Harnessing the Potential of Killers and Altruists within the Microbial Community: A Possible Alternative to Antibiotic Therapy?
Abstract
1. Introduction
2. Kin Killing in Lactobacillales: Fratricide in S. pneumoniae and Enterococcal “Siblicide”
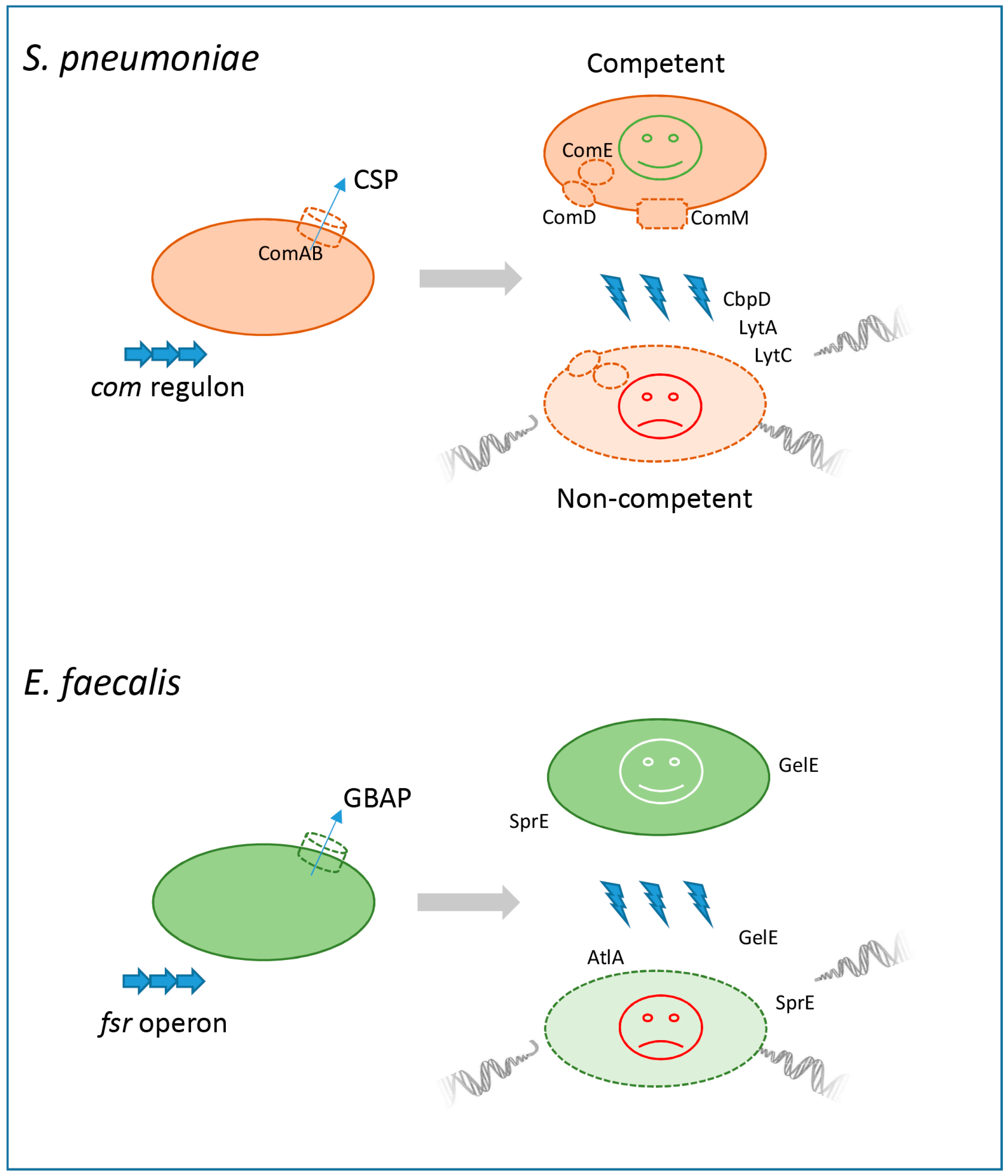
2.1. Fratricide in Streptococcus
2.2. Enterococcal “Siblicide”
3. Sibling Killing in Bacillales: Cannibalism in Bacillus subtilis and Induced Suicide in Paenibacillus dendritiformis
3.1. Cannibalism in Bacillus subtilis
3.2. “Forced Suicide” in Paenibacillus dendritiformis
4. Why do Bacteria Kill their Siblings?
5. Allolysis as Weekdays of the Bacterial “Multicellular” Life
6. The Inter-Relationships within the Microbial Community—Unfolding the Puzzle
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gilmore, M.S.; Haas, W. The selective advantage of microbial fratricide. Proc. Natl. Acad. Sci. USA 2005, 102, 8401–8402. [Google Scholar] [CrossRef]
- Claverys, J.P.; Prudhomme, M.; Martin, B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 2006, 60, 451–475. [Google Scholar] [CrossRef]
- Bayles, K.W. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 2007, 5, 721. [Google Scholar] [CrossRef]
- Claverys, J.P.; Havarstein, L.S. Cannibalism and fratricide: Mechanisms and raisons d’etre. Nat. Rev. Microbiol. 2007, 5, 219. [Google Scholar] [CrossRef]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85. [Google Scholar] [CrossRef]
- Prozorov, A.A.; Danilenko, V.N. Allolysis in bacteria. Microbiology 2011, 80, 1–9. [Google Scholar] [CrossRef]
- Ibáñez de Aldecoa, A.L.; Zafra, O.; González-Pastor, J.E. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef]
- Veening, J.W.; Blokesch, M. Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat. Rev. Microbiol. 2017, 15, 621. [Google Scholar] [CrossRef]
- Popp, P.F.; Mascher, T. Coordinated cell death in isogenic bacterial populations: Sacrificing some for the benefit of many? J. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Guiral, S.; Mitchell, T.J.; Martin, B.; Claverys, J.P. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: Genetic requirements. Proc. Natl. Acad. Sci. USA 2005, 102, 8710–8715. [Google Scholar] [CrossRef]
- Håvarstein, L.S.; Martin, B.; Johnsborg, O.; Granadel, C.; Claverys, J.P. New insights into the pneumococcal fratricide: Relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 2006, 59, 1297–1307. [Google Scholar] [CrossRef]
- Claverys, J.P.; Martin, B.; Håvarstein, L.S. Competence-induced fratricide in streptococci. Mol. Microbiol. 2007, 64, 1423–1433. [Google Scholar] [CrossRef]
- Gonzalez-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by sporulating bacteria. Science 2003, 301, 510–513. [Google Scholar] [CrossRef]
- Sedgley, C.M.; Clewell, D.B.; Flannagan, S.E. Plasmid pAMS1-encoded, bacteriocin-related “siblicide” in Enterococcus faecalis. J. Bacteriol. 2009, 191, 3183–3188. [Google Scholar] [CrossRef]
- Hwang, Y.C.; Flannagan, S.E.; Clewell, D.B.; Sedgley, C.M. Bacteriocin-related siblicide in clinical isolates of enterococci. Probiotics Antimicrob. Protein 2011, 3, 57–61. [Google Scholar] [CrossRef]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef]
