Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site Description and Livestock Management
2.2. Study Groups and Sampling Protocol
2.3. Fecal Sample Collection
2.4. Antimicrobial Susceptibility Screening
2.5. Characterization of Antibiotic Resistance Genes
2.6. Detection of E. coli Virulence Genes and Phylogenetic Grouping
2.7. Statistical Analysis
3. Results
3.1. Antimicrobial Resistance Proportions and Resistance Phenotypes
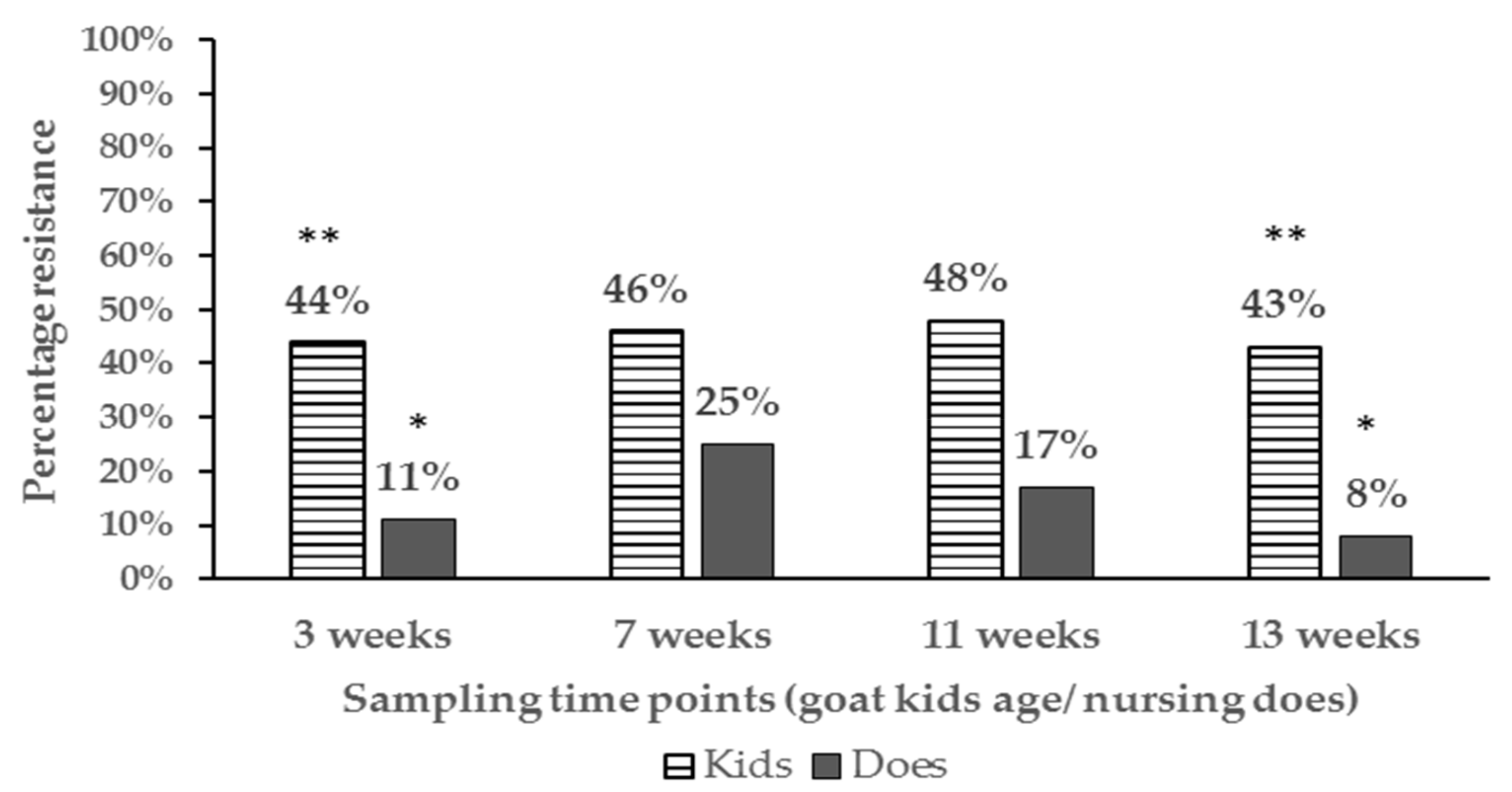
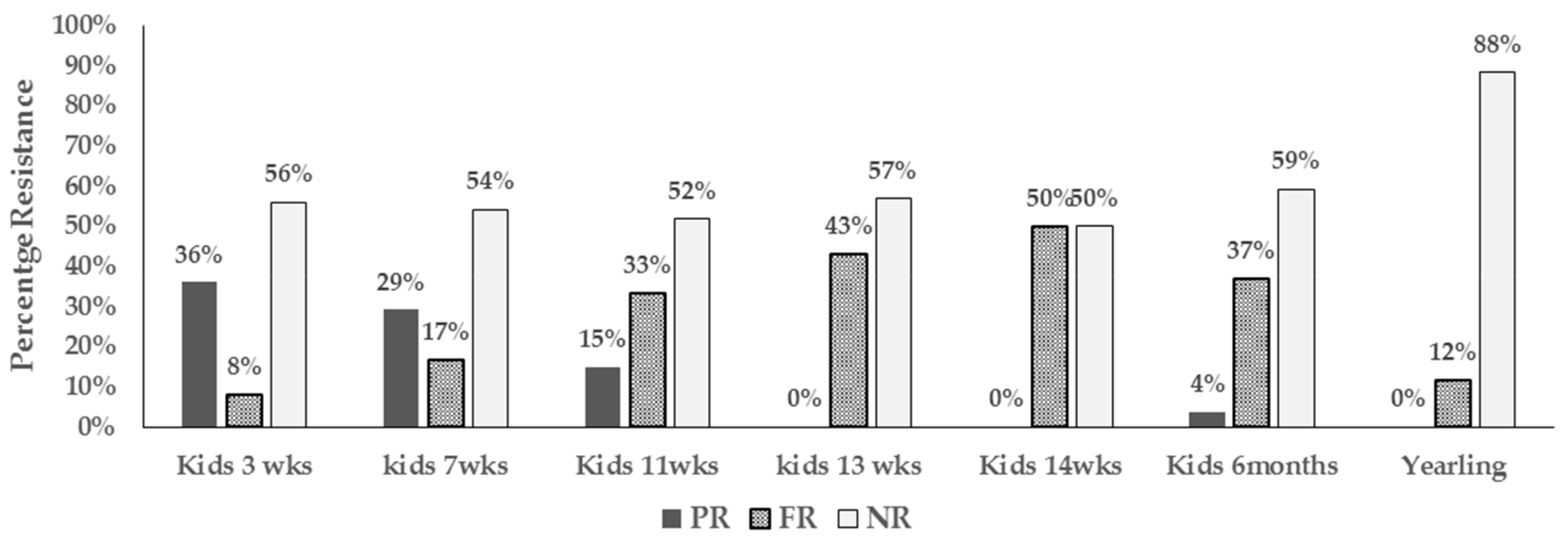
3.2. Proportions of Resistant E. coli Isolates and Phenotypes Among Age Groups and at Different Farm Location
3.3. Antibiotic Resistance Mechanisms
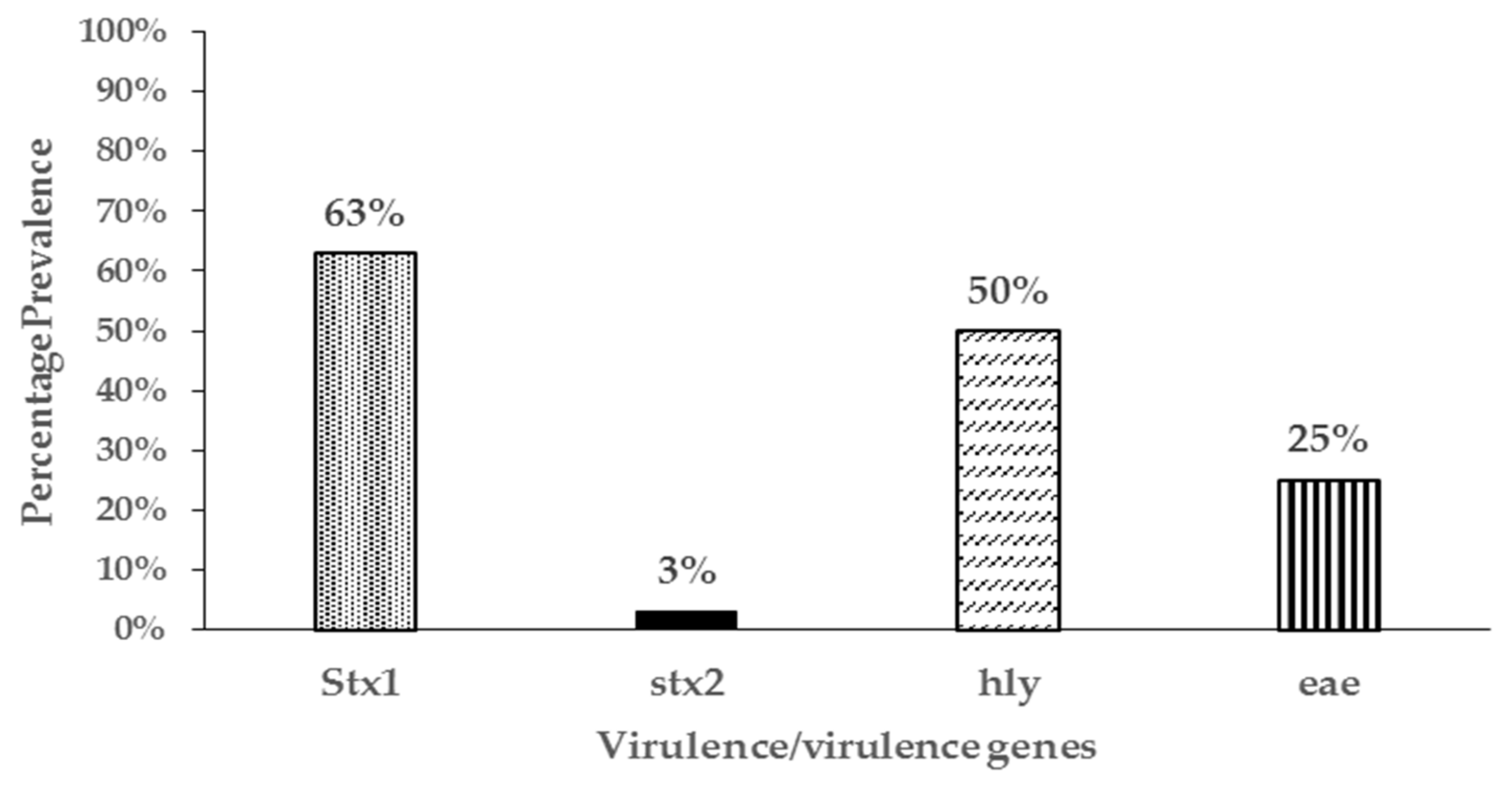
3.4. Virulence Genes and Phylogenetic Grouping of Resistant Isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Name | Sequence (5’ 3’) | Target Gene(s) or Region | Amplicon Size | Ref |
|---|---|---|---|---|
| TEM-F | TTCTTGAAGACGAAAGGGC | blaTEM | 1150 | [48] |
| TEM-R | ACGCTCAGTGGAACGAAAAC | |||
| SHV-SE | ACTCAAGGATGTATTGTG | blaSHV | 747 | [57] |
| SHV-AS | TTAGGGTTGCCAGTGCTCG | |||
| TEM-C | ATCAGCAATAAACCAGC | blaTEM | 516 | [58] |
| TEM-H | CCCCGAAGAACGTTTTC | |||
| HE605 | TTTCGTGTCGCCCTTATTCC | blaTEM | 690 | [59] |
| HE606 | CCGGCTCCAGATTTATCAGC | |||
| TEM-164.SE | TCGCCGCATACACTATTCTCAGAATGA | blaTEM | 447 | [57] |
| TEM-165.AS | ACGCTCACCGGCTCCAGATTTAT | |||
| Tet A-F | GTAATTCTGAGCACTGTCGC | tetA | 937 | [48] |
| Tet A-R | CTGTCCTGGACAACATTGCTT | |||
| Tet B-F | CTCAGTATTCCAAGCCTTTG | tetB | 416 | [48] |
| Tet B-R | CTAAGCACTTGTCTCCTGTT | |||
| Tet C-F | TCTAACAATGCGCTCATCGT | tetC | 570 | [48] |
| Tet C-R | GGTTGAAGGCTCTCAAGGGC | |||
| Tet D-F | ATTACACTGCTGGACGCGAT | tetD | 1104 | [48] |
| Tet D-R | CTGATCAGCAGACAGATTGC | |||
| Tet E-F | GTGATGATGGCACTGGTCAT | tetE | 1,179 | [48] |
| Tet E-R | CTCTGCTGTACATCGCTCTT | |||
| Str A-R | ATGGTGGACCCTAAAACTCT | streptA/B | 893 | [13] |
| Str B-F | CGTCTAGGATCGAGACAAAG | |||
| strA-F | CTTGGTGATAAGGCAATTC | strept A | 548 | [40] |
| strA-R | CCAATCGCAGATAGAAGGC | |||
| strA-F | GTCAAGGGATTGAAACC | streptA/B | 509 | [40] |
| strB-R | GGATCGTAGAACATATTGGC | |||
| AadA F | GTGGATGGCGGCCTGAAGCC | aadA | 525 | [40] |
| AadA R | AATGCCCAGTCGGCAGCG | |||
| Sul1-F | TGGTGACGGTGTTCGGCATTC | sul1 | 789 | [48] |
