Molecular Characterization and Moxifloxacin Susceptibility of Clostridium difficile
Abstract
1. Introduction
2. Results
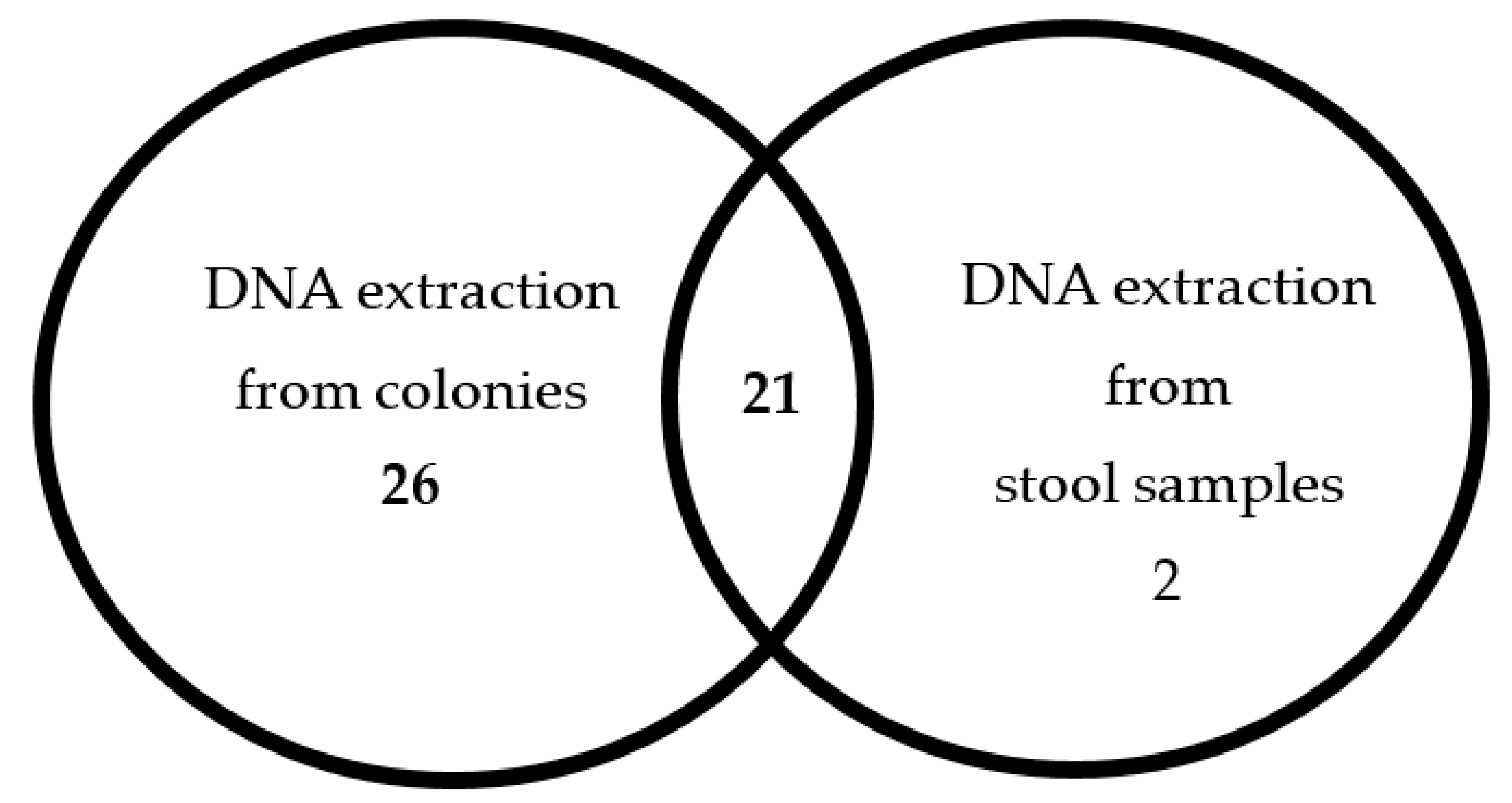
2.1. A Comparison Between the Two Methods for DNA Extraction
2.2. Genotypes Detected by Genotype CDiff
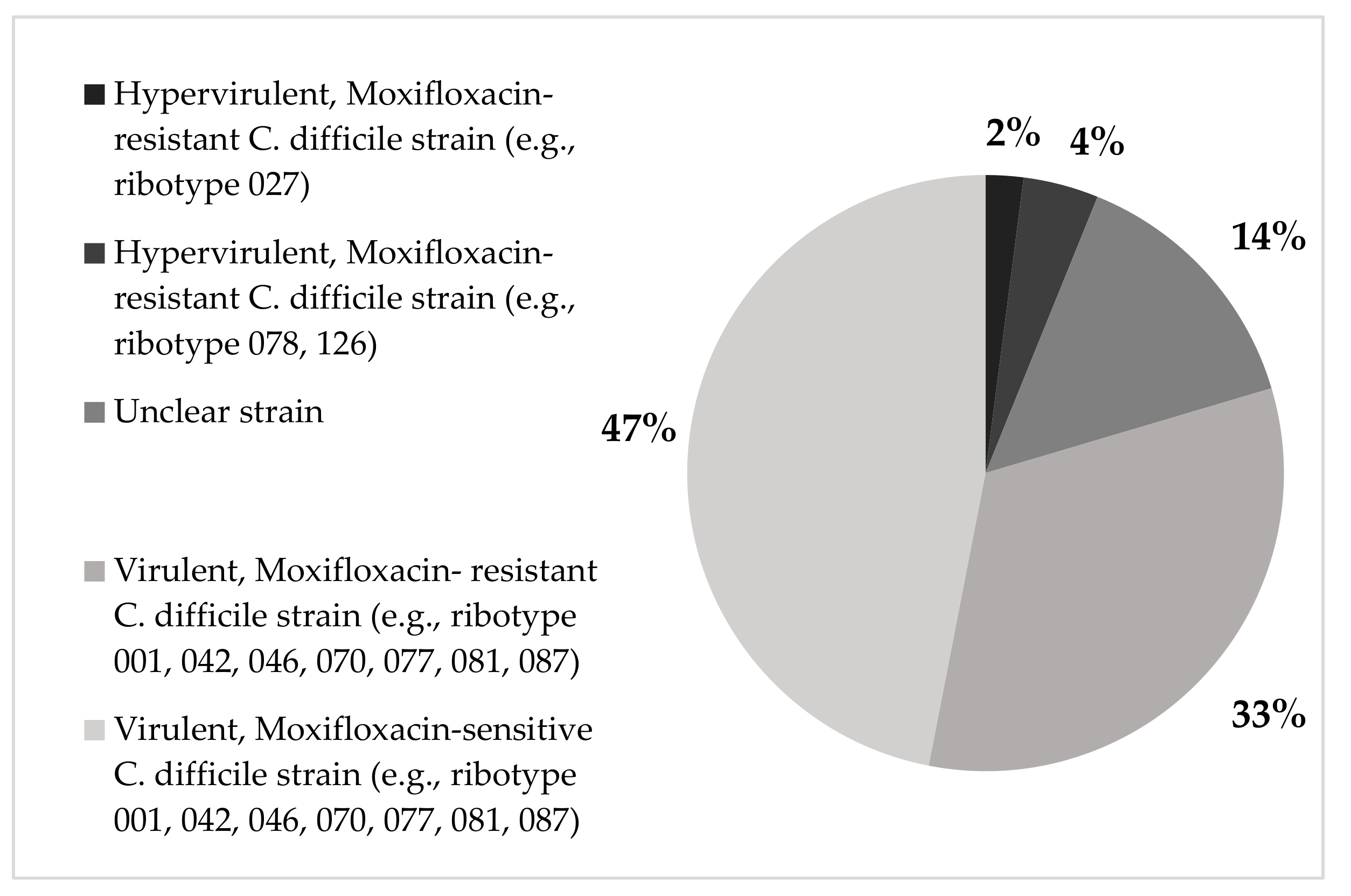
2.3. The Correlation of C. difficile Strains with Infection Acquisition and Disease Severity
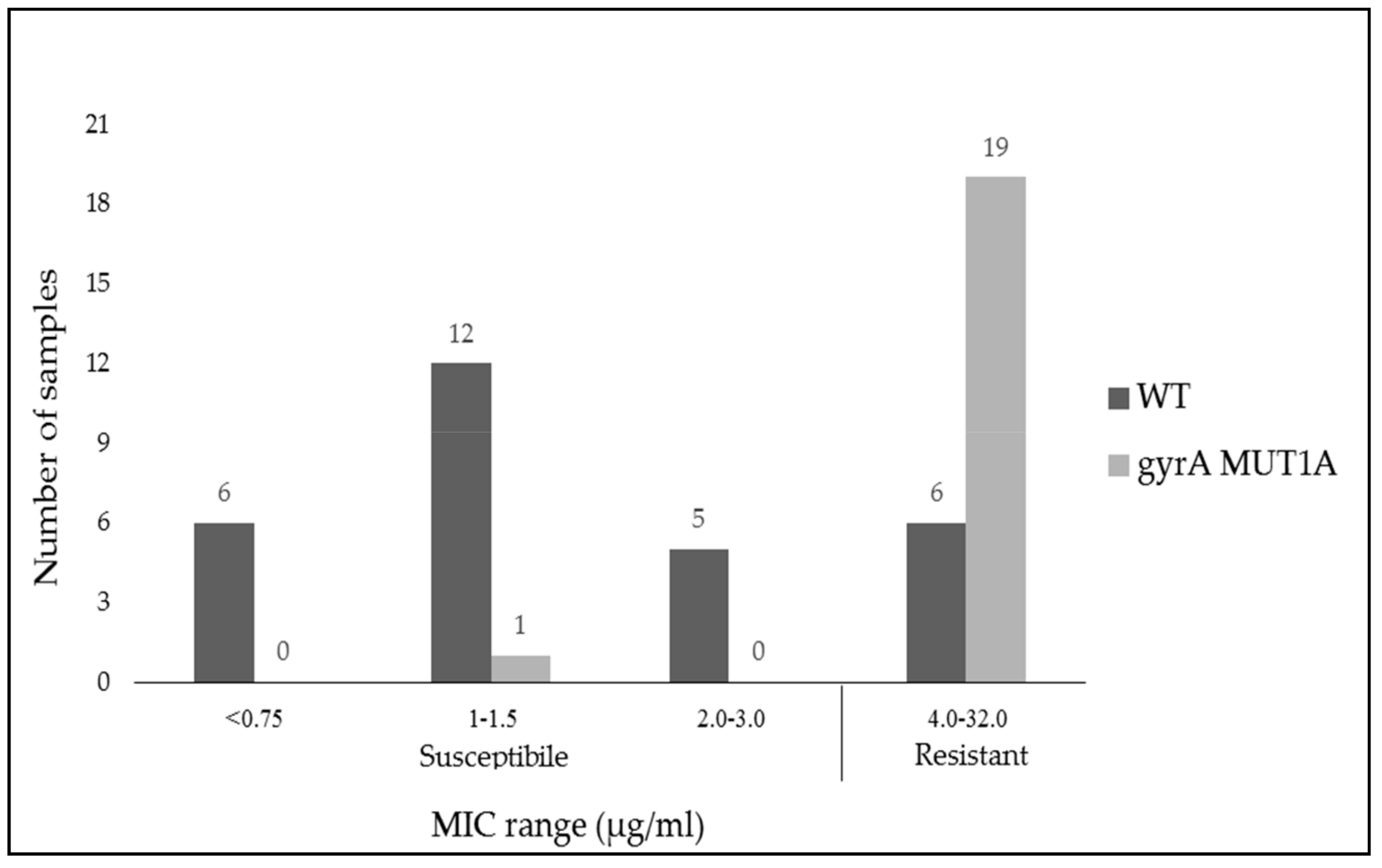
2.4. Comparison of Antibiotic Susceptibility to Moxifloxacin by MIC Breakpoints (Phenotype) and Gene Expression (Genotype)
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Disease Severity Score and Epidemiologic and Clinical Data Collection
4.3. C. difficile Susceptibility to Moxifloxacin
4.4. DNA Extraction
4.5. Molecular Identification Using GenoType CDiff kit
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartlett, J.G.; Moon, N.; Chang, T.W.; Taylor, N.; Onderdonk, A.B. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 1978, 75, 778–782. [Google Scholar] [CrossRef]
- Jones, A.M.; Kuijper, E.J.; Wilcox, M.H. Clostridium difficile: A European perspective. J. Infect. 2013, 66, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P.; LaMont, J.T. Clostridium difficile–More difficult than ever. N. Engl. J. Med. 2008, 35, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Balsells, E.; Shi, T.; Leese, C.; Lyell, I.; Burrows, J.; Wiuff, C.; Campbell, H.; Kyaw, M.H.; Nair, H. Global burden of Clostridium difficile infections: A systematic review and meta-analysis. J. Glob. Health. 2019, 9, 010407. [Google Scholar] [CrossRef] [PubMed]
- Lyerly, D.M.; Saum, K.E.; MacDonald, D.K.; Wilkins, T.D. Effects of Clostridium difficile toxins given intragastrically to animals. Infect. Immun. 1985, 47, 349–352. [Google Scholar] [PubMed]