- Thomas, V.C.; Hancock, L.E. Suicide and fratricide in bacterial biofilms. Int. J. Artif. Organs 2009, 32, 537–544. [Google Scholar] [CrossRef]
- Beer, A.; Ariel, G.; Kalisman, O.; Helmanc, Y.; Sirota-Madi, A.; Zhang, H.P.; Florin, E.L.; Payne, S.M.; Ben-Jacob, E.; Swinney, H.L. Lethal protein produced in response to competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. USA 2010, 107, 6258–6263. [Google Scholar] [CrossRef]
- Taylor, J.D.; Taylor, G.; Hare, S.A.; Matthews, S.J. Structures of the DfsB protein family suggest a cationic, helical sibling lethal factor peptide. J. Mol. Biol. 2016, 428, 554–560. [Google Scholar] [CrossRef]
- Slager, J.; Kjos, M.; Attaiech, L.; Veening, J.W. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 2014, 157, 395–406. [Google Scholar] [CrossRef]
- Dash, M.; Dash, H.R.; Das, S. Genetic regulation of allolysis in response to sub-lethal antibiotic stress in Streptococcus pneumoniae. Nusant. Biosci. 2014, 6, 111–117. [Google Scholar] [CrossRef]
- Prudhomme, M.; Berge, M.; Martin, B.; Polard, P. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet. 2016, 12, e1006113. [Google Scholar] [CrossRef]
- Fontaine, L.; Wahl, A.; Flichard, M.; Mignolet, J.; Hols, P. Regulation of competence for natural transformation in streptococci. Genet. Evol. 2014, 33, 343–360. [Google Scholar] [CrossRef]
- Johnston, C.; Campo, N.; Berge, M.J.; Polard, P.; Claverys, J.P. Streptococcus pneumoniae, le transformiste. Trends Microbiol. 2014, 22, 113. [Google Scholar] [CrossRef]
- Li, Y.H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- Lin, J.; Zhu, L.; Lau, G.W. Disentangling competence for genetic transformation and virulence in Streptococcus pneumoniae. Curr. Genet. 2016, 62, 97–103. [Google Scholar] [CrossRef]
- Peterson, S.N.; Sung, C.K.; Cline, R.; Desai, B.V.; Snesrud, E.C.; Luo, P.; Walling, J.; Li, H.; Mintz, M.; Tsegaye, G.; et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 2004, 51, 1051–1070. [Google Scholar] [CrossRef]
- Mirouze, N.; Dubnau, D. Chance and necessity in Bacillus subtilis development. Microbiol Spectr. 2013, 1, 1–34. [Google Scholar] [CrossRef]
- Wei, H.; Havarstein, L.S. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl. Environ. Microbiol. 2012, 78, 5897–5905. [Google Scholar] [CrossRef]
- Berg, K.H.; Birnstad, T.J.; Johnsborg, O.; Havarstein, L.S. Properties and biological role of streptococcal fratricins. Appl. Environ. Microbiol. 2012, 78, 3515–3522. [Google Scholar] [CrossRef]
- Thomas, V.C.; Hiromasa, Y.; Harms, N.; Thurlow, L.; Tomich, J.; Hancock, L.E. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 2009, 72, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, O.E.; Rozen, D.E.; May, R.M.; Levin, B.R. Oscillations in continuous culture populations of Streptococcus pneumoniae: Population dynamics and the evolution of clonal suicide. Proc. R. Soc. B 2009, 276, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, M.; Claverys, J.P. Release of DNA into the medium by competent Streptococcus pneumoniae: Kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 2004, 54, 783–794. [Google Scholar] [CrossRef]