| Sul1-R | GCGAGGGTTTCCGAGAAGGCC | |||
| Sul2-F | CGGCATCAACATAACC | sul2 | 722 | [48] |
| Sul2-R | GTGTGCGGATGAAGTCAG | |||
| Sul3-F | GAGCAAGATTTTTGGAATCG | sul3 | 792 | [48] |
| Sul3-R. | CATCTGCAGCTAACCTAGGGCTTTGGA | |||
| GyrA-F | TACACCGGTCAACATTGAGG | gyr | 648 | [48] |
| Gyra-R. | TTAATGATTGCCGCCGTCGG | |||
| CmlA.F | TGTCATTTACGGCATACTCG | cml | 455 | [60] |
| CmlA.R | ATCAGGCATCCCATTCCCAT | |||
| aac(3)IV F | TGCTGGTCCACAGCTCCTTC | aac(3)IV | 653 | [61] |
| aac(3)IVR | CGGATGCAGGAAGATCAA | |||
| Stx1-a | TCTCAGTGGGCGTTCTTATG | stx1 | 338 | [62] |
| Stx1-b | TACCCCCTCAACTGCTAATA | |||
| Stx2-a | GCGGTTTTATTTGCATTAGC | stx2 | 115 | [62] |
| Stx2-b | TCCCGTCAACCTTCACTGTA | |||
| EAE-a | ATGCTTAGTGCTGGTTTAGG | eae | 248 | [62] |
| EAE-b | GCCTTCATCATTTCGCTTTC | |||
| HlyA-a | AGCTGCAAGTGCGGGTCTG | hly | 569 | [62] |
| HlyA-b | TACGGGTTATGCCTGCAAGTTCAC | |||
| ChuA.1 | GACGAACCA ACGGTCAGGAT | chuA | 279 | [33] |
| ChuA.2 | TGCCGCCAGTACC AAAGACA | |||
| YjaA.1 | TGAAGTGTCAGGAGACGCTG | yjaA | 211 | [33] |
| YjaA.2 | ATGGAGAATGCGTTCCTCAAC | |||
| TspE4C2.1 | GAGTAATGTCGGGGCATTCA | tspE4.C2 | 152 | [33] |
| TspE4C2.2 | CGCGCCAACAAAGTATTACG | |||
| EC16S-a | CCCCCTGGACGAAGACTGAC | E. coli 16s | 401 | [62] |
| EC16S-b | ACCGCTGGCAACAAAGGATA |
Appendix B
| Comparison Groups | Chi Square Value | P Value |
|---|---|---|
| Kids vs does 3 weeks | 5.282 | P = 0.0215 |
| Kids vs does 7 weeks | 2.03 | P = 0.1543 |
| Kids vs does 11 weeks | 3.287 | P = 0.0698 |
| Kids vs does 13 weeks | 5.151 | P = 0.0232 |
| Kids 7 weeks vs kids six months | 2.611 | P = 0.1061 |
| Kids 7 weeks vs kids one year | 9.734 | P = 0.0018 |
| Goats on pasture vs facility | 8.968 | P = 0.0027 |
| Tetracycline resistance on facility vs pasture | 57.654 | P < 0.0001 |
| Ampicillin resistance on pasture vs facility | 14.742 | P = 0.0001 |
References
- Bengtsson, B.; Greko, C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Ups. J. Med. Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Schroeder, C.M.; Meng, J.; White, D.G.; McDermott, P.F.; Wagner, D.D.; Yang, H.; Simjee, S.; DebRoy, C.; Walker, R.D. Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals. J. Antimicrob. Chemother. 