Phage Therapy with a Focus on the Human Microbiota
Abstract
1. Introduction
2. Bacteriophages as Therapeutic Agents
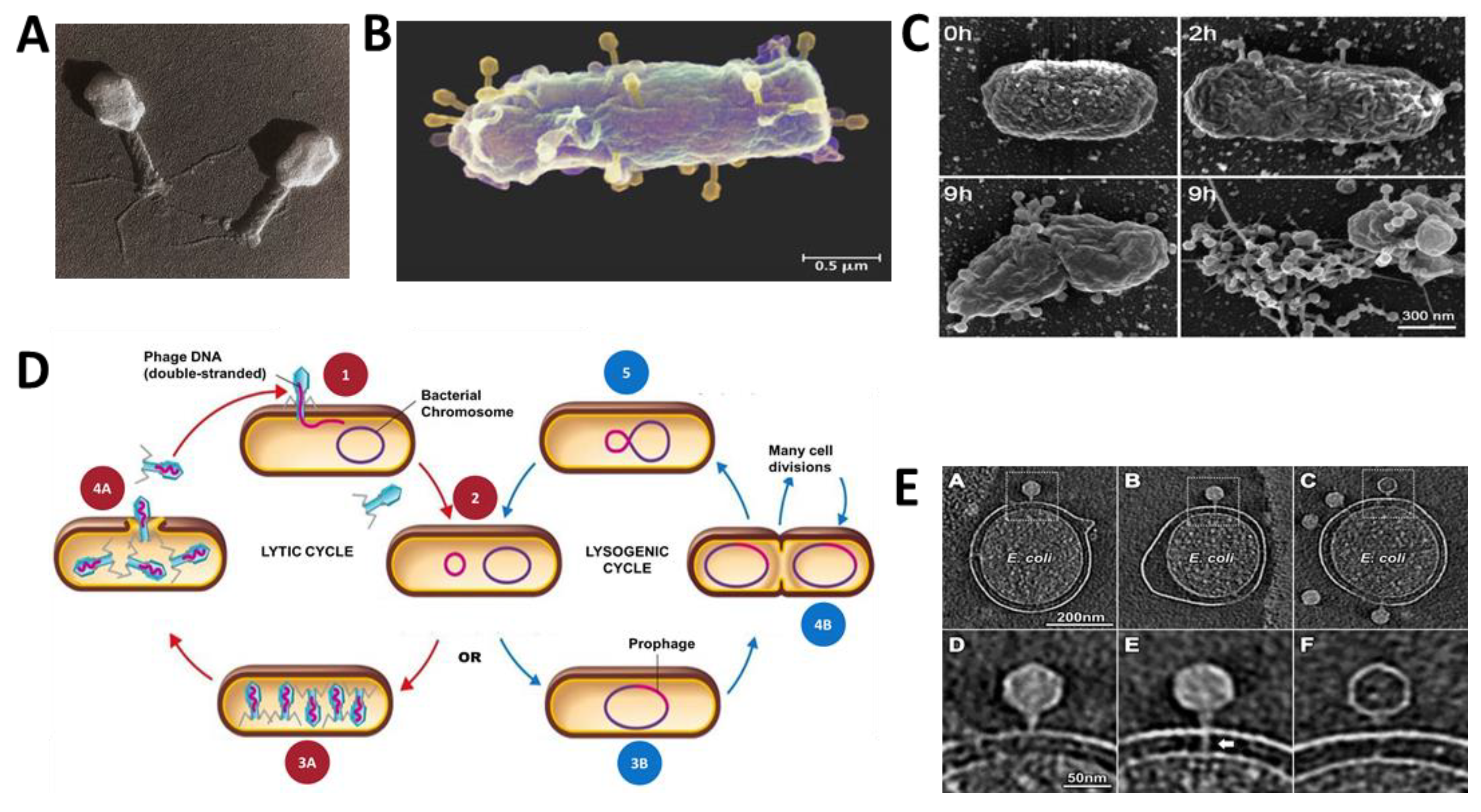
2.1. Phage Biology
2.2. History of Phage Therapy
2.3. Phage Therapy Compared to Antibiotic Therapy
3. Phage Therapy: Where Are We Now?
3.1. Advantages and Challenges of Phage Therapy
- -
- Specificity: Bacteriophages are generally very specific in their host range, which promises less harm to our microbiota. However, this specificity also means that for most clinically relevant infections, a mixture of different phage will have to be used to address the polymicrobial nature of the infection [60].
- -
- -
- Active on-site propagation: bacteriophages increase in concentration in situ as they propagate in the presence of bacterial host (infectious agents). Unlike antibiotics, which often require frequent doses to kill the bacteria efficiently, only one bacteriophage is theoretically needed to target a single corresponding host bacterium (single-hit kinetics) [65]. It is possible to administer a single low-dose of phage, which will then propagate itself, given the existing bacterial density as an active therapy, resulting in continued bacterial adsorption and killing [66].
- -
- Low inherent toxicity: bacteriophages are primarily composed of nucleic acid and proteins that have been studied to be non-toxic with the use of highly purified phage preparations [67].
- -
- Enormous diversity: Phage is found in diverse abundances across the biosphere and therefore isolating and purifying new phages, necessary to target a known pathogenic bacteria, is achievable even when bacteria evolve resistance [68].
- -
- Formulation and application versatility: Multiple strains of phages can be added together into a “phage cocktail” to target multiple bacteria of interest with a broader killing spectrum [69]. Furthermore, mode of administration (liquid, ointment [70], powder [71], oral tablets [72]) could also vary in accordance with each unique situation.