- Voth, D.E.; Ballard, J.D. Clostridium difficile Toxins: Mechanism of Action and Role in Disease. Clin. Microbiol. Rev. 2005, 18, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Curry, S.R.; Marsh, J.W.; Muto, C.A.; O’Leary, M.M.; Pasculle, A.W.; Harrison, L.H. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 2007, 45, 215–221. [Google Scholar] [CrossRef]
- Gerding, D.N.; Johnson, S.; Rupnik, M.; Aktories, K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014, 5, 15–27. [Google Scholar] [CrossRef]
- Warny, M.; Pepin, J.; Fang, A.; Killgore, G.; Thompson, A.; Brazier, J.; Frost, E.; McDonald, L.C. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005, 366, 1079–1084. [Google Scholar] [CrossRef]
- O’Connor, J.R.; Johnson, S.; Gerding, D.N. Clostridium difficile Infection Caused by the Epidemic BI/NAP1/027 Strain. Gastroenterology 2009, 136, 1913–1924. [Google Scholar] [CrossRef]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence of Clostridium difficile Infection Due to a New Hypervirulent Strain, Polymerase Chain Reaction Ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Adler, A.; Miller-Roll, T.; Bradenstein, R.; Block, C.; Mendelson, B.; Parizade, M.; Paitan, Y.; Schwartz, D.; Peled, N.; Carmeli, Y.; et al. A national survey of the molecular epidemiology of Clostridium difficile in Israel: The dissemination of the ribotype 027 strain with reduced susceptibility to vancomycin and metronidazole. Diagn. Microbiol. Infect. Dis. 2015, 83, 21–24. [Google Scholar] [CrossRef]
- Johanesen, P.A.; Mackin, K.E.; Hutton, M.L.; Awad, M.M.; Larcombe, S.; Amy, J.M.; Lyras, D. Disruption of the Gut Microbiome: Clostridium difficile infection and the threat of antibiotic resistance. Genes 2015, 6, 1347–1360. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 2011, 8.1, 17–26. [Google Scholar] [CrossRef]