2005, 56, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Poeta, P.; Senz, Y.; Vinu, L.; Coelho, A.C.; Matos, M.; Rojo-Bezares, B.; Rodrigues, J.; Torres, C. Mechanisms of antibiotic resistance in Escherichia coli isolates recovered from wild animals. Microb. Drug. Resi. 2008, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Bartoloni, A.; Paradisi, F.; Rossolini, G.M. Antibiotic resistance in the absence of antimicrobial use: Mechanisms and implications. Expert Rev. Anti-Infect. Ther. 2008, 6, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Bartoloni, A.; Riccobono, E.; Fernandez, C.; Mantella, A.; Magnelli, D.; Mannini, D.; Strohmeyer, M.; Bartalesi, F.; Rodriguez, H. Quinolone resistance in absence of selective pressure: The experience of a very remote community in the Amazon forest. PLoS Negl. Trop. Dis. 2012, 6, e1790. [Google Scholar] [CrossRef] [PubMed]
- Berge, A.C.; Hancock, D.D.; Sischo, W.M.; Besser, T.E. Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J. Am. Vet. Med. Assoc. 2010, 236, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- De Verdier, K.; Nyman, A.; Greko, C.; Bengtsson, B. Antimicrobial resistance and virulence factors in Escherichia coli from Swedish dairy calves. Acta Vet. Scand. 2012, 54, 2. [Google Scholar] [CrossRef]
- Duse, A.; Waller, K.P.; Emanuelson, U.; Unnerstad, H.E.; Persson, Y.; Bengtsson, B. Risk factors for antimicrobial resistance in fecal Escherichia coli from preweaned dairy calves. J. Dairy Sci. 2015, 98, 500–516. [Google Scholar] [CrossRef]
- Ho, P.-L.; Wong, R.C.; Lo, S.W.; Chow, K.-H.; Wong, S.S.; Que, T.-L. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 2010, 59, 702–707. [Google Scholar] [CrossRef]
- Hordijk, J.; Mevius, D.J.; Kant, A.; Bos, M.E.; Graveland, H.; Bosman, A.B.; Hartskeerl, C.M.; Heederik, D.J.; Wagenaar, J.A. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: A longitudinal approach. J. Antimicrob. Chemother. 2013, 68, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Kikuvi, G.; Schwarz, S.; Ombui, J.; Mitema, E.; Kehrenberg, C. Streptomycin and chloramphenicol resistance genes in Escherichia coli isolates from cattle, pigs, and chicken in Kenya. Microb. Drug Resi. 2007, 13, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003, 69, 6489–6494. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, N.; Caniça, M.; Capelo-Martinez, J.L.; Brito, F.; Igrejas, G.; Poeta, P. High prevalence of antimicrobial-resistant Escherichia coli from animals at slaughter: A food safety risk. J. Sci. Food Agric. 2013, 93, 517–526. [Google Scholar] [CrossRef]