- -
- Narrow potential for antibiotic cross-resistance: The mechanism of bacterial resistance to antibiotics and phage are entirely different [61,73]. Thus, bacteria that are resistant to antibiotics may be targeted and treated with the use of phage therapy, and vice-versa, presenting combination therapies as an attractive strategy [74].
- -
- -
- Low environmental impact: bacteriophages are natural components of the environment that can be naturally evolved (as opposed to genetic modification), thus easing public acceptance of phage therapy. Furthermore, because this natural product is composed primarily of proteins and nucleic acids, unused phage materials can easily be inactivated and discarded [63].
- -
3.2. Outstanding Challenges of Phage Therapy
- -
- Narrow host-range: phages generally have a narrow host-range, which limits exactly which strain(s) of bacteria can be targeted. This is a challenge because most infections in the body are polymicrobial. Thus it is imperative to employ effective and efficient phage cocktails, curated from a combination of multiple selected phages to develop a broader host spectrum [80].
- -
- Even though phage therapy was employed in Western medicine before antibiotics took over, it currently does not have the approval for human administration from the Food and Drug Administration (FDA) or the European Medicines Agency (EMA) due to an increase in regulatory standards, partially as a result of the increased understanding within the scientific community about the potential effect of drugs on the human microbiome and partially due to concerns about phage resistance and the role of phages in bacterial genome evolution. To obtain regulatory approvals, current research is heavily exploring roadblocks through numerous animal studies and clinical trials [81,82,83]. The FDA has, however, approved the use of phages for food decontamination [84], dietary supplements [85], and environmental prophylaxis. Phage is allowed only on a compassionate care basis for human therapeutic use [86,87].
- -
- Phages as drivers of evolution: unlike common pharmaceuticals, bacteriophages are DNA/RNA-containing protein-based biological agents that have the potential to interact with the body’s immune system and other microbial cells in the body and can actively replicate and evolve within the body. This evolution can, in turn, result in the evolution of the commensal host bacterial communities and possibly even impact the composition of the niche microbiome [88].
- -
- Phage selection criteria: The criteria for the selection of therapeutic phage is not well determined. Most reports to date have focused on phage host range, but factors such as the ability of phage to infect stationary phase bacteria [89], phage enzymes, stability to serum inactivation, and mutation rate have been shown to be important and deserve more investigation beyond proof-of-concept reports [68,90].
- -
- Lack of well-curated public phage libraries: there is a serious lack of well-curated, well-characterized libraries for therapeutic phage. Various research labs in academia, industry, and army research centers have small to mid-sized collections that, except for very few cases, are considered proprietary and are not shared with the broader scientific community. This need was acknowledged by the community recently and led the DSMZ (German Collection of Microorganisms and Cell Cultures) to start the Phage Call Project and encourage researchers to deposit characterized phages to their public phage library. This effort, although valuable, is progressing very slowly, with only a few phages per strain currently in the library. The slow pace of progress in this regard can partially be attributed to the vast diversity of phages around the globe, partially to the lack of high throughput methods for rapid isolation and characterization of new phage, and partially to the lack of a mechanism to acknowledge phage as the intellectual property of the discovering lab, leading to the unwillingness of researchers to share so as to maintain competitive advantage.
3.3. Clinical Trials and Current Phage Therapy
4. Phage-Host Evolutionary Dynamics and Diversification
5. Bacteriophages and the Microbiota
5.1. Introduction to the Microbiota
5.2. Bacteriophages as Part of the Human Microbiota
5.3. Microbiome Therapy with Bacteriophages
5.4. Possible Interactions of Therapeutic Phage and Niche Microbiota
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- D’Herelle, F.; Malone, R.H.; Lahiri, M.N. Studies on Asiatic cholera. Indian Med. Res. Mem. 1927, 14, 195–219. [Google Scholar]
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharm. 2017, 133, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Summers, W.C. The strange history of phage therapy. Bacteriophage 2012, 2, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Michail, S.; Durbin, M.; Turner, D.; Griffiths, A.M.; Mack, D.R.; Hyams, J.; Leleiko, N.; Kenche, H.; Stolfi, A.; Wine, E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 2011, 18, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Abramson, S.B. The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol. 2011, 7, 569. [Google Scholar] [CrossRef]