- Steinmoen, H.; Knutsen, E.; Håvarstein, L.S. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 2002, 99, 7681–7686. [Google Scholar] [CrossRef]
- Steinmoen, H.; Teigen, A.; Havarstein, L.S. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J. Bacteriol. 2003, 185, 7176–7183. [Google Scholar] [CrossRef]
- Eldholm, V.; Johnsborg, O.; Straume, D.; Ohnstad, H.S.; Berg, K.H.; Hermoso, J.A.; Håvarstein, L.S. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol. Microbiol. 2010, 76, 905–917. [Google Scholar] [CrossRef]
- Straume, D.; Stamsas, G.A.; Salehian, Z.; Havarstein, L.S. Overexpression of the fratricide immunity protein ComM leads to growth inhibition and morphological abnormalities in Streptococcus pneumoniae. Microbiology 2017, 163, 9–21. [Google Scholar] [CrossRef]
- Johnsborg, O.; Eldholm, V.; Bjørnstad, M.L.; Håvarstein, L.S. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol. 2008, 69, 245–253. [Google Scholar] [CrossRef]
- Romero, P.; Lopez, R.; Garcıa, E. Key role of amino acid residues in the dimerization and catalytic activation of the autolysin LytA, an important virulence factor in Streptococcus pneumoniae. J. Biol. Chem. 2007, 282, 17729–17737. [Google Scholar] [CrossRef]
- Morales, M.; Martın-Galiano, A.J.; Domenech, M.; Garcıa, E. Insights into the evolutionary relationships of LytA autolysin and Ply pneumolysin-like genes in Streptococcus pneumoniae and related streptococci. Genome Biol. Evol. 2015, 7, 2747–2761. [Google Scholar] [CrossRef]
- Garcia, P.; Gonzalez, M.P.; Garcia, E.; Garcia, J.L.; Lopez, R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 1999, 33, 128–138. [Google Scholar] [CrossRef]
- Pérez-Dorado, I.; González, A.; Morales, M.; Sanles, R.; Striker, W.; Vollmer, W.; Mobashery, S.; García, J.L.; Martínez-Ripoll, M.; García, P.; et al. Insights into pneumococcal fratricide from the crystal structures of the modular killing factor LytC. Nat. Struct. Mol. Biol. 2010, 17, 576. [Google Scholar] [CrossRef]
- Perez-Dorado, I.; Galan-Bartual, S.; Hermoso, J.A. Pneumococcal surface proteins: When the whole is greater than the sum of its parts. Mol. Oral Microbiol. 2012, 27, 221–245. [Google Scholar] [CrossRef]
- Eldholm, V.; Johnsborg, O.; Haugen, K.; Ohnstad, H.S.; Havarstein, L.S. Fratricide in Streptococcus pneumoniae: Contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 2009, 155, 2223–2234. [Google Scholar] [CrossRef]
- Kausmally, L.; Johnsborg, O.; Lunde, M.; Knutsen, E.; Håvarstein, L.S. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 2005, 187, 4338–4345. [Google Scholar] [CrossRef]
- Shanker, E.; Federle, M.J. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 2017, 8, 15. [Google Scholar] [CrossRef]
- Kjos, M.; Miller, E.; Slager, J.; Lake, F.B.; Gericke, O.; Roberts, I.S.; Rozen, D.E.; Veening, J.W. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 2016, 12, e1005422. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Kochan, T.J.; Storck, D.N.; Dawid, S. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 2016, 12, e1005413. [Google Scholar] [CrossRef]
