Put a Bow on It: Knotted Antibiotics Take Center Stage
Abstract
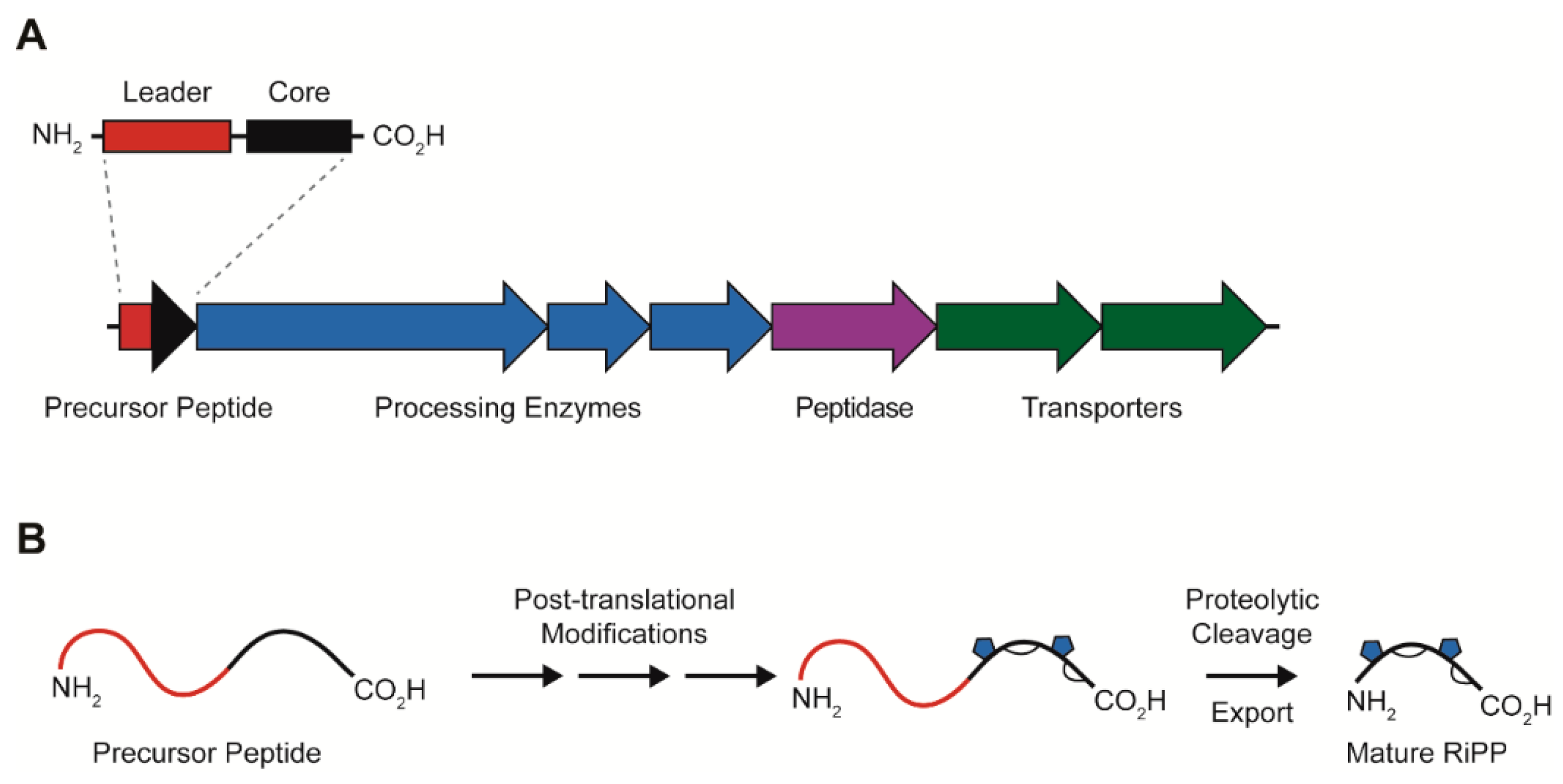
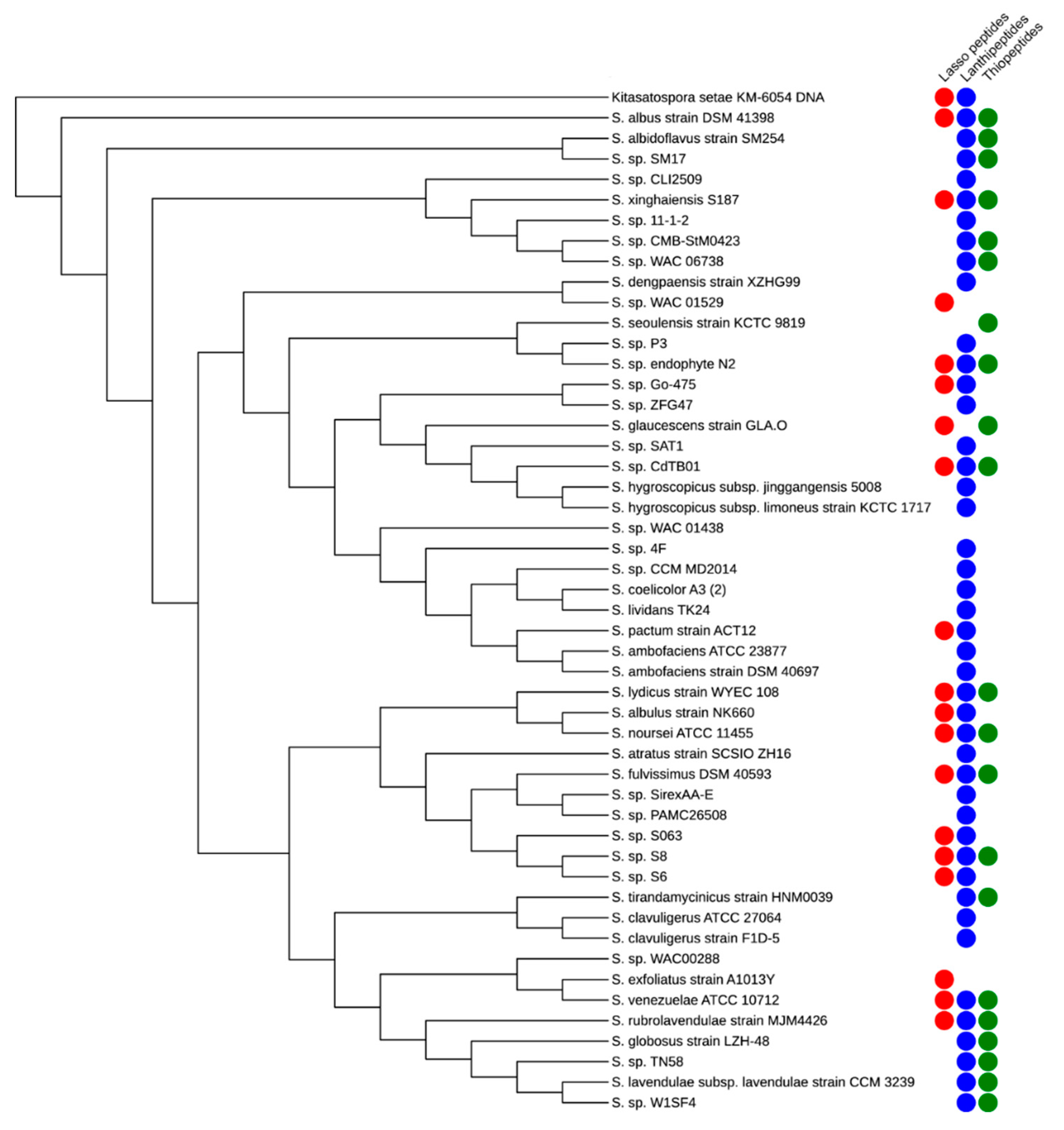
1. Introduction
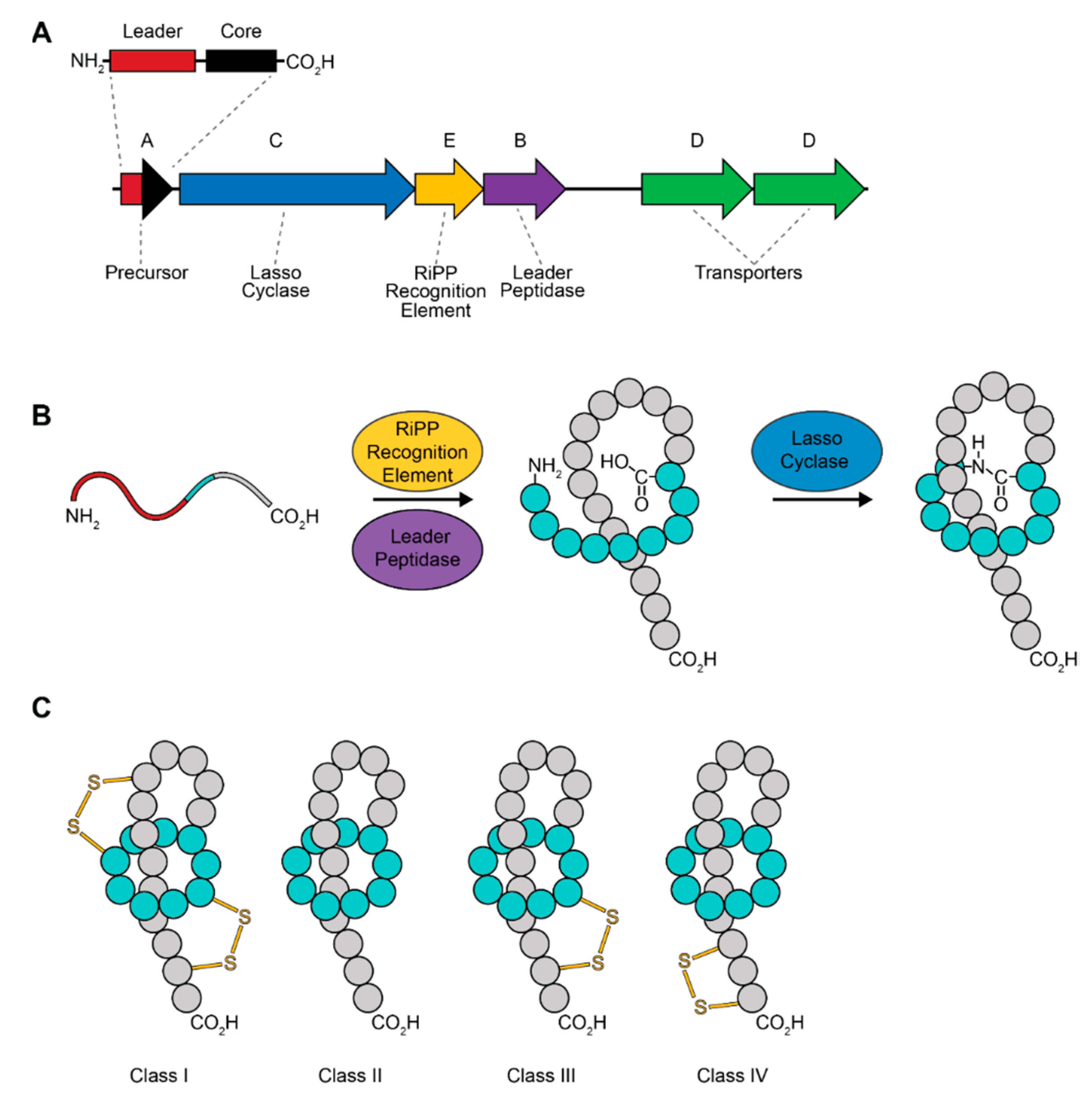
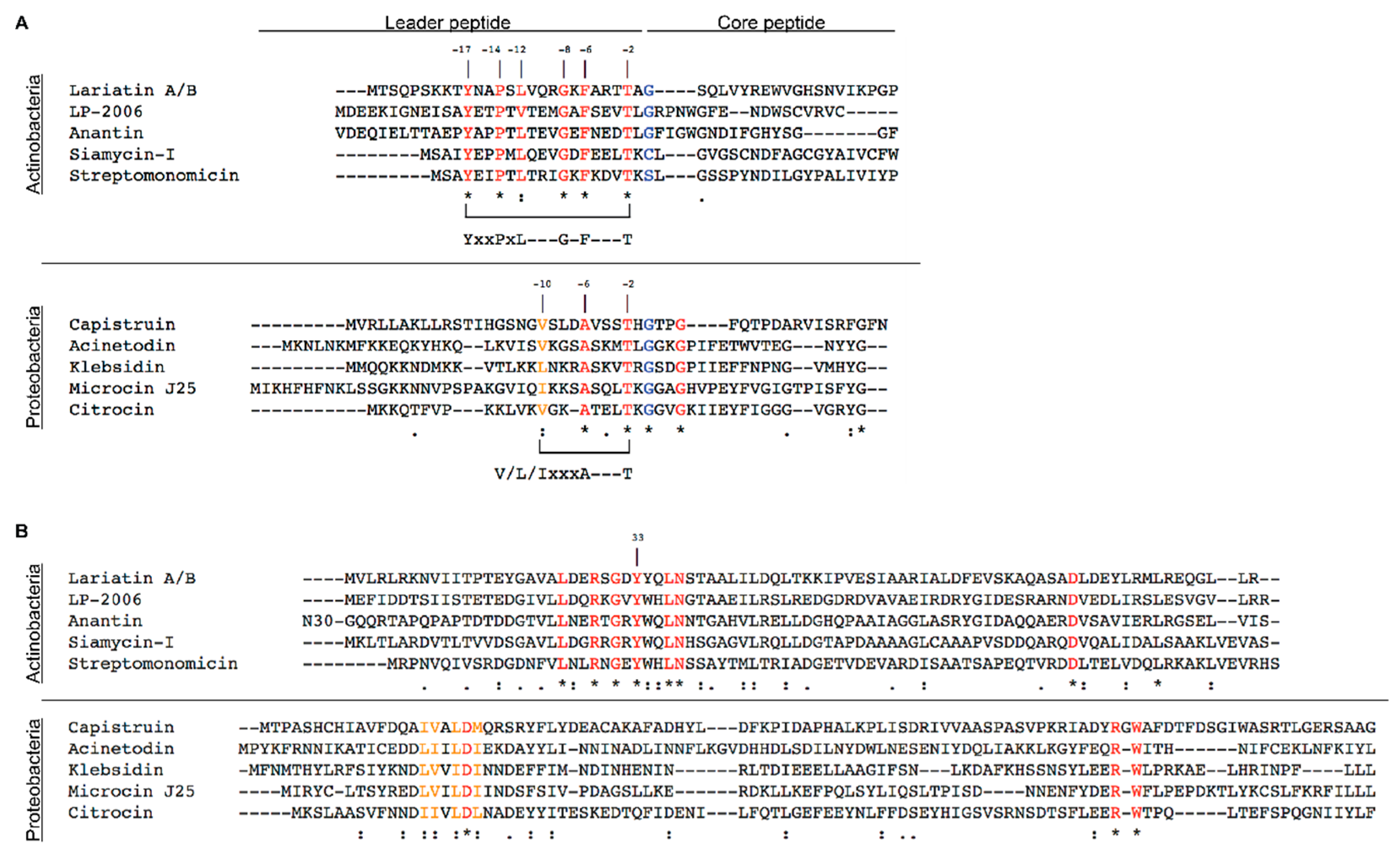
2. Lasso Peptides
Lasso Peptide Biosynthesis
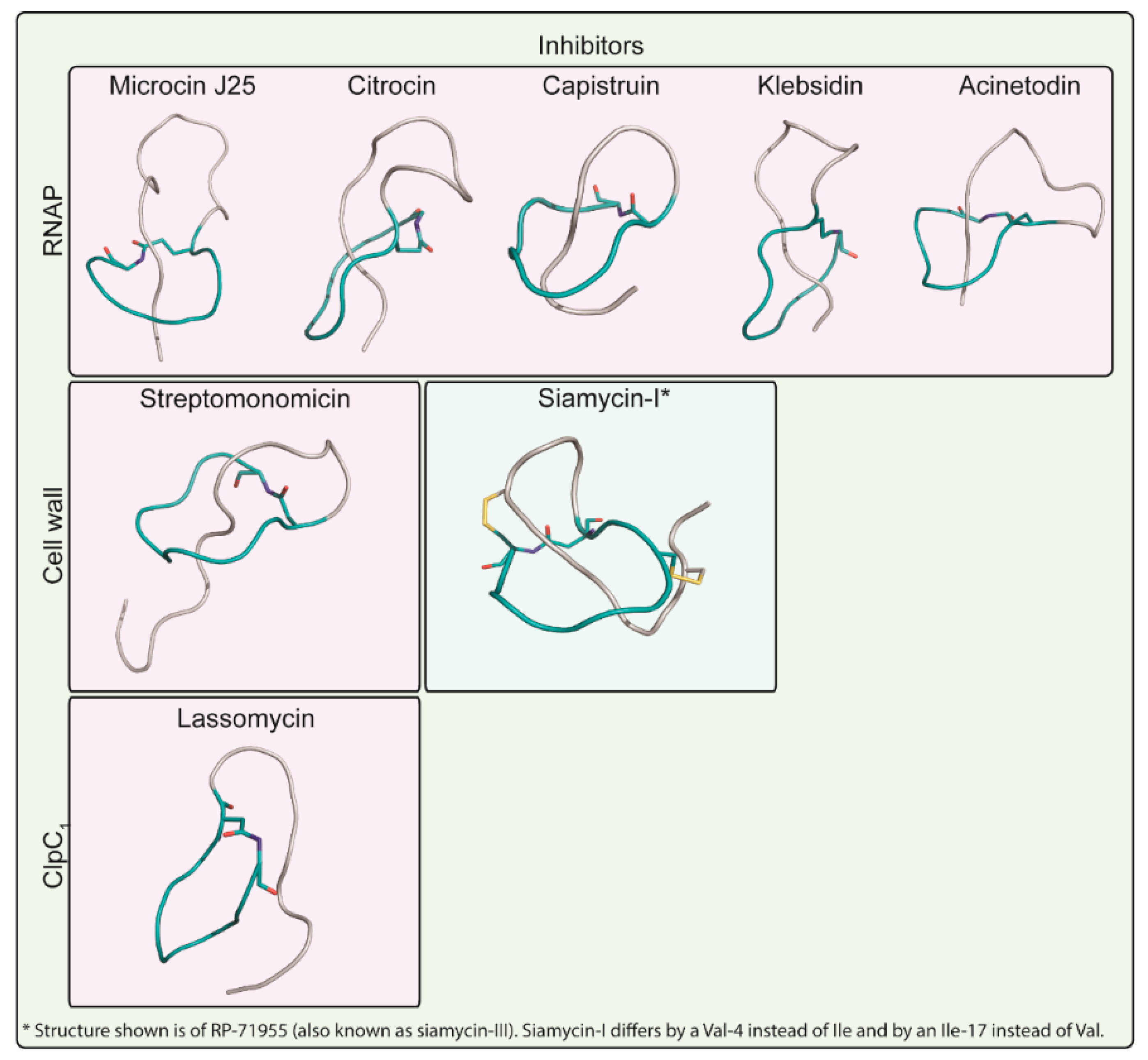
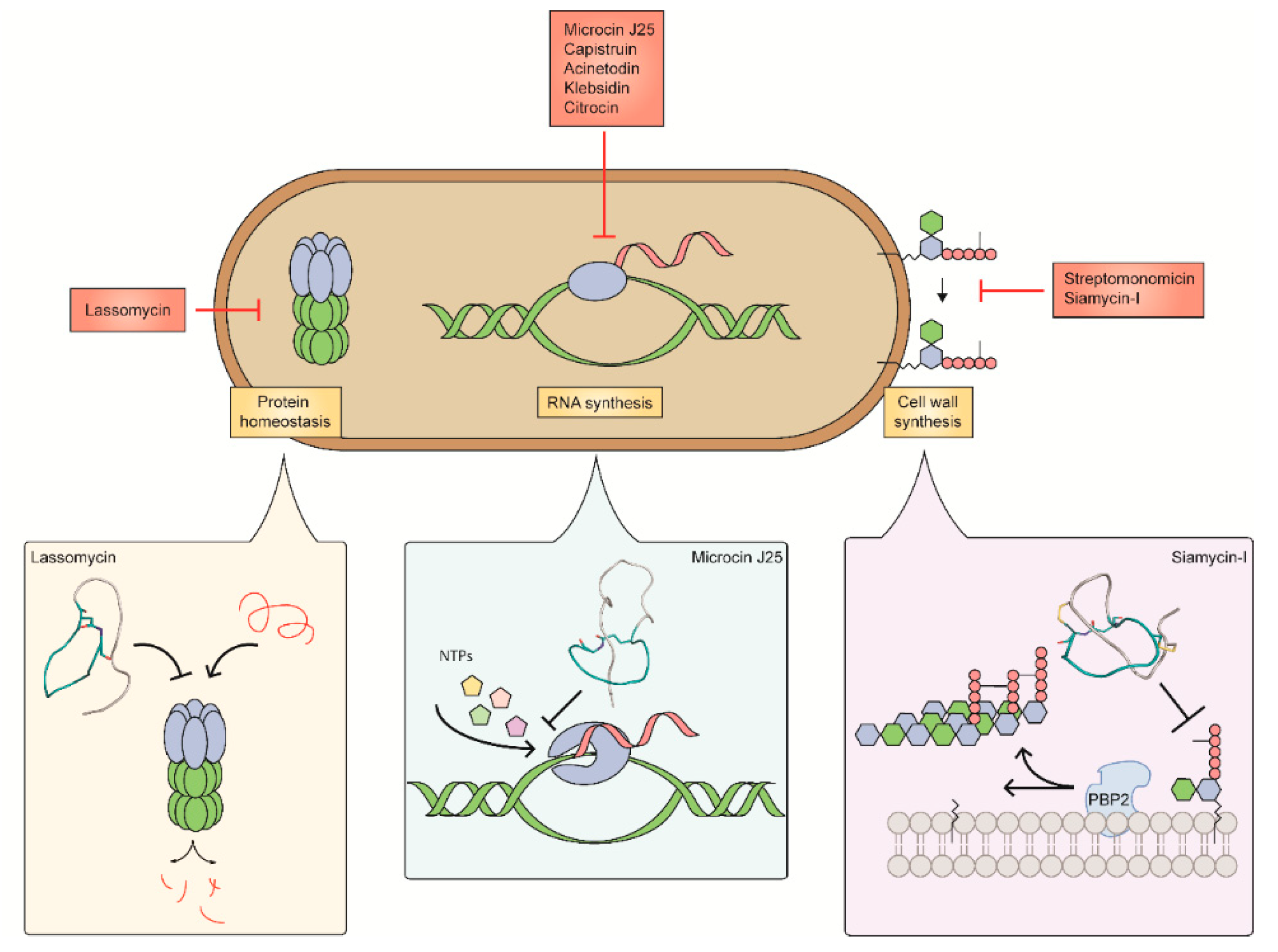
3. Targets of Lasso Peptides
3.1. Inhibitors of Peptidoglycan Biosynthesis
3.1.1. Siamycin-I
3.1.2. Streptomonomicin
3.1.3. Lariatin A/B
3.2. Inhibitors of RNA Synthesis
3.2.1. Microcin J25
3.2.2. Capistruin
3.2.3. Acinetodin and Klebsidin
3.2.4. Citrocin
3.3. Inhibitors of the ClpC1P1P2 Protease
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baltz, R.H. Gifted Microbes for Genome Mining and Natural Product Discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F. Recent Advances in Understanding Streptomyces. F1000Research 2016, 5, 2795. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.; Guimaraes, D.O.; De Frias, U.A.; Pupo, M.T.; Seepe, P.; Feng, Z.; Brady, S.F. Global Biogeographic Sampling of Bacterial Secondary Metabolism. Elife 2015, 4, e05048. [Google Scholar] [CrossRef] [PubMed]
- Rappe, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Mattick, A.T.R.; HIRSCH, A. Further Observations on an Inhibitory Substance (Nisin) from Lactic Streptococci. Lancet 1947, 250, 5–8. [Google Scholar] [CrossRef]
- Highland, J.H.; Howard, G.A.; Ochsner, E.; Stoffler, G.; Hasenbank, R.; Gordon, J. Identification of a Ribosomal Protein Necessary for Thiostrepton Binding to Escherichia Coli Ribosomes. J. Biol. Chem. 1975, 250, 1141–1145. [Google Scholar]
- Olivera, B.M.; Gray, W.R.; Zeikus, R.; McIntosh, J.M.; Varga, J.; Rivier, J.; De Santos, V.; Cruz, L.J. Peptide Neurotoxins from Fish-Hunting Cone Snails. Science 1985, 230, 1338–1343. [Google Scholar] [CrossRef]
- Schmidtko, A.; Lötsch, J.; Freynhagen, R.; Geisslinger, G. Ziconotide for Treatment of Severe Chronic Pain. Lancet 2010, 375, 1569–1577. [Google Scholar] [CrossRef]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, Natural Antimicrobials for Food Preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-Q.; Wilkinson, B. Drug Discovery beyond the ‘Rule-of-Five’. Curr. Opin. Biotechnol. 2007, 18, 478–488. [Google Scholar] [CrossRef]
- Knerr, P.J.; Van Der Donk, W.A. Discovery, Biosynthesis, and Engineering of Lantipeptides. Annu. Rev. Biochem. 2012, 81, 479–505. [Google Scholar] [CrossRef] [PubMed]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; Van Der Donk, W.A. Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef] [PubMed]