- Goudarzi, M.; Seyedjavadi, S.S.; Goudarzi, H.; Mehdizadeh, A.E.; Nazeri, S. Clostridium difficile infection: epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica 2014, 2014, 916826. [Google Scholar]
- Lessa, F.C.; Gould, C.V.; McDonald, L.C. Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 2012, 55 (Suppl. 2), S65–S70. [Google Scholar] [CrossRef]
- McFarland, L.V. Antibiotic-associated diarrhea: Epidemiology, trends and treatment. Future. Microbiol. 2008, 3, 563–578. [Google Scholar] [CrossRef]
- Shen, A. Clostridium difficile toxins: mediators of inflammation. J. Innate. Immun. 2012, 4, 149–158. [Google Scholar] [CrossRef]
- Davies, A.H.; Roberts, A.K.; Shone, C.C.; Acharya, K.R. Super toxins from a super bug: Structure and function of Clostridium difficile toxins. Biochem. J. 2011, 436, 517–526. [Google Scholar] [CrossRef]
- Baines, S.D.; Wilcox, M.H. Antimicrobial Resistance and Reduced Susceptibility in Clostridium difficile: Potential Consequences for Induction, Treatment, and Recurrence of C. difficile Infection. Antibiotics 2015, 4, 267–298. [Google Scholar] [CrossRef]
- Youngster, I.; Sauk, J.; Pindar, C.; Wilson, R.G.; Kaplan, J.L.; Smith, M.B.; Alm, E.J.; Gevers, D.; Russell, G.H.; Hohmann, E.L. Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: A randomized, open-label, controlled pilot study. Clin. Infect. Dis. 2014, 58, 1515–1522. [Google Scholar] [CrossRef]
- Spigaglia, P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther. Adv. Infect. Dis. 2016, 3, 23–42. [Google Scholar] [CrossRef]
- Snydman, D.R.; McDermott, L.A.; Jacobus, N.V.; Thorpe, C.; Stone, S.; Jenkins, S.G.; Goldstein, E.J.C.; Patel, R.; Forbes, B.A.; Mirrett, S.; et al. US-based national sentinel surveillance study for the epidemiology of Clostridium difficile-associated diarrheal isolates and their susceptibility to fidaxomicin. Antimicrob. Agents. Chemother. 2015, 59, 6437–6443. [Google Scholar] [CrossRef]