- Sengeløv, G.; Halling-Sørensen, B.; Aarestrup, F.M. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 2003, 95, 91–101. [Google Scholar] [CrossRef]
- Smith, J.; Drum, D.; Dai, Y.; Kim, J.; Sanchez, S.; Maurer, J.; Hofacre, C.; Lee, M. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 2007, 73, 1404–1414. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741. [Google Scholar] [CrossRef]
- Witte, W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 2000, 16, 19–24. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Ding, L.J.; Fan, M.Z. Resistance patterns and detection of aac (3)-IV gene in apramycin-resistant Escherichia coli isolated from farm animals and farm workers in northeastern of China. Res. Vet. Sci. 2009, 87, 449–454. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Menzies, P. Antimicrobial resistance and small ruminant veterinary practice. Vet. Clin. North. Am. Food Anim. Pract. 2011, 27, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Motoi, Y.; Sato, M.; Maruyama, A.; Watanabe, H.; Fukumoto, Y.; Shimamoto, T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007, 73, 6686–6690. [Google Scholar] [CrossRef] [PubMed]
- Roug, A.; Byrne, B.A.; Conrad, P.A.; Miller, W. Zoonotic fecal pathogens and antimicrobial resistance in county fair animals. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Sayah, R.S.; Kaneene, J.B.; Johnson, Y.; Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic-and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 2005, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B: Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Ward, M.J. Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef]
- Espie, E.; Vaillant, V.; Mariani-Kurkdjian, P.; Grimont, F.; Martin-Schaller, R.; De Valk, H.; Vernozy-Rozand, C. Escherichia coli O157 outbreak associated with fresh unpasteurized goats’ cheese. Epidemiol. Infect. 2006, 134, 143–146. [Google Scholar] [CrossRef]
- Jacob, M.; Foster, D.; Rogers, A.; Balcomb, C.; Shi, X.; Nagaraja, T. Evidence of non-O157 Shiga toxin–Producing Escherichia coli in the feces of meat goats at a US slaughter plant. J. Food Prot. 2013, 76, 1626–1629. [Google Scholar] [CrossRef]