- Just-Baringo, X.; Albericio, F.; Álvarez, M. Thiopeptide Antibiotics: Retrospective and Recent Advances. Mar. Drugs 2014, 12, 317–351. [Google Scholar] [CrossRef] [PubMed]
- Weber, W.; Fischli, W.; Hochuli, E.; Kupfer, E.; Weibel, E.K. Anantin - A Peptide Antagonist of the Atrial Natiuretic Factor (ANF). J. Antibiot. (Tokyo) 1991, 44, 164–171. [Google Scholar] [CrossRef]
- Maksimov, M.O.; Pan, S.J.; James Link, A. Lasso Peptides: Structure, Function, Biosynthesis, and Engineering. Nat. Prod. Rep. 2012, 29, 996. [Google Scholar] [CrossRef]
- Salomón, R.A.; Farías, R.N.; Salomn, R.A.; Farias, R.N. Microcin 25, a Novel Antimicrobial Peptide Produced by Escherichia Coli. J. Bacteriol. 1992, 174, 7428–7435. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Zimmermann, M.; Zhu, S.; Steuber, H.; Harms, K.; Xie, X.; Marahiel, M.A. Xanthomonins I–III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring. Angew. Chem. Int. Ed. 2014, 53, 2230–2234. [Google Scholar] [CrossRef]
- Zimmermann, M.; Hegemann, J.D.; Xie, X.; Marahiel, M.A. The Astexin-1 Lasso Peptides: Biosynthesis, Stability, and Structural Studies. Cell Chem. Biol. 2013, 20, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.D.; Zimmermann, M.; Zhu, S.; Klug, D.; Marahiel, M.A. Lasso Peptides from Proteobacteria: Genome Mining Employing Heterologous Expression and Mass Spectrometry. Biopolymers 2013, 100, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.D. Factors Governing the Thermal Stability of Lasso Peptides. Chem. Bio Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A.A. New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Knappe, T.A.; Linne, U.; Xie, X.; Marahiel, M.A. The Glucagon Receptor Antagonist BI-32169 Constitutes a New Class of Lasso Peptides. FEBS. Lett. 2010, 584, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Caulosegnins I−III: A Highly Diverse Group of Lasso Peptides Derived from a Single Biosynthetic Gene Cluster. J. Am. Chem. Soc. 2013, 135, 210–222. [Google Scholar] [CrossRef]
- Li, Y.; Zirah, S.; Rebuffat, S. Lasso Peptides: Bacterial Strategies to Make and Maintain Bioactive Entangled Scaffolds; Springer: New York, NY, USA, 2015. [Google Scholar]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Duquesne, S.; Destoumieux-Garzó, D.; Zirah, S.V.; Goulard, C.; Peduzzi, J.; Rebuffat, S.; Destoumieux-Garzón, D.; Zirah, S.V.; Goulard, C.; Peduzzi, J.; et al. Two Enzymes Catalyze the Maturation of a Lasso Peptide in Escherichia Coli. Chem. Biol. 2007, 14, 793–803. [Google Scholar] [CrossRef]