- Tkhawkho, L.; Nitzan, O.; Pastukh, N.; Brodsky, D.; Jackson, K.; Peretz, A. Antimicrobial susceptibility of Clostridium difficile isolates in Israel. J. Glob. Antimicrob. Resist. 2017, 10, 161–164. [Google Scholar] [CrossRef]
- Pepin, J.; Saheb, N.; Coulombe, M.A.; Alary, M.E.; Corriveau, M.P.; Authier, S.; Leblanc, M.; Rivard, G.; Bettez, M.; Primeau, V.; et al. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: A cohort study during an epidemic in Quebec. Clin. Infect. Dis. 2005, 41, 1254–1260. [Google Scholar] [CrossRef]
- Eckert, C.; Burghoffer, B.; Lalande, V.; Barbut, F. Evaluation of the chromogenic agar chromID C. difficile. J. Clin. Microbiol. 2013, 51, 1002–1004. [Google Scholar] [CrossRef]
- René, P.; Frenette, C.P.; Schiller, I.; Dendukuri, N.; Brassard, P.; Fenn, S.; Loo, V.G. Comparison of eight commercial enzyme immunoassays for the detection of Clostridium difficile from stool samples and effect of strain type. Diagn. Microbiol. Infect. Dis. 2012, 73, 94–96. [Google Scholar] [CrossRef]
- Florea, D.; Oţelea, D.; Rafila, A.; Bădicuț, I.; Popoiu, M.; Popescuet, G.A. Clinical utility of the GeneXpert assay for the diagnosis of Clostridium difficile infections. BMC. Infect. Dis. 2014, 14 (Suppl. 7), O34. [Google Scholar]
- Goldenberg, S.D.; Cliff, P.R.; French, G.L. Glutamate dehydrogenase for laboratory diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 2010, 48, 3050–3051. [Google Scholar] [CrossRef]
- Moyenuddin, M.; Williamson, J.; Ohl, C.A. Clostridium difficile-associated diarrhea: Current strategies for diagnosis and therapy. Curr. Gastroenterol. Rep. 2002, 4, 279–286. [Google Scholar] [CrossRef]