- Wang, C.Y.; Dawid, S. Mobilization of bacteriocins during competence in streptococci. Trends Microbiol. 2018, 26, 389. [Google Scholar] [CrossRef]
- Gómez-Mejia, A.; Gámez, G.; Hammerschmidt, S. Streptococcus pneumoniae two-component regulatory systems: The interplay of the pneumococcus with its environment. Int. J. Med. Microbiol. 2018, 308, 722–737. [Google Scholar] [CrossRef]
- Bergé, M.J.; Mercy, C.; Mortier-Barrière, I.; Van Nieuwenhze, M.S.; Brun, Y.V.; Grangeasse, C.; Polard, P.; Campo, N. A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat. Commun. 2017, 8, 1621. [Google Scholar] [CrossRef]
- Flannagan, S.E.; Clewell, D.B.; Sedgley, C.M. A ‘‘retrocidal” plasmid in Enterococcus faecalis: Passage and protection. Plasmid 2008, 59, 217–230. [Google Scholar] [CrossRef]
- Dunny, G.M.; Hancock, L.E.; Shankar, N. Enterococcal biofilm structure and role in colonization and disease. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Engelberg-Kulka, H.; Hazan, R. Cannibals defy starvation and avoid sporulation. Science 2003, 301, 467–468. [Google Scholar] [CrossRef]
- Lopez, D.; Vlamakis, H.; Kolter, R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 2009, 33, 152–163. [Google Scholar] [CrossRef]
- Lopez, D.; Kolter, R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 2010, 34, 134–149. [Google Scholar] [CrossRef]
- Schultz, D.; Wolynes, P.G.; Ben Jacob, E.; Onuchic, J.N. Deciding fate in adverse times: Sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2009, 106, 21027–21034. [Google Scholar] [CrossRef]
- Fujita, M.; González-Pastor, J.E.; Losick, R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005, 187, 1357–1368. [Google Scholar] [CrossRef]
- Veening, J.W.; Hamoen, L.W.; Kuipers, O.P. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 2005, 56, 1481–1494. [Google Scholar] [CrossRef]
- Pérez Morales, T.G.; Ho, T.D.; Liu, W.T.; Dorrestein, P.C.; Ellermeier, C.D. Production of the cannibalism toxin SDP is a multistep process that requires SdpA and SdpB. J. Bacteriol. 2013, 195, 3244–3251. [Google Scholar] [CrossRef]
- Liu, W.T.; Yang, Y.L.; Xu, Y.; Lamsa, A.; Haste, N.M.; Yang, J.Y. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2010, 107, 16286–16290. [Google Scholar] [CrossRef]
- Ellermeier, C.D.; Hobbs, E.C.; González-Pastor, J.E.; Losick, R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 2006, 124, 549–559. [Google Scholar] [CrossRef]
- Lamsa, A.; Liu, W.T.; Dorrestein, P.C.; Pogliano, K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 2012, 84, 486–500. [Google Scholar] [CrossRef]
- Povolotsky, T.L.; Orlova, E.; Tamang, D.G.; Saier, M.H. Defense against cannibalism: The SdpI family of bacterial immunity/signal transduction proteins. J. Membr. Biol. 2010, 235, 145–162. [Google Scholar] [CrossRef]