- Yan, K.-P.; Li, Y.; Zirah, S.; Goulard, C.; Knappe, T.A.; Marahiel, M.A.; Rebuffat, S. Dissecting the Maturation Steps of the Lasso Peptide Microcin J25 in Vitro. Chem. Bio Chem. 2012, 13, 1046–1052. [Google Scholar] [CrossRef]
- Burkhart, B.J.; Hudson, G.A.; Dunbar, K.L.; Mitchell, D.A. A Prevalent Peptide-Binding Domain Guides Ribosomal Natural Product Biosynthesis. Nat. Chem. Biol. 2015, 11, 564–570. [Google Scholar] [CrossRef]
- DiCaprio, A.J.; Firouzbakht, A.; Hudson, G.A.; Mitchell, D.A. Enzymatic Reconstitution and Biosynthetic Investigation of the Lasso Peptide Fusilassin. J. Am. Chem. Soc. 2019, 141, 290–297. [Google Scholar] [CrossRef]
- Gavrish, E.; Sit, C.S.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium Tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Zhu, S.; Fage, C.D.; Hegemann, J.D.; Mielcarek, A.; Yan, D.; Linne, U.; Marahiel, M.A. The B1 Protein Guides the Biosynthesis of a Lasso Peptide. Sci. Rep. 2016, 6, 35604. [Google Scholar] [CrossRef]
- Zong, C.; Cheung-Lee, W.L.; Elashal, H.E.; Raj, M.; Link, A.J. Albusnodin: An Acetylated Lasso Peptide from: Streptomyces Albus. Chem. Commun. 2018, 54, 1339–1342. [Google Scholar] [CrossRef]
- Inokoshi, J.; Matsuhama, M.; Miyake, M.; Ikeda, H.; Tomoda, H. Molecular Cloning of the Gene Cluster for Lariatin Biosynthesis of Rhodococcus Jostii K01-B0171. Appl. Microbiol. Biotechnol. 2012, 95, 451–460. [Google Scholar] [CrossRef]
- Daniel-Ivad, M.; Hameed, N.; Tan, S.; Dhanjal, R.; Socko, D.; Pak, P.; Gverzdys, T.; Elliot, M.A.; Nodwell, J.R. An Engineered Allele of AfsQ1 Facilitates the Discovery and Investigation of Cryptic Natural Products. ACS Chem. Biol. 2017, 12, 628–634. [Google Scholar] [CrossRef]
- Metelev, M.; Tietz, J.I.; Melby, J.O.; Blair, P.M.; Zhu, L.; Livnat, I.; Severinov, K.; Mitchell, D.A. Structure, Bioactivity, and Resistance Mechanism of Streptomonomicin, an Unusual Lasso Peptide from an Understudied Halophilic Actinomycete. Chem. Biol. 2015, 22, 241–250. [Google Scholar] [CrossRef]
- Knappe, T.A.; Linne, U.; Zirah, S.; Rebuffat, S.; Xie, X.; Marahiel, M.A. Isolation and Structural Characterization of Capistruin, a Lasso Peptide Predicted from the Genome Sequence of Burkholderia Thailandensis E264. J. Am. Chem. Soc. 2008, 130, 11446–11454. [Google Scholar] [CrossRef]
