparison of epidemiological, clinical, and biological features of invasive aspergillosis in neutropenic and nonneutropenic patients: A 6-year survey. Clin. Infect. Dis. 2006, 43, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Kononenko, I. Machine learning for medical diagnosis: History, state of the art and perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef]
- Chai, L.Y.; van de Veerdonk, F.; Marijnissen, R.J.; Cheng, S.C.; Khoo, A.L.; Hectors, M.; Lagrou, K.; Vonk, A.G.; Maertens, J.; Joosten, L.A.; et al. Anti-Aspergillus human host defence relies on type 1 T helper (Th1), rather than type 17 T helper (Th17), cellular immunity. Immunology 2010, 130, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hartemink, K.J.; Paul, M.A.; Spijkstra, J.J.; Girbes, A.R.; Polderman, K.H. Immunoparalysis as a cause for invasive aspergillosis? Intensive Care Med. 2003, 29, 2068–2071. [Google Scholar] [CrossRef] [PubMed]
- Bellelli, V.; d’Ettorre, G.; Celani, L.; Borrazzo, C.; Ceccarelli, G.; Venditti, M. Clinical significance of lymphocytopenia in patients hospitalized with pneumonia caused by influenza virus. Crit. Care 2019, 23, 330. [Google Scholar] [CrossRef] [PubMed]
- Crum-Cianflone, N.F. Invasive Aspergillosis Associated with Severe Influenza Infections. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2016; Volume 3. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, N.; Huang, X.; Xiong, S.; Feng, Y.; Zhang, Y.; Li, M.; Zhan, Q. Invasive pulmonary aspergillosis in patients with influenza infection: A retrospective study and review of the literature. Clin. Respir. J. 2019, 13, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Lionakis, M.S.; Kontoyiannis, D.P. Glucocorticoids and invasive fungal infections. Lancet 2003, 362, 1828–1838. [Google Scholar] [CrossRef]
- Ng, T.T.; Robson, G.D.; Denning, D.W. Hydrocortisone-enhanced growth of Aspergillus spp.: Implications for pathogenesis. Microbiology 1994, 140 Pt 9, 2475–2479. [Google Scholar] [CrossRef]
- De Pauw, B.; Walsh, T.J.; Donnelly, J.P.; Stevens, D.A.; Edwards, J.E.; Calandra, T.; Pappas, P.G.; Maertens, J.; Lortholary, O.; Kauffman, C.A.; et al. European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group; National Institute of Allergy and Infectious Diseases Mycoses StudyGroup (EORTC/MSG) Consensus Group. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 2008, 46, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Schauwvlieghe, A.F.A.D.; Rijnders, B.J.A.; Philips, N.; Verwijs, R.; Vanderbeke, L.; Van Tienen, C.; Lagrou, K.; Verweij, P.E.; Van de Veerdonk, F.L.; Gommers, D.; et al. Dutch-Belgian Mycosis study group. Invasive aspergillosis in patients admitted to the Intensive care unit with severe influenza: A retrospective cohort study. Lancet Respir. Med. 2018, 6, 782–792. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).