- Khan, A.A.; Shrivastava, A.; Khurshid, M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochim. Biophys. Acta Rev. Cancer 2012, 1826, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.; Abdolmaleky, H.M.; Zhou, J.R. Microbiome, inflammation, epigenetic alterations, and mental diseases. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 651–660. [Google Scholar] [CrossRef]
- Ubeda, C.; Pamer, E.G. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012, 33, 459–466. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Döring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef]
- Shin, H.; Price, K.; Albert, L.; Dodick, J.; Park, L.; Dominguez-Bello, M.G. Changes in the eye microbiota associated with contact lens wearing. MBio 2016, 7, e00198-16. [Google Scholar] [CrossRef]
- Dudek, N.K.; Sun, C.L.; Burstein, D.; Kantor, R.S.; Goltsman DS, A.; Bik, E.M.; Thomas, B.C.; Banfield, J.F.; Relman, D.A. Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome. Curr. Biol. 2017, 27, 3752–3762. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Forney, L.J.; Ravel, J. Vaginal Microbiome: Rethinking Health and Disease. Annu. Rev. Microbiol. 2012, 66, 371–389. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; Van De Ven, T.G. Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2862–2871. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G. Formation of biofilms under phage predation: Considerations concerning a biofilm increase. Biofouling 2013, 29, 457–468. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Van De Ven, T.G.; Tufenkji, N. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl. Environ. Microbiol. 2013, 79, 6110–6116. [Google Scholar] [CrossRef]
- Bobay, L.M.; Ochman, H. Biological species in the viral world. Proc. Natl. Acad. Sci. USA 2018, 115, 6040–6045. [Google Scholar] [CrossRef]
- Armon, R. Soil Bacteria and Bacteriophages. In Biocommunication in Soil Microorganisms; Witzany, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 67–112. [Google Scholar]
- Perez Sepulveda, B.; Redgwell, T.; Rihtman, B.; Pitt, F.; Scanlan, D.J.; Millard, A. Marine phage genomics: The tip of the iceberg. FEMS Microbiol. Lett. 2016, 363, fnw158. [Google Scholar] [CrossRef]
- Navarro, F.; Muniesa, M. Phages in the Human Body. Front. Microbiol. 2017, 8, 566. [Google Scholar] [CrossRef]
- Calendar, R.L. The Bacteriophages, 2nd ed.; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Hatfull, G.F. Bacteriophage genomics. Curr. Opin. Microbiol. 2008, 11, 447–453. [Google Scholar] [CrossRef]
- Ackermann, H.W. Chapter 1—Bacteriophage Electron Microscopy. In Advances in Virus Research; Łobocka, M., Szybalski, W.T., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 82, pp. 1–32. [Google Scholar]
- Grose, J.H.; Casjens, S.R. Understanding the enormous diversity of bacteriophages: The tailed phages that infect the bacterial family Enterobacteriaceae. Virology 2014, 468, 421–443. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Dy, R.L.; Richter, C.; Salmond, G.P.; Fineran, P.C. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Simmonds, P.; Aiewsakun, P. Virus classification—Where do you draw the line? Arch. Virol. 2018, 163, 2037–2046. [Google Scholar] [CrossRef]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with ‘Lytic or lysogenic’. FEMS Microbiol. Lett. 2016, 363, fnw047. [Google Scholar] [CrossRef]
- Campbell, A. The future of bacteriophage biology. Nat. Rev. Genet. 2003, 4, 471. [Google Scholar] [CrossRef]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. The Bacteriophage T7 Virion Undergoes Extensive Structural Remodeling During Infection. Science 2013, 339, 576–579. [Google Scholar] [CrossRef]
- Dou, C.; Xiong, J.; Gu, Y.; Yin, K.; Wang, J.; Hu, Y.; Zhou, D.; Fu, X.; Qi, S.; Zhu, X.; et al. Structural and functional insights into the regulation of the lysis–lysogeny decision in viral communities. Nat. Microbiol. 2018, 3, 1285–1294. [Google Scholar] [CrossRef]
- Goerke, C.; Köller, J.; Wolz, C. Ciprofloxacin and Trimethoprim Cause Phage Induction and Virulence Modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 171–177. [Google Scholar] [CrossRef]
- Parkinson, J.S. Classic Spotlight: The Discovery of Bacterial Transduction. J. Bacteriol. 2016, 198, 2899–2900. [Google Scholar] [CrossRef]
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2004. [Google Scholar]
- Chatterjee, S.; Rothenberg, E. Interaction of bacteriophage l with its E. coli receptor, LamB. Viruses 2012, 4, 3162–3178. [Google Scholar] [CrossRef]
- Straus, S.K.; Bo, H.E. Filamentous Bacteriophage Proteins and Assembly. Subcell. Biochem. 2018, 88, 261–279. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Therapy: Various Perspectives on How to Improve the Art. In Host-Pathogen Interactions: Methods and Protocols; Medina, C., López-Baena, F.J., Eds.; Springer: New York, NY, USA, 2018; pp. 113–127. [Google Scholar]
- Monteiro, R.; Pires, D.P.; Costa, A.R.; Azeredo, J. Phage Therapy: Going Temperate? Trends Microbiol. 2018. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Calendar, R. The Bacteriophages; Oxford University Press: Oxford, UK, 2006; p. 746. [Google Scholar]
- Yap, M.L.; Rossmann, M.G. Structure and function of bacteriophage T4. Future Microbiol. 2014, 9, 1319–1327. [Google Scholar] [CrossRef]
- Biozentrum, University of Basel/Sceince Photo Library, TEM Of T4 Bacteriophage. 2013. Available online: https://www.sciencephoto.com/media/249780/view (accessed on 18 June 2019).