- Metelev, M.; Arseniev, A.; Bushin, L.B.; Kuznedelov, K.; Artamonova, T.O.; Kondratenko, R.; Khodorkovskii, M.; Seyedsayamdost, M.R.; Severinov, K. Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter Gyllenbergii and Klebsiella Pneumoniae. ACS Chem. Biol. 2017, 12, 814–824. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Parry, M.E.; Cartagena, A.J.; Darst, S.A.; James Link, A. Discovery and Structure of the Antimicrobial Lasso Peptide Citrocin. J. Biol. Chem. 2019, 294, 6822–6830. [Google Scholar] [CrossRef]
- Sumida, T.; Dubiley, S.; Wilcox, B.; Severinov, K.; Tagami, S. Structural Basis of Leader Peptide Recognition in Lasso Peptide Biosynthesis Pathway. ACS Chem. Biol. 2019, 14, 1619–1627. [Google Scholar] [CrossRef]
- Li, Y.; Ducasse, R.; Zirah, S.; Blond, A.; Goulard, C.; Lescop, E.; Giraud, C.; Hartke, A.; Guittet, E.; Pernodet, J.-L.; et al. Characterization of Sviceucin from Streptomyces Provides Insight into Enzyme Exchangeability and Disulfide Bond Formation in Lasso Peptides. ACS Chem. Biol. 2015, 10, 2641–2649. [Google Scholar] [CrossRef]
- Elsayed, S.S.; Trusch, F.; Deng, H.; Raab, A.; Prokes, I.; Busarakam, K.; Asenjo, J.A.; Andrews, B.A.; van West, P.; Bull, A.T.; et al. Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces Leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert. J. Org. Chem. 2015, 80, 10252–10260. [Google Scholar] [CrossRef]
- Koos, J.D.; Link, A.J. Heterologous and in Vitro Reconstitution of Fuscanodin, a Lasso Peptide from Thermobifida Fusca. J. Am. Chem. Soc. 2019, 141, 928–935. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A Bioinformatics Resource for Deciphering Chemical Structures of RiPPs Based on Prediction of Cleavage and Cross-Links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef]
- Santos-Aberturas, J.; Chandra, G.; Frattaruolo, L.; Lacret, R.; Pham, T.H.; Vior, N.M.; Eyles, T.H.; Truman, A.W. Uncovering the Unexplored Diversity of Thioamidated Ribosomal Peptides in Actinobacteria Using the RiPPER Genome Mining Tool. Nucleic Acids Res. 2019, 47, 4624–4637. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Edgar, R.E.; Dejong, C.A.; Merwin, N.J.; Rees, P.N.; Magarvey, N.A. Genomic Charting of Ribosomally Synthesized Natural Product Chemical Space Facilitates Targeted Mining. Proc. Natl. Acad. Sci. USA 2016, 113, E6343–E6351. [Google Scholar] [CrossRef]