- Vu-Khac, H.; Cornick, N.A. Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 2008, 126, 356–363. [Google Scholar] [CrossRef]
- Tobias, J.; Vutukuru, S.-R. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol. Res. 2012, 167, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Zarazaga, M.; Briñas, L.; Ruiz-Larrea, F.; Torres, C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 2003, 51, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007, 1, 56. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, D.V.; Shaw, D.J.; Knight, H.I.; Davison, H.C.; Pearce, M.C.; Low, J.C.; Gunn, G.J.; Woolhouse, M.E. Age-related decline in carriage of ampicillin-resistant Escherichia coli in young calves. Appl. Environ. Microbiol. 2004, 70, 6927–6930. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Lucchetti, C.; Bartoloni, A.; Bartalesi, F.; Mantella, A.; Gamboa, H.; Carattoli, A.; Paradisi, F.; Rossolini, G.M. Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 2007, 51, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Edrington, T.; Farrow, R.; Carter, B.; Islas, A.; Hagevoort, G.; Callaway, T.; Anderson, R.; Nisbet, D. Age and diet effects on fecal populations and antibiotic resistance of a multi-drug resistant Escherichia coli in dairy calves. Agric. Food Anal. Bacteriol. 2012, 2, 162–174. [Google Scholar]
- Hoyle, D.; Davison, H.; Knight, H.; Yates, C.; Dobay, O.; Gunn, G.; Amyes, S.; Woolhouse, M. Molecular characterisation of bovine faecal Escherichia coli shows persistence of defined ampicillin resistant strains and the presence of class 1 integrons on an organic beef farm. Vet. Microbiol. 2006, 115, 250–257. [Google Scholar] [CrossRef]
- Langlois, B.E.; Dawson, K.A.; Leak, I.; Aaron, D.K. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988, 54, 1341–1344. [Google Scholar]
- Srinivasan, V.; Nam, H.-M.; Sawant, A.A.; Headrick, S.I.; Nguyen, L.T.; Oliver, S.P. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 2008, 55, 184–193. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007, 1, 191. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Kim, C. Prevalence of antimicrobial resistance (AMR) in bacteria isolated from farm animals, wildlife, and food samples in the eastern United States between 2007 and 2013. EC Nutrition 2017, 7, 264–274. [Google Scholar]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.; Shapir, N.; Sadowsky, M.J. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 2004, 70, 2503–2507. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Sáenz, Y.; Coelho, A.C.; Matos, M.; Vinué, L.; Rodrigues, J.; Torres, C. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol. 2008, 127, 97–105. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Distribution of the bla TEM gene and bla TEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 2011, 66, 745–751. [Google Scholar] [CrossRef]
- Briñas, L.; Zarazaga, M.; Sáenz, Y.; Ruiz-Larrea, F.; Torres, C. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 2002, 46, 3156–3163. [Google Scholar] [CrossRef]
- Olesen, I.; Hasman, H.; Møller Aarestrup, F. Prevalence of β-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb. Drug. Resi. 2004, 10, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Gillespie, B.E.; Lewis, M.J.; Nguyen, L.T.; Headrick, S.I.; Schukken, Y.H.; Oliver, S.P. Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet. Microbiol. 2007, 124, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Fuente, R.D.L.; García, S.; Orden, J.A.; Ruiz-Santa-Quiteria, J.A.; Díez, R.; Cid, D. Prevalence and characteristics of attaching and effacing strains of Escherichia coli isolated from diarrheic and healthy sheep and goats. J. Am. Vet. Med. Assoc. 2002, 63, 262–266. [Google Scholar]
- Zhao, S.; Blickenstaff, K.; Bodeis-Jones, S.; Gaines, S.; Tong, E.; McDermott, P. Comparison of the prevalences and antimicrobial resistances of Escherichia coli isolates from different retail meats in the United States, 2002 to 2008. Appl. Environ. Microbiol. 2012, 78, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Cortés, C.; De la Fuente, R.; Blanco, J.; Blanco, M.; Blanco, J.; Dhabi, G.; Mora, A.; Justel, P.; Contreras, A.; Sanchez, A. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 2005, 110, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010, 10, 161. [Google Scholar] [CrossRef]