- Yong, E. Viruses in the gut protect from infection. Nature 2013. [Google Scholar] [CrossRef]
- Sabehi, G.; Shaulov, L.; Silver, D.H.; Yanai, I.; Harel, A.; Lindell, D. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc. Natl. Acad. Sci. USA 2012, 109, 2037–2042. [Google Scholar] [CrossRef]
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. Lancet 1915, 2, 1241–1243. [Google Scholar] [CrossRef]
- d’Herelle, F. An invisible antagonist microbe of dysentery bacillus. C. R. Hebd. Seances Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Duckworth, D.H. Who discovered bacteriophage? Bacteriol. Rev. 1976, 40, 793. [Google Scholar] [PubMed]
- Summers, W.C. How Bacteriophage Came to Be Used by the Phage Group. J. Hist. Biol. 1993, 26, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- d’Herelle, F.; Smith, G.H. The Bacteriophage and Its Behavior; Williams & Wilkins: Baltimore, MA, USA, 1926. [Google Scholar]
- Eaton, M.D.; Bayne-Jones, S. Bacteriophage therapy: Review of the principles and results of the use of bacteriophage in the treatment of infections. J. Am. Med. Assoc. 1934, 103, 1769–1776. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Kuchment, A. They’re Not a Panacea: Phage Therapy in the Soviet Union and Georgia. In The Forgotten Cure: The Past and Future of Phage Therapy; Kuchment, A., Ed.; Springer: New York, NY, USA, 2012; pp. 53–62. [Google Scholar]
- Kutateladze, M.; Adamia, R. Phage therapy experience at the Eliava Institute. Med. Mal. Infect. 2008, 38, 426–430. [Google Scholar] [CrossRef]
- Stone, R. Bacteriophage therapy: Stalin’s forgotten cure. Science 2002, 298, 728–731. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Salmasian, H.; Cohen, B.; Abrams, J.A.; Larson, E.L. Receipt of Antibiotics in Hospitalized Patients and Risk for Clostridium difficile Infection in Subsequent Patients Who Occupy the Same Bed. JAMA Intern. Med. 2016, 176, 1801–1808. [Google Scholar] [CrossRef]
- Weledji, E.P.; Assob, J.C.; Nsagha, D.S.; Weledji, E.K. Pros, cons and future of antibiotics. New Horiz. Transl. Med. 2017, 4, 9–14. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- El Haddad, L.; Harb, C.P.; A Gebara, M.; A Stibich, M.; Chemaly, R.F. A Systematic and Critical Review of Bacteriophage Therapy Against Multidrug-resistant ESKAPE Organisms in Humans. Clin. Infect. Dis. 2018, 69, 167–178. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Kristian, S.A.; Timmer, A.M.; Liu, G.Y.; Lauth, X.; Sal-Man, N.; Rosenfeld, Y.; Shai, Y.; Gallo, R.L.; Nizet, V. Impairment of innate immune killing mechanisms by bacteriostatic antibiotics. FASEB J. 2007, 21, 1107–1116. [Google Scholar] [CrossRef]
- Moldovan, R.; Chapman-McQuiston, E.; Wu, X.L. On kinetics of phage adsorption. Biophys. J. 2007, 93, 303–315. [Google Scholar] [CrossRef]
- Payne, R.; Jansen, V.A. Phage therapy: The peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharm. Ther. 2000, 68, 225–230. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and Safety Requirements for Sustainable Phage Therapy Products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef]
- Gill, J.J.; Hyman, P. Phage Choice, Isolation, and Preparation for Phage Therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Tanji, Y.; Shimada, T.; Fukudomi, H.; Miyanaga, K.; Nakai, Y.; Unno, H. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 2005, 100, 280–287. [Google Scholar] [CrossRef]
- Cheng, M.; Zhang, L.; Zhang, H.; Li, X.; Wang, Y.; Xia, F.; Wang, B.; Cai, R.; Guo, Z.; Zhang, Y.; et al. An Ointment Consisting of the Phage Lysin LysGH15 and Apigenin for Decolonization of Methicillin-Resistant Staphylococcus aureus from Skin Wound. Viruses 2018, 10, 244. [Google Scholar] [CrossRef]
- Leung, S.S.Y.; Parumasivam, T.; Gao, F.G.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Britton, W.J.; Kutter, E.; Chan, H.K. Production of Inhalation Phage Powders Using Spray Freeze Drying and Spray Drying Techniques for Treatment of Respiratory Infections. Pharm. Res. 2016, 33, 1486–1496. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.; Garton, N.J.; Stapley, A.G.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): β-Lactam and Quinolone Antibiotics Stimulate Virulent Phage Growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Abedon, S.T. Ecology of anti-biofilm agents I: Antibiotics versus bacteriophages. Pharmaceuticals 2015, 8, 525–558. [Google Scholar] [CrossRef]
- Harper, D.R.; Parracho, H.M.R.T.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and Biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Abedon, S.T. Ecology of anti-biofilm agents II: Bacteriophage exploitation and biocontrol of biofilm bacteria. Pharmaceuticals 2015, 8, 559–589. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Zdzisław, P.; Paweł, R.; Marlena, K.; Elżbieta, W.; et al. Chapter 3—Clinical Aspects of Phage Therapy. In Advances in Virus Research; Łobocka, M., Szybalski, W., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 83, pp. 73–121. [Google Scholar]