- Katahira, R.; Shibata, K.; Yamasaki, M.; Matsuda, Y.; Yoshida, M. Solution Structure of Endothelin B Receptor Selective Antagonist RES-701-1 Determined by 1H NMR Spectroscopy. Bioorg. Med. Chem. 1995, 3, 1273–1280. [Google Scholar] [CrossRef]
- Um, S.; Kim, Y.J.; Kwon, H.; Wen, H.; Kim, S.H.; Kwon, H.C.; Park, S.; Shin, J.; Oh, D.C. Sungsanpin, a Lasso Peptide from a Deep-Sea Streptomycete. J. Nat. Prod. 2013, 76, 873–879. [Google Scholar] [CrossRef]
- Pan, S.J.; Rajniak, J.; Maksimov, M.O.; Link, A.J. The Role of a Conserved Threonine Residue in the Leader Peptide of Lasso Peptide Precursors. Chem. Commun. 2012, 48, 1880. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Uchida, R.; Takakusagi, Y.; Matsumoto, A.; Jiang, C.; Takahashi, Y.; Arai, M.; Kobayashi, S.; Matsumoto, M.; Inokoshi, J.; et al. Lariatins, Novel Anti-Mycobacterial Peptides with a Lasso Structure, Produced by Rhodococcus Jostii K01-B0171. J. Antibiot. (Tokyo) 2007, 60, 357–363. [Google Scholar] [CrossRef]
- Shao, M.; Ma, J.; Li, Q.; Ju, J.; Shao, M.; Ma, J.; Li, Q.; Ju, J. Identification of the Anti-Infective Aborycin Biosynthetic Gene Cluster from Deep-Sea-Derived Streptomyces Sp. SCSIO ZS0098 Enables Production in a Heterologous Host. Mar. Drugs 2019, 17, 127. [Google Scholar] [CrossRef]
- Kimura, K.-I.; Kanou, F.; Takahashi, H.; Esumi, Y.; Uramoto, M.; Yoshihama, M. Propeptin, a New Inhibitor of Polyl Endopeptidase Produced by Microbispora. J. Antibiot. (Tokyo) 1997, 50, 373–378. [Google Scholar] [CrossRef]
- Potterat, O.; Stephan, H.; Metzger, J.W.; Gnau, V.; Zähner, H.; Jung, G. Aborycin—A Tricyclic 21-Peptide Antibiotic Isolated from Streptomyces Griseoflavus. Liebigs Ann. der Chem. 1994, 1994, 741–743. [Google Scholar] [CrossRef]
- Mukhopadhyay, J.; Sineva, E.; Knight, J.; Levy, R.M.; Ebright, R.H. Antibacterial Peptide Microcin J25 Inhibits Transcription by Binding within and Obstructing the RNA Polymerase Secondary Channel. Mol. Cell 2004, 14, 739–751. [Google Scholar] [CrossRef]
- Tan, S.; Ludwig, K.C.; Mueller, A.; Schneider, T.; Nodwell, J.R. The Lasso Peptide Siamycin-I Targets Lipid II at the Gram-Positive Cell Surface. ACS Chem. Biol. 2019, 14, 966–974. [Google Scholar] [CrossRef]
- Müller, A.; Klöckner, A.; Schneider, T. Targeting a Cell Wall Biosynthesis Hot Spot. Nat. Prod. Rep. 2017, 34, 909–932. [Google Scholar] [CrossRef]