- Rosenberg, G.; Steinberg, N.; Oppenheimer-Shaanan, Y.; Olender, T.; Doron, S.; Ben-Ari, J.; Sirota-Madi, A.; Bloom-Ackermann, Z.; Kolodkin-Gal, I. Not so simple, not so subtle: The interspecies competition between Bacillus simplex and Bacillus subtilis and its impact on the evolution of biofilms. NPJ Biofilms Microbiomes 2016, 2, 15027. [Google Scholar] [CrossRef]
- López, D.; Vlamakis, H.; Losick, R.; Kolter, R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 2009, 74, 609–618. [Google Scholar] [CrossRef]
- Tcherpakov, M.; Ben-Jacob, E.; Gutnick, D.L. Paenibacillus dendritiformis sp. nov., proposal for a new pattern-forming species and its localization within a phylogenetic cluster. Int. J. Syst. Bacteriol. 1999, 49, 239–246. [Google Scholar] [CrossRef]
- Sirota-Madi, A.; Olender, T.; Helman, Y.; Brainis, I.; Finkelshtein, A.; Roth, D.; Hagai, E.; Leshkowitz, D.; Brodsky, L.; Galatenko, V.; et al. Genome sequence of the pattern-forming social bacterium Paenibacillus dendritiformis C454 Chiral Morphotype. J. Bacteriol. 2012, 194, 2127–2128. [Google Scholar] [CrossRef]
- Beer, A.; Zhang, H.P.; Florin, E.L.; Payne, S.M.; Ben-Jacob, E.; Swinney, H.L. Deadly competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. USA 2009, 106, 428–433. [Google Scholar] [CrossRef]
- Shen, P.; Lees, J.A.; Bee, C.G.W.; Brown, S.P.; Weiser, J.N. Pneumococcal quorum sensing drives an asymmetric owner–intruder competitive strategy during carriage via the competence regulon. Nat. Microbiol. 2019, 4, 198–208. [Google Scholar] [CrossRef]
- Andrukov, B.G.; Somova, L.M.; Timchenko, N.F. Strategy of programmed cell death in prokaryotes. Russ. J. Infect. Immun. 2015, 5, 15–26. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Ilio, C.D.; Laurenzi, V.D. Die for the community: An overview of programmed cell death in bacteria. Cell Death Dis. 2015, 6, e1609. [Google Scholar] [CrossRef]
- Koksharova, O.A. Bacteria and phenoptosis. Biochem. (Mosc.) 2013, 78, 963–970. [Google Scholar] [CrossRef]
- Peeters, S.H.; de Jonge, M.I. For the greater good: Programmed cell death in bacterial communities. Microbiol. Res. 2018, 207, 161–169. [Google Scholar] [CrossRef]
- Starokadomskyy, P.; Dmytruk, K.V. A bird’s-eye view of autophagy. Autophagy 2013, 9, 1121–1126. [Google Scholar] [CrossRef]
- Oleskin, A.V. Biosocial phenomena in unicellular organisms (exemplified by data concerning Prokaryota). J. Gen. Biol. 2009, 70, 225–238. (In Russian) [Google Scholar]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K. Quorum sensing: A prospective therapeutic target for bacterial diseases. BioMed Res. Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Wall, D. Kin recognition in bacteria. Annu. Rev. Microbiol. 2016, 70, 143–160. [Google Scholar] [CrossRef]
- Berg, K.H.; Ohnstad, H.S.; Havarstein, L.S. LytF, a novel competence-regulated murein hydrolase in the genus streptococcus. J. Bacteriol. 2009, 194, 627–635. [Google Scholar] [CrossRef]
- Cullin, N.; Merrit, J.; Kreth, J. Beyond cell division: The ecological roles of autolysins in oral biofilm communities. Curr. Oral Health Rep. 2017, 4, 14–21. [Google Scholar] [CrossRef]
- Liu, Y.; Burne, R.A. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J. Bacteriol. 2011, 193, 2826–2837. [Google Scholar] [CrossRef]