- Miedzybrodzki, R.; Fortuna, W.; Weber-Dabrowska, B.; Górski, A. Phage therapy of staphylococcal infections (including MRSA) may be less expensive than antibiotic treatment. Postepy Hig. Med. Dosw. 2007, 61, 461–465. [Google Scholar]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Alam Sarker, S.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef]
- Rose, T.; Verbeken, G.; De Vos, D.; Merabishvili, M.; Vaneechoutte, M.; Lavigne, R.; Jennes, S.; Zizi, M.; Pirnay, J.P. Experimental phage therapy of burn wound infection: Difficult first steps. Int. J. Burn. Trauma 2014, 4, 66–73. [Google Scholar]
- Smithyman, A.; Speck, P. Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol. Lett. 2015, 363, 1–5. [Google Scholar] [CrossRef]
- Sarhan, W.A.; Azzazy, H.M. Phage approved in food, why not as a therapeutic? Expert Rev. Anti Infect. Ther. 2015, 13, 91–101. [Google Scholar] [CrossRef]
- Aleshkin, A.V.; Rubalskii, E.O.; Volozhantsev, N.V.; Verevkin, V.V.; Svetoch, E.A.; Kiseleva, I.A.; Bochkareva, S.S.; Borisova, O.Y.; Popova, A.V.; Bogun, A.G.; et al. A small-scale experiment of using phage-based probiotic dietary supplement for prevention of E. coli traveler’s diarrhea. Bacteriophage 2015, 5, e1074329. [Google Scholar] [CrossRef][Green Version]
- Fish, R.; Kutter, E.; Wheat, G.; Blasdel, B.; Kutateladze, M.; Kuhl, S. Compassionate Use of Bacteriophage Therapy for Foot Ulcer Treatment as an Effective Step for Moving Toward Clinical Trials. Methods Mol. Biol. 2018, 1693, 159–170. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Koskella, B.; Brockhurst, M.A. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef]
- Bryan, D.; El-Shibiny, A.; Hobbs, Z.; Porter, J.; Kutter, E.M. Bacteriophage T4 Infection of Stationary Phase, E. coli: Life after Log from a Phage Perspective. Front. Microbiol. 2016, 7, 1391. [Google Scholar] [CrossRef]
- Sunderland, K.S.; Yang, M.; Mao, C. Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine. Angew. Chem. Int. Ed. 2017, 1964–1992. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2018. [Google Scholar] [CrossRef]
- McCallin, S.; Alam Sarker, S.; Barretto, C.; Sultana, S.; Berger, B.; Huq, S.; Krause, L.; Bibiloni, R.; Schmitt, B.; Reuteler, G.; et al. Safety analysis of a Russian phage cocktail: From MetaGenomic analysis to oral application in healthy human subjects. Virology 2013, 443, 187–196. [Google Scholar] [CrossRef]
- Nahum, G.G.; Uhl, K.; Kennedy, D.L. Antibiotic Use in Pregnancy and Lactation: What Is and Is Not Known About Teratogenic and Toxic Risks. Obs. Gynecol. 2006, 107, 1120–1138. [Google Scholar] [CrossRef]
- Bacteriophage. Available online: https://www.microgen.ru/en/products/bakteriofagi/ (accessed on 26 November 2019).
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Chan, B.K.; E Turner, P.; Kim, S.; Mojibian, H.R.; A Elefteriades, J.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Criscuolo, E.; Spadini, S.; Lamanna, J.; Ferro, M.; Burioni, R. Bacteriophages and Their Immunological Applications against Infectious Threats. J. Immunol. Res. 2017, 2017, 13. [Google Scholar] [CrossRef]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef]
- Standard Treatment Associated with Phage Therapy Versus Placebo for Diabetic Foot Ulcers Infected by S. aureus. Available online: https://clinicaltrials.gov/ct2/show/NCT02664740 (accessed on 17 June 2019).
- AmpliPhi Biosciences Corporation. Individual Patient Expanded Access for AB-SA01, An Investigational Anti-Staphylococcus aureus Bacteriophage Therapeutic. Available online: https://clinicaltrials.gov/ct2/show/NCT03395769 (accessed on 26 June 2019).
- Corporation AmpliPhi Biosciences. Individual Patient Expanded Access for AB-PA01, an Investigational Anti-Pseudomonas Aeruginosa Bacteriophage Therapeutic. Available online: https://clinicaltrials.gov/ct2/show/NCT03395743 (accessed on 18 June 2019).
- Safety and Efficacy of EcoActive on Intestinal Adherent Invasive E coli in Patients with Inactive Crohn’s Disease. Available online: https://clinicaltrials.gov/ct2/show/NCT03808103 (accessed on 18 June 2019).
- Experimental Phage Therapy of Bacterial Infections. Available online: https://clinicaltrials.gov/ct2/show/NCT00945087 (accessed on 19 June 2019).
- Langlet, J.; Gaboriaud, F.; Gantzer, C.; Duval, J.F.L. Impact of chemical and structural anisotropy on the electrophoretic mobility of spherical soft multilayer particles: The case of bacteriophage MS2. Biophys. J. 2008, 94, 3293–3312. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.; Änggård, E.; Harper, D. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Leitner, L.; Sybesma, W.; Chanishvili, N.; Goderdzishvili, M.; Chkhotua, A.; Ujmajuridze, A.; Schneider, M.P.; Sartori, A.; Mehnert, U.; Bachmann, L.M.; et al. Bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomized, placebo-controlled, double-blind clinical trial. BMC Urol. 2017, 17, 90. [Google Scholar] [CrossRef]
- A Prospective, Randomized, Double-Blind Controlled Study of WPP-201 for the Safety and Efficacy of Treatment of Venous Leg Ulcers (WPP-201). Available online: https://clinicaltrials.gov/ct2/show/NCT00663091 (accessed on 18 June 2019).