- Constantine, K.L.; Friedrichs, M.S.; Detlefsen, D.; Nishio, M.; Tsunakawa, M.; Furumai, T.; Ohkuma, H.; Oki, T.; Hill, S.; Bruccoleri, R.E.; et al. High-Resolution Solution Structure of Siamycin II: Novel Amphipathic Character of a 21-Residue Peptide That Inhibits HIV Fusion. J. Biomol. NMR 1995, 5, 271–286. [Google Scholar] [CrossRef]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New Insights into the WalK/WalR (YycG/YycF) Essential Signal Transduction Pathway Reveal a Major Role in Controlling Cell Wall Metabolism and Biofilm Formation in Staphylococcus Aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef]
- Dubrac, S.; Bisicchia, P.; Devine, K.M.; Msadek, T.A. Matter of Life and Death: Cell Wall Homeostasis and the WalKR (YycGF) Essential Signal Transduction Pathway. Mol. Microbiol. 2008, 70, 1307–1322. [Google Scholar] [CrossRef]
- Howden, B.P.; McEvoy, C.R.E.; Allen, D.L.; Chua, K.; Gao, W.; Harrison, P.F.; Bell, J.; Coombs, G.; Bennett-Wood, V.; Porter, J.L.; et al. Evolution of Multidrug Resistance during Staphylococcus Aureus Infection Involves Mutation of the Essential Two Component Regulator WalKR. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Tiyanont, K.; Doan, T.; Lazarus, M.B.; Fang, X.; Rudner, D.Z.; Walker, S. Imaging Peptidoglycan Biosynthesis in Bacillus Subtilis with Fluorescent Antibiotics. Proc. Natl. Acad. Sci. USA 2006, 103, 11033–11038. [Google Scholar] [CrossRef]
- Lin, P.F.; Samanta, H.; Bechtold, C.M.; Deminie, C.A.; Patick, A.K.; Alam, M.; Riccardi, K.; Rose, R.E.; White, R.J.; Colonno, R.J. Characterization of Siamycin I, a Human Immunodeficiency Virus Fusion Inhibitor. Antimicrob. Agents Chemother. 1996, 40, 133–138. [Google Scholar] [CrossRef]
- Nakayama, J.; Tanaka, E.; Kariyama, R.; Nagata, K.; Nishiguchi, K.; Mitsuhata, R.; Uemura, Y.; Tanokura, M.; Kumon, H.; Sonomoto, K. Siamycin Attenuates Fsr Quorum Sensing Mediated by a Gelatinase Biosynthesis-Activating Pheromone in Enterococcus Faecalis. J. Bacteriol. 2007, 189, 1358–1365. [Google Scholar] [CrossRef]
- Ma, P.; Nishiguchi, K.; Yuille, H.M.; Davis, L.M.; Nakayama, J.; Phillips-Jones, M.K. Anti-HIV Siamycin I Directly Inhibits Autophosphorylation Activity of the Bacterial FsrC Quorum Sensor and Other ATP-Dependent Enzyme Activities. FEBS Lett. 2011, 585, 2660–2664. [Google Scholar] [CrossRef]
- Inokoshi, J.; Koyama, N.; Miyake, M.; Shimizu, Y.; Tomoda, H. Structure-Activity Analysis of Gram-Positive Bacterium-Producing Lasso Peptides with Anti-Mycobacterial Activity. Sci. Rep. 2016, 6, 30375. [Google Scholar] [CrossRef]