- Doern, G.V.; Coughlin, R.T.; Wu, L. Laboratory diagnosis of Clostridium difficile associated gastrointestinal disease: comparison of a monoclonal antibody enzyme immunoassay for toxins A and B with a monoclonal antibody enzyme immunoassay for toxin A only and two cytotoxicity assays. J. Clin. Microbiol. 1992, 30, 2042–2046. [Google Scholar]
- Rajabally, N.; Kullin, B.; Ebrahim, K.; Brock, T.; Weintraub, A.; Whitelaw, A.; Bamford, C.; Watermeyer, G.; Thomson, S.; Abratt, V.; et al. Comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African centre. J. Med. Microbiol. 2016, 65, 320–327. [Google Scholar] [CrossRef]
- Jin, D.; Luo, Y.; Huang, C.; Cai, J.; Ye, J.; Zheng, Y.; Wang, L.; Zhao, P.; Liu, A.; Fang, W.; et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J. Clin. Microbiol. 2017, 55, 801–810. [Google Scholar] [CrossRef]
- McDonald, L.C.; Killgore, G.E.; Thompson, A.; Owens, R.C.; Kazakova, S.V.; Sambol, S.P.; Johnson, S.; Gerding, D.N. An Epidemic, Toxin Gene–Variant Strain of Clostridium difficile. N. Engl. J. Med. 2005, 353, 2433–2441. [Google Scholar] [CrossRef]
- MacCannell, D.R.; Louie, T.J.; Gregson, D.B.; Laverdiere, M.; Labbe, A.C.; Laing, F.; Henwick, S. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 2006, 44, 2147–2152. [Google Scholar] [CrossRef]
- Freeman, J.; Bauer, M.P.; Baines, S.D.; Corver, J.; Fawley, W.N.; Goorhuis, B.; Kuijper, E.J.; Wilcox, M.H. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 2010, 23, 529–549. [Google Scholar] [CrossRef]
- Khanna, S.; Pardi, D.S.; Aronson, S.L.; Kammer, P.P.; Orenstein, R.; St Sauver, J.L.; Harmsen, W.S.; Zinsmeister, A.R. The epidemiology of community-acquired Clostridium difficile infection: A population-based study. Am. J. Gastroenterol. 2012, 107, 89–95. [Google Scholar] [CrossRef]