- Fernández, L.; Rodriguez, A.; García, P. Phage or foe: An insight into the impact of viral predation on microbial communities. ISME J. 2018, 12, 1171–1179. [Google Scholar] [CrossRef]
- Scanlan, P.D. Bacteria-Bacteriophage Coevolution in the Human Gut: Implications for Microbial Diversity and Functionality. Trends Microbiol. 2017, 25, 614–623. [Google Scholar] [CrossRef]
- Janzen, D.H. When is it Co-evolution? Evolution 1980, 34, 611–612. [Google Scholar] [CrossRef]
- Mizoguchi, K.; Morita, M.; Fischer, C.R.; Yoichi, M.; Tanji, Y.; Unno, H. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 2003, 69, 170–176. [Google Scholar] [CrossRef]
- Hynes, A.P.; Rousseau, G.M.; Agudelo, D.; Goulet, A.; Amigues, B.; Loehr, J.; Romero, D.A.; Fremaux, C.; Horvath, P.; Doyon, Y.; et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 2018, 9, 2919. [Google Scholar] [CrossRef]
- Levin, B.R.; Bull, J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004, 2, 166–173. [Google Scholar] [CrossRef]
- Azam, A.H.; Tanji, Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 2121–2131. [Google Scholar] [CrossRef]
- Lopez Pascua, L.; Gandon, S.; Buckling, A. Abiotic heterogeneity drives parasite local adaptation in coevolving bacteria and phages. J. Evol. Biol. 2012, 25, 187–195. [Google Scholar] [CrossRef]
- Mavrich, T.N.; Hatfull, G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017, 2, 17112. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology: Phage cocktails. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 78, pp. 1–23. [Google Scholar]
- Avrani, S.; Lindell, D. Convergent evolution toward an improved growth rate and a reduced resistance range in Prochlorococcus strains resistant to phage. Proc. Natl. Acad. Sci. USA 2015, 112, E2191–E2200. [Google Scholar] [CrossRef]
- Yang, H.; Schmitt-Wagner, D.; Stingl, U.; Brune, A. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 2005, 7, 916–932. [Google Scholar] [CrossRef]
- Schrag, S.J.; Mittler, J.E. Host-parasite coexistence: The role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 1996, 148, 348–377. [Google Scholar] [CrossRef]
- Heilmann, S.; Sneppen, K.; Krishna, S. Coexistence of phage and bacteria on the boundary of self-organized refuges. Proc. Natl. Acad. Sci. USA 2012, 109, 12828–12833. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Buckling, A.; Rainey, P.B.; Brockhurst, M. Spatial heterogeneity and the stability of host-parasite coexistence. J. Evol. Biol. 2006, 19, 374–379. [Google Scholar] [CrossRef]
- De Sordi, L.; Khanna, V.; Debarbieux, L. The Gut Microbiota Facilitates Drifts in the Genetic Diversity and Infectivity of Bacterial Viruses. Cell Host Microbe 2017, 22, 801–808. [Google Scholar] [CrossRef]
- Duerkop, B.A. Bacteriophages shift the focus of the mammalian microbiota. PLoS Path. 2018, 14, e1007310. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; A Reyes, J.; A Shah, S.; Leleiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027. [Google Scholar] [CrossRef]
- Hill, J.M.; Clement, C.; Pogue, A.I.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. Pathogenic microbes, the microbiome, and Alzheimer’s disease (AD). Front. Aging Neurosci. 2014, 6, 127. [Google Scholar]
- Love, B.L.; Mann, J.R.; Hardin, J.W.; Lu, Z.K.; Cox, C.; Amrol, D.J. Antibiotic prescription and food allergy in young children. Allergy Asthma Clin. Immunol. Off. J. Can. Soc. Allergy Clin. Immunol. 2016, 12, 41. [Google Scholar] [CrossRef]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2012, 152, 39–50. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 1283. [Google Scholar] [CrossRef]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Turroni, F.; Tremblay, D.; Ferrario, C.; Mancabelli, L.; Duranti, S.; Ward, D.V.; Ossiprandi, M.C.; Moineau, S.; et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ. Microbiol. 2016, 18, 2196–2213. [Google Scholar] [CrossRef]