- Salomon, R.A.; Farias, R.N. The FhuA Protein Is Involved in Microcin 25 Uptake. J. Bacteriol. 1993, 175, 7741–7742. [Google Scholar] [CrossRef][Green Version]
- Salomon, R.A.; Farias, R.N. The Peptide Antibiotic Microcin 25 Is Imported through the TonB Pathway and the SbmA Protein. J. Bacteriol. 1995, 177, 3323–3325. [Google Scholar] [CrossRef]
- Delgado, M.A.A.; Rintoul, M.R.R.; Farias, R.N.; Salomon, R.A.; Farías, R.N.; Salomón, R.A. Escherichia Coli RNA Polymerase Is the Target of the Cyclopeptide Antibiotic Microcin J25. J. Bacteriol. 2001, 183, 4543–4550. [Google Scholar] [CrossRef]
- Pavlova, O.; Mukhopadhyay, J.; Sineva, E.; Ebright, R.H.; Severinov, K. Systematic Structure-Activity Analysis of Microcin J25. J. Biol. Chem. 2008, 283, 25589–25595. [Google Scholar] [CrossRef]
- Braffman, N.R.; Piscotta, F.J.; Hauver, J.; Campbell, E.A.; Link, A.J.; Darst, S.A. Structural Mechanism of Transcription Inhibition by Lasso Peptides Microcin J25 and Capistruin. Proc. Natl. Acad. Sci. USA 2019, 116, 1273–1278. [Google Scholar] [CrossRef]
- Malinen, A.M.; Turtola, M.; Parthiban, M.; Vainonen, L.; Johnson, M.S.; Belogurov, G.A. Active Site Opening and Closure Control Translocation of Multisubunit RNA Polymerase. Nucleic Acids Res. 2012, 40, 7442–7451. [Google Scholar] [CrossRef][Green Version]
- Kuznedelov, K.; Semenova, E.; Knappe, T.A.; Mukhamedyarov, D.; Srivastava, A.; Chatterjee, S.; Ebright, R.H.; Marahiel, M.A.; Severinov, K. The Antibacterial Threaded-Lasso Peptide Capistruin Inhibits Bacterial RNA Polymerase. J. Mol. Biol. 2011, 412, 842–848. [Google Scholar] [CrossRef]
| Lasso Peptide | Producer Strain | Antibiotic Activity | No Antibiotic Activity | ||
|---|---|---|---|---|---|
| CLASS I | |||||
| Siamycin-I ¥ | Streptomyces sp. | B. subtilis+ S. epidermis+ S. saprophyticus+ | S. aureus+ E. faecalis+ | A. baumannii− B. cepacian− P. aeruginosa− | E. coli− K. pneumoniae− |
| Aborycin ‡ | Streptomyces griseoflavus TU4072 Streptomyces sp. 9440 | B. brevis+ E. fallinarum+ B. subtilis+ S. viridochromeogenes+ | E. faecalis+ S. aureus+ B. thuringiensis+ P. saccharophilia− | A. baumannii− M. luteus+ C. perfringens+ S. typhirium− | E. coli− V. alginolyticus− K. pneumoniae− |
| CLASS II | |||||
| Acinetodin ¥ | Acinetobacter gyllenbergii (clinical gut isolate) | E. coli * − | A. baumannii− B. subtilis+ S. aureus+ | K. pneumoniae− P. aeruginosa− | |