- Monstein, H.J.; Östholm-Balkhed, Å.; Nilsson, M.; Nilsson, M.; Dornbusch, K.; Nilsson, L. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Colom, K.; Pérez, J.; Alonso, R.; Fernández-Aranguiz, A.; Lariño, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA–1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Commensal Escherichia coli of healthy humans: A reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 2010, 59, 1331–1339. [Google Scholar] [CrossRef]
- Sáenz, Y.; Brinas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Boerlin, P.; Travis, R.; Gyles, C.L.; Reid-Smith, R.; Lim, N.J.H.; Nicholson, V.; McEwen, S.A.; Friendship, R.; Archambault, M. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 2005, 71, 6753–6761. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Clark, C.G.; Rodgers, F.G. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157: H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 2002, 40, 3613–3619. [Google Scholar] [CrossRef] [PubMed]


| Resistance Phenotype | Number of Isolates (n = 136) |
|---|---|
| Tetracycline(Tet) | 50 |
| Streptomycin(Strept) | 25 |
| Ampicillin(Amp) | 17 |
| Nalidixic acid(Nal) | 6 |
| Chloramphenicol (Chl) | 2 |
| Amikacin(Ak) | 2 |
| Amoxycillin/clavulanate(Amc) | 1 |
| Tobramycin(Tob) | 1 |
| Gentamicin(GN) | 1 |
| Sulfamethoxazole-trimethoprim(Sxt) | 0 |
| Meropenem | 0 |
| Ciprofloxacin | 0 |
| Tet/Strept | 12 |
| Amp/Amc | 3 |
| Tet/Amp | 1 |
| Tet/Nal | 2 |
| Tet/Chl | 1 |
| Amp/Chl | 2 |
| Tet/Amc | 1 |
| Tob/Strept | 1 |
| Chl/Tob | 1 |
| Amp/Nal | 1 |
| Ak/Tet | 1 |
| Amp/strept | 1 |
| Amc/Ak/Nal | 1 |
| Nal/Strept/Tet | 1 |
| Amp/Nal/Sxt | 1 |
| Amc/Chl/Tet/Strept | 1 |
| No resistance | 272 |
| Antimicrobial Agent | n = 136 |
|---|---|
| Ampicillin | 26 |
| Streptomycin | 41 |
| Gentamicin | 1 |
| Tetracycline | 70 |
| Amikacin | 4 |
| Ciprofloxacin | 0 |
| Amoxycillin/Clavulanate | 7 |
| Meropenem | 0 |
| Choramphenicol | 7 |
| Tobramycin | 3 |
| Sulfamethoxazole-trimethoprim | 1 |
| Nalidixic acid | 12 |
| Sampling Group | Isolate Phenotype | Number of Antibiotics Resistant to: | ||||
|---|---|---|---|---|---|---|
| Sensitive | Resistant | Total | 1 | 2 | >2 | |
| Kids 3wks (P) | 14 | 11 | 25 | 10 | 1 | 0 |
| Kids 7wks (P) | 13 | 11 | 24 | 11 | 0 | 0 |
| Kids 11wks (F) | 14 | 13 | 27 | 11 | 2 | 0 |
| kids 13wks (F) | 33 | 25 | 58 | 24 | 1 | 0 |
| Kids 14wks (F) | 11 | 11 | 22 | 9 | 2 | 0 |
| Kids 6months (F/P) | 32 | 22 | 54 | 14 | 7 | 1 |
| Kids 1 year (P) | 38 | 5 | 43 | 3 | 1 | 1 |