- Allen, H.K.; Looft, T.; Bayles, D.O.; Humphrey, S.; Levine, U.Y.; Alt, D.; Stanton, T.B. Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2011, 2, e00260-11. [Google Scholar] [CrossRef]
- Duerkop, B.A.; Clements, C.V.; Rollins, D.; Rodrigues, J.L.; Hooper, L.V. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. USA 2012, 109, 17621–17626. [Google Scholar] [CrossRef]
- Rajpal, D.K.; Brown, J.R. Modulating the human gut microbiome as an emerging therapeutic paradigm. Sci. Prog. 2013, 96, 224–236. [Google Scholar] [CrossRef]
- Paule, A.; Frezza, D.; Edeas, M. Microbiota and Phage Therapy: Future Challenges in Medicine. Med. Sci. 2018, 6, 86. [Google Scholar] [CrossRef]
- Viertel, T.M.; Ritter, K.; Horz, H.P. Viruses versus bacteria--novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J. Antimicrob. Chemother. 2014, 69, 2326–2336. [Google Scholar] [CrossRef]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and immunomodulation. Future Microbiol. 2017, 12, 905–914. [Google Scholar] [CrossRef]
- Kuramitsu, H.K.; He, X.; Lux, R.; Anderson, M.H.; Shi, W. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 2007, 71, 653–670. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15. [Google Scholar] [CrossRef]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814. [Google Scholar] [CrossRef]
- Galtier, M.; De Sordi, L.; Maura, D.; Arachchi, H.; Volant, S.; Dillies, M.A.; Debarbieux, L. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environ. Microbiol. 2016, 18, 2237–2245. [Google Scholar] [CrossRef]
- Reyes, A.; Wu, M.; McNulty, N.P.; Rohwer, F.L.; Gordon, J.I. Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. USA 2013, 110, 20236–20241. [Google Scholar] [CrossRef]
- Weiss, M.; Denou, E.; Bruttin, A.; Serra-Moreno, R.; Dillmann, M.L.; Brüssow, H. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology 2009, 393, 16–23. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef]
- Langlet, J.; Gaboriaud, F.; Gantzer, C. Effects of pH on plaque forming unit counts and aggregation of MS2 bacteriophage. J. Appl. Microbiol. 2007, 103, 1632–1638. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Evans, D.F.; Pye, G.; Bramley, R.; Clark, A.G.; Dyson, T.J.; Hardcastle, J.D. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 1988, 29, 1035–1041. [Google Scholar] [CrossRef]
- Dressman, J.B.; Berardi, R.R.; Dermentzoglou, L.C.; Russell, T.L.; Schmaltz, S.P.; Barnett, J.L.; Jarvenpaa, K.M. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm. Res. 1990, 7, 756–761. [Google Scholar] [CrossRef]
- Dini, C.; Islan, G.A.; De Urraza, P.J.; Castro, G.R. Novel Biopolymer Matrices for Microencapsulation of Phages: Enhanced Protection Against Acidity and Protease Activity. Macromol. Biosci. 2012, 12, 1200–1208. [Google Scholar] [CrossRef]
- Leung, V.; Szewczyk, A.; Chau, J.; Hosseinidoust, Z.; Groves, L.; Hawsawi, H.; Anany, H.; Griffiths, M.W.; Ali, M.M.; Filipe, C.D.M. Long-Term Preservation of Bacteriophage Antimicrobials Using Sugar Glasses. ACS Biomater. Sci. Eng. 2017, 4, 3802–3808. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Wojciechowska, R.; Górski, A. Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog. 2017, 9, 44. [Google Scholar] [CrossRef]
- Barr, J.J. A bacteriophages journey through the human body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef]
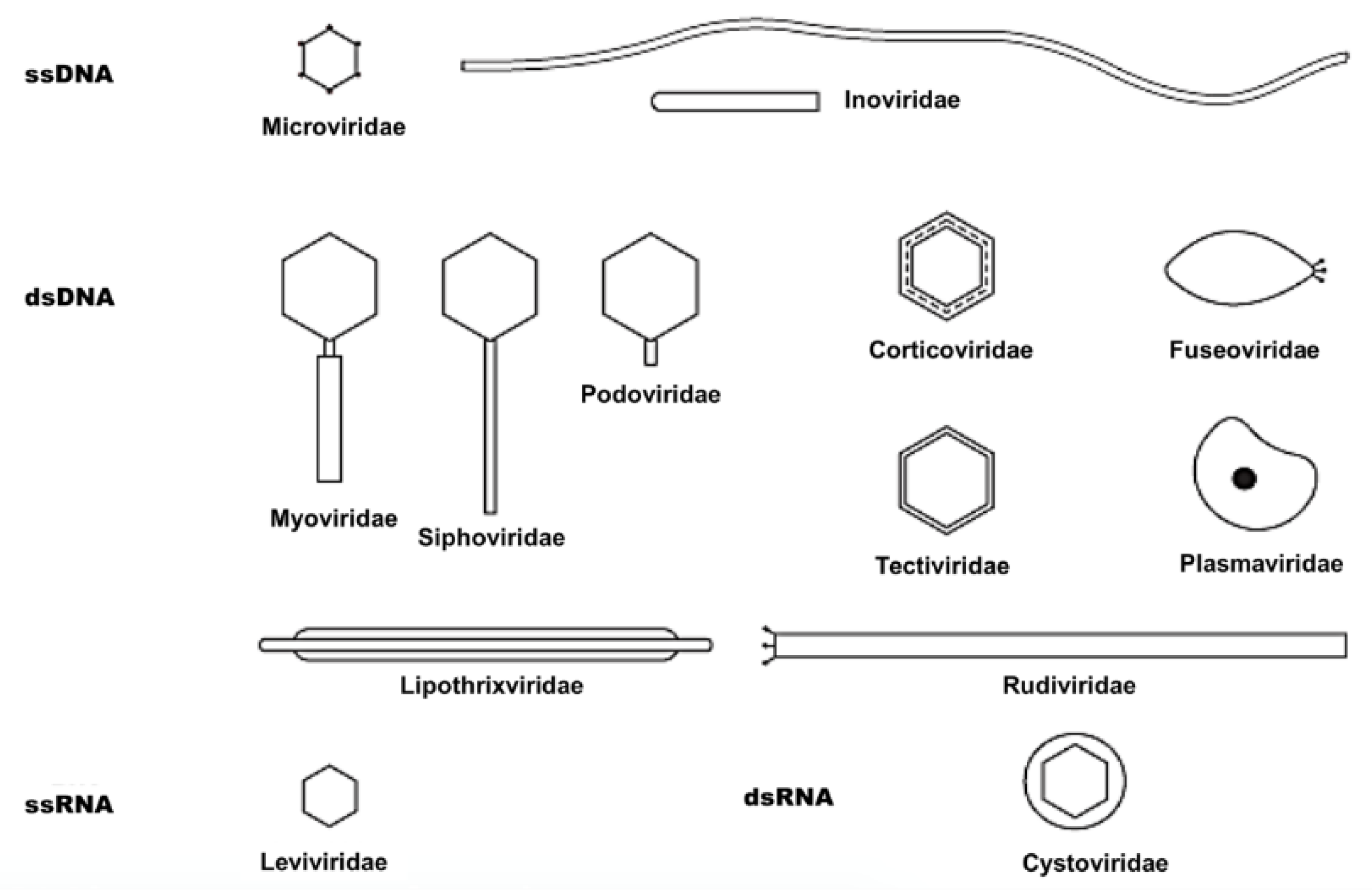
| Symmetry | Nucleic Acid | Order and Families | Genera | Members | Particulars |
|---|---|---|---|---|---|
| Binary (tailed) | DNA, ds, L | Caudovirales | 15 | 4950 | |
| Myoviridae | 6 | 1243 | Tail contractile | ||
| Siphoviridae | 6 | 3011 | Tail long, noncontractile | ||
| Podoviridae | 3 | 696 | Tail short | ||
| Cubic | DNA, ss, C | Microviridae | 4 | 40 | |
| ds, C, T | Corticoviridae | 1 | 3 | Complex capsid, lipids | |