| Anantin B2 ‡ | Streptomyces coerulescens | B. subtilis+ E. coli− | A. baumannii− B. anthracis+ M. smegatis+ N. meningitidis− L. monocyte-genes+ | E. coli− P. aeruginosa− K. pneumoniae¯ S. aureus+ E. faecium+ | |
| Astexin-1 ¥ | Asticcacaulis excentricus CB48 | C. crescentus− | B. thailandensis− V. harveyi− E. coli− | S. newport− V. fischeri− | |
| Capistruin ¥ | Burkholderia thailandensis E264 | Burkholderia sp. − E. coli − P. aeruginosa − | A. viridans+ Pseudomonas sp. − B. megaterium + | K. pneumoniae− S. enterica¯ S. aureus+ | |
| Citrocin ¥ | Citrobacter braakii ATCC 51113 | Citrobacter sp. − E. coli ¯ | P. aeruginosa− S. marcescens− | ||
| Klebsidin ¥ | Klebsiella pneumoniae (clinical gut isolate) | E. coli * − K. pneumoniae − | A. baumannii− P. aeruginosa− | B. subtilis+ S. aureus+ | |
| Lariatin A/B ¥ | Rhodoccocus jostii K01-B0171 | M. smegatis+ M. tuberculosis‡ + | P. aeruginosa− B. subtilis+ S. aureus+ | E. coli− X. campestris− M. luteus+ | |
| Lasssomycin ‡ | Lentzea kentuckyensis | M. tuberculosis+ Mycobacterium spp. + | B. anthracis+ K. pneumoniae− C. difficile+ Lactobacillus sp. + | E. faecalis+ S. mutans+ E. coli− S. aureus+ | |
| Microcin J25 ¥ | Escherichia coli | E. coli− S. flexneri− | S. newport− | P. mendocina L. acidophilus+ B. subtilis+ S. enterica− | K. pneumoniae− S. typhirium− |
| Propeptin 1/2 ¥ | Microbispora sp. SNA-115 | M. phlei+ X. orzyae− | P. aeruginosa− | N/A | |
| Streptomono -micin ‡ | Streptomonospora alba YIM | B. subtilis+ L. monocyte− genes+ | B. anthracis+ B. cereus+ S. aureus+ | E. coli− | P. aeruginosa− |
| CLASS IV | |||||
| LP2006 ‡ | Nocardiopsis alba | B. anthracis+ B. subtilis+ | E. faecalis+ M. smegatis+ | A. baumannii N. meningitidis− E. coli− P. aeruginosa− | K. pneumoniae− S. aureus+ L. monocyte− genes+ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, S.; Moore, G.; Nodwell, J. Put a Bow on It: Knotted Antibiotics Take Center Stage. Antibiotics 2019, 8, 117. https://doi.org/10.3390/antibiotics8030117
Tan S, Moore G, Nodwell J. Put a Bow on It: Knotted Antibiotics Take Center Stage. Antibiotics. 2019; 8(3):117. https://doi.org/10.3390/antibiotics8030117
Chicago/Turabian StyleTan, Stephanie, Gaelen Moore, and Justin Nodwell. 2019. "Put a Bow on It: Knotted Antibiotics Take Center Stage" Antibiotics 8, no. 3: 117. https://doi.org/10.3390/antibiotics8030117
APA StyleTan, S., Moore, G., & Nodwell, J. (2019). Put a Bow on It: Knotted Antibiotics Take Center Stage. Antibiotics, 8(3), 117. https://doi.org/10.3390/antibiotics8030117