- Miller-Roll, T.; Na’amnih, W.; Cohen, D.; Carmeli, Y.; Adler, A. Molecular types and antimicrobial susceptibility patterns of Clostridium difficile isolates in different epidemiological settings in a tertiary care center in Israel. Diagn. Microbiol. Infect. Dis. 2016, 86, 450–454. [Google Scholar] [CrossRef]
- Taori, S.K.; Wroe, A.; Hardie, A.; Gibb, A.P.; Poxton, I.R. A prospective study of community-associated Clostridium difficile infections: The role of antibiotics and co-infections. J. Infect. 2014, 69, 134–144. [Google Scholar] [CrossRef]
- Cloud, J.; Noddin, L.; Pressman, A.; Hu, M.; Kelly, C. Clostridium difficile Strain NAP-1 Is Not Associated with Severe Disease in a Nonepidemic Setting. Clin. Gastroenterol. Hepatol. 2009, 7, 868–873. [Google Scholar] [CrossRef]
- Sirard, S.; Valiquette, L.; Fortier, L.C. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J. Clin. Microbiol. 2011, 49, 4040–4046. [Google Scholar] [CrossRef]
- Walk, S.T.; Micic, D.; Jain, R.; Lo, E.S.; Trivedi, I.; Liu, E.W.; Almassalha, L.M.; Ewing, S.A.; Ring, C.; Galecki, A.T.; et al. Clostridium difficile ribotype does not predict severe infection. Clin. Infect. Dis. 2012, 55, 1661–1668. [Google Scholar] [CrossRef]
- Spigaglia, P.; Barbanti, F.; Mastrantonio, P.; Brazier, J.S.; Barbut, F.; Delmée, M.; Kuijper, E.; Poxton, I.R.; European Study Group on Clostridium difficile (ESGCD). Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J. Med. Microbiol. 2008, 57, 784–789. [Google Scholar] [CrossRef]
- Barbut, F.; Mastrantonio, P.; Delmée, M.; Brazier, J.; Kuijper, E.; Poxton, I.; European Study Group on Clostridium difficile (ESGCD). Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin. Microbiol. Infect. 2007, 13, 1048–1057. [Google Scholar] [CrossRef]
- Huang, H.; Wu, S.; Wang, M.; Zhang, Y.; Fang, H.; Palmgren, A.C.; Weintraub, A.; Nord, C.E. Clostridium difficile infections in a Shanghai hospital: Antimicrobial resistance, toxin profiles and ribotypes. Int. J. Antimicrob. Agents. 2009, 33, 339–342. [Google Scholar] [CrossRef]