| ds, L | Tectiviridae | 1 | 18 | Internal lipoprotein vesicle | |
| RNA, ss, L | Leviviridae | 2 | 39 | ||
| ds, L, S | Cystoviridae | 1 | 1 | Envelope, lipids | |
| Helical | DNA, ss, C | Inoviridae | 2 | 57 | Filaments or rods |
| ds, L | Lipothrixviridae | 1 | 6 | Envelope, lipids | |
| ds, L | Rudiviridae | 1 | 2 | Resembles TMV | |
| Pleomorphic | DNA, ds, C, T | Plasmaviridae | 1 | 6 | Envelope, lipids, no capsid |
| ds, C, T | Fuselloviridae | 1 | 8 | Spindle-shaped, no capsid |
| Trial Title | Condition/Infection | Intervention | Status | Country | Ref. |
|---|---|---|---|---|---|
| Standard Treatment Associated with Phage Therapy Versus Placebo for Diabetic Foot Ulcers Infected by S. aureus | Diabetic Foot, Staphylococcal Infections | PhagoPied: Topical anti-Staphylococcus bacteriophage therapy | Not Yet Recruiting | France | [92] |
| Individual Patient Expanded Access for AB-SA01, an Investigational Anti- S. aureus Bacteriophage Therapeutic | MDR Staphylococcus aureus infections | AB-SA01 (3- phage cocktail) | In Progress | USA | [93] |
| Individual Patient Expanded Access for AB-PA01, an Investigational Anti-Pseudomonas aeruginosa Bacteriophage Therapeutic | Pseudomonas aeruginosa infections (incl. MDR stains) | AB-PA01 (4-phage cocktail) | In Progress | USA | [94] |
| Safety and Efficacy of EcoActive on Intestinal Adherent Invasive E. coli in Patients With Inactive Crohn’s Disease | Crohn’s Disease | EcoActive (collection of bacteriophages) | Recruiting | USA | [95] |
| Analysis of changes in inflammatory markers in patients treated with bacterial viruses | Wide-range, non-healing postoperative wounds or bone, upper respiratory tract, genital or urinary tract infections whom extensive antibiotic therapy failed | oral, rectal and/or topical bacteriophage lysates/purified phage formulations/phage cocktails | Completed | Poland | [96] |
| Evaluation of Phage Therapy for the Treatment of Escherichia coli and Pseudomonas aeruginosa Wound infections in Burned Patients | Wound infection | PhagoBurn: E. coli phages cocktail (15-phage cocktail), aeruginosa Phages cocktail (13-phage cocktail) | Completed | Belgium, France, Switzerland | [91] |
| Bacteriophage Effects on Pseudomonas aeruginosa | Cystic Fibrosis (CF) | Mucophages (10-phage cocktail) | Completed | France | [97] |
| Therapeutic bacteriophage preparation in chronic otits due to antibiotic-resistant Pseudomonas aeruginosa | Antibiotic-resistant Pseudomonas aeruginosa in chronic otitis | Biophage-PA | Completed | United Kingdom | [98] |
| Antibacterial Treatment against Diarrhea in Oral Rehydration Solution | ETEC and EPEC Diarrhea | Oral T4 phage cocktail | Completed | Bangladesh | [81] |
| Bacteriophages for treating Urinary Tract Infections in Patients Undergoing Transurethral Resection of the Prostate | Urinary Tract Infections (UTI) | Intravesical instillation- PYO phage | Completed | Georgia | [99] |
| A Prospective, Randomized, Double-Blind Controlled Study of WPP-201 for the Safety and Efficacy of Treatment of Venous Leg Ulcers | Venous Leg Ulcers | WPP-201 (8-phage cocktail) | Completed | USA | [100] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Divya Ganeshan, S.; Hosseinidoust, Z. Phage Therapy with a Focus on the Human Microbiota. Antibiotics 2019, 8, 131. https://doi.org/10.3390/antibiotics8030131
Divya Ganeshan S, Hosseinidoust Z. Phage Therapy with a Focus on the Human Microbiota. Antibiotics. 2019; 8(3):131. https://doi.org/10.3390/antibiotics8030131
Chicago/Turabian StyleDivya Ganeshan, Sharita, and Zeinab Hosseinidoust. 2019. "Phage Therapy with a Focus on the Human Microbiota" Antibiotics 8, no. 3: 131. https://doi.org/10.3390/antibiotics8030131
APA StyleDivya Ganeshan, S., & Hosseinidoust, Z. (2019). Phage Therapy with a Focus on the Human Microbiota. Antibiotics, 8(3), 131. https://doi.org/10.3390/antibiotics8030131