| Nursing does (3wks) (P) | 16 | 2 | 18 | 2 | 0 | 0 |
| Nursing does (7wks) (P) | 15 | 5 | 20 | 5 | 0 | 0 |
| Nursing does (11wks) (F) | 10 | 2 | 12 | 1 | 1 | 0 |
| Nursing does (13wks) (F) | 11 | 1 | 12 | 1 | 0 | 0 |
| Adult does ** (P) | 32 | 12 | 44 | 8 | 4 | 0 |
| Other goats * (F) | 33 | 16 | 49 | 6 | 8 | 2 |
| Total | 272 | 136 | 408 | 105 | 27 | 4 |
| Antimicrobial Agent | Animal Location | |||
|---|---|---|---|---|
| Near facility (n = 212) | % | Pasture (n = 196) | % | |
| Ampicillin | 4* | 2% | 22** | 11% |
| Streptomycin | 18 | 9% | 23 | 12% |
| Gentamicin | 1 | 0.5% | 0 | 0% |
| Tetracycline | 65* | 31% | 5** | 2.5% |
| Amikacin | 4 | 2% | 0 | 0% |
| Ciprofloxacin | 0 | 0% | 0 | 0% |
| Amoxycillin/Clavulate | 4 | 2% | 3 | 1.5% |
| Meropenem | 0 | 0% | 0 | 0% |
| Choramphenical | 3 | 1.4% | 4 | 2% |
| Tobramycin | 1 | 0.5% | 2 | 1% |
| Sulfamethoxazole-trimethoprim | 0 | 0% | 1 | 0.5% |
| Nalidixic acid | 8 | 4% | 4 | 2% |
| Total resistant isolates | 85** | 40% | 51* | 26% |
| Phenotype of Resistance | Number of Isolates with This Phenotype | Gene Detected | Number of Isolates |
|---|---|---|---|
| Ampicillin | 26 | blaTEM | 12 |
| Amoxycillin/clavulanate | 7 | blaSHV | 1 |
| Tetracycline | 70 | tet(B) | 69 |
| Streptomycin | 41 | aadA strB/strA | 18 10 |
| Sulfamethoxazole-trimethoprim | 1 | sul2 | 1 |
| Chloramphenicol | 7 | - | - |
| Tobramycin | 3 | aac (3)/aac (3) | 1 |
| Amikacin | 4 | - | - |
| Gentamicin | 1 | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndegwa, E.; Almehmadi, H.; Chyer, K.; Kaseloo, P.; Ako, A.A. Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study. Antibiotics 2019, 8, 136. https://doi.org/10.3390/antibiotics8030136
Ndegwa E, Almehmadi H, Chyer K, Kaseloo P, Ako AA. Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study. Antibiotics. 2019; 8(3):136. https://doi.org/10.3390/antibiotics8030136
Chicago/Turabian StyleNdegwa, Eunice, Hanin Almehmadi, Kim Chyer, Paul Kaseloo, and Ankrah A. Ako. 2019. "Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study" Antibiotics 8, no. 3: 136. https://doi.org/10.3390/antibiotics8030136
APA StyleNdegwa, E., Almehmadi, H., Chyer, K., Kaseloo, P., & Ako, A. A. (2019). Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study. Antibiotics, 8(3), 136. https://doi.org/10.3390/antibiotics8030136