- Rodriguez, A.M.; Callahan, J.E.; Fawcett, P.; Ge, X.; Xu, P.; Kitten, T. Physiological and molecular characterization of genetic competence in Streptococcus sanguinis. Mol. Oral Microbiol. 2011, 26, 99–116. [Google Scholar] [CrossRef]
- Xu, Y.; Kreth, J. Role of LytF and AtlS in eDNA release by Streptococcus gordonii. PLoS ONE 2013, 8, e62339. [Google Scholar] [CrossRef]
- Salvadori, G.; Junges, R.; Morrison, D.A.; Petersen, F.C. Competence in Streptococcus pneumoniae and close commensal relatives: Mechanisms and implications. Front. Cell. Infect. Microbiol. 2019, 9, 94. [Google Scholar] [CrossRef]

| Microorganism | Regulation | Toxin | Role | |
|---|---|---|---|---|
| S. pneumoniae | ComX-dependent | CbpD | Murein hydrolase choline-binding protein D | Fratricine |
| ComX-dependent | LytA | N-acetylmuramoyl-L-alanine amidase | Autolysin | |
| non-CSP-regulated | LytC | Muramidase (lysozyme) | Autolysin | |
| ComM | Membrane protein | Immunity | ||
| ComX-dependent | CibAB | Two-peptide bacteriocin | Auxiliary role in fratricide | |
| CibC | Putative transmembrane protein | Immunity | ||
| E. faecalis | GBAP-dependent | GelE | Zinc metalloprotease (gelatinase) | Pro-lysis role |
| AtlA | N-actyl glucosaminidase | Autolysin | ||
| GBAP-dependent | SprE | Serine protease | Putative immunity function | |
| MC4-1 (product of bacA gene) | Plasmid-encoded class IIa bacteriocin | Bacteriocin | ||
| BacB | Putative bacteriocin immunity protein | Immunity | ||
| Bacillus subtilis | SpoOA-regulated | SkfA | Bacteriocin-like pro-peptide | Bacteriocin |
| SkfEF | ABC transporter | Putative immunity function | ||
| SpoOA-regulated | SdpC | Toxic protein | Toxin | |
| SdpI | Immunity | |||
| Paenibacillus dendritiformis | Slf | Peptide sibling-lethal factor | Toxin | |
| Subtilysin | Serine protease | Auxiliary role | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikryannikova, L.N.; Kurbatov, L.K.; Soond, S.M.; Zamyatnin, A.A., Jr. Harnessing the Potential of Killers and Altruists within the Microbial Community: A Possible Alternative to Antibiotic Therapy? Antibiotics 2019, 8, 230. https://doi.org/10.3390/antibiotics8040230
Ikryannikova LN, Kurbatov LK, Soond SM, Zamyatnin AA Jr. Harnessing the Potential of Killers and Altruists within the Microbial Community: A Possible Alternative to Antibiotic Therapy? Antibiotics. 2019; 8(4):230. https://doi.org/10.3390/antibiotics8040230
Chicago/Turabian StyleIkryannikova, Larisa N., Leonid K. Kurbatov, Surinder M. Soond, and Andrey A. Zamyatnin, Jr. 2019. "Harnessing the Potential of Killers and Altruists within the Microbial Community: A Possible Alternative to Antibiotic Therapy?" Antibiotics 8, no. 4: 230. https://doi.org/10.3390/antibiotics8040230
APA StyleIkryannikova, L. N., Kurbatov, L. K., Soond, S. M., & Zamyatnin, A. A., Jr. (2019). Harnessing the Potential of Killers and Altruists within the Microbial Community: A Possible Alternative to Antibiotic Therapy? Antibiotics, 8(4), 230. https://doi.org/10.3390/antibiotics8040230