- Linder, J.A.; Huang, E.S.; Steinman, M.A.; Gonzales, R.; Stafford, R.S. Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 2005, 118, 259–268. [Google Scholar] [CrossRef]
- Ackermann, G.; Tang-Feldman, Y.J.; Schaumann, R.; Henderson, J.P.; Rodloff, A.C.; Silva, J.; Cohen, S.H. Antecedent use of fluoroquinolones is associated with resistance to moxifloxacin in Clostridium difficile. Clin. Microbiol. Infect. 2003, 9, 526–530. [Google Scholar] [CrossRef][Green Version]
- Spigaglia, P.; Barbanti, F.; Louie, T.; Barbut, F.; Mastrantonio, P. Molecular analysis of the gyrA and gyrB quinolone resistance-determining regions of fluoroquinolone-resistant Clostridium difficile mutants selected in vitro. Antimicrob. Agents Chemother. 2009, 53, 2463–2468. [Google Scholar] [CrossRef]
- Velazquez-Gomez, I.; Rocha-Rodriguez, R.; Toro, D.; Gutierrez-Nuñez, J.J.; Gonzalez, G.; Saavedra, S. A severity score index for Clostridium difficile infection. Infect. Dis. Clin. Pract. 2008, 16, 376–378. [Google Scholar] [CrossRef]
- Hamo, Z.; Azrad, M.; Nitzan, O.; Sagie, A.; Tkhawkho, L.; Binyamin, D.; Peretz, A. Role of Single Procalcitonin Test on Admission as a Biomarker for Predicting the Severity of Clostridium difficile Infection. Front. Microbiol. 2017, 8, 2532. [Google Scholar] [CrossRef]
| Result | GenoType CDiff (n = 49) | |
|---|---|---|
| Culture n (%) | Stool n (%) | |
| Positive | 47 (96) | 23 (47) |
| Negative Percent of Agreement (compared with GeneXpert Results) | 2 (4) | 26 (53) |
| 95.9 [95% CI *: 86.0, 99.5] | 46.9 [95% CI: 32.5, 61.7] | |
| C. difficile Strain | Infection Acquisition | Disease Severity | |||||
|---|---|---|---|---|---|---|---|
| Community (Total N = 21) N (%) | Hospital (Total N =28) N (%) | p-Value | Mild (Total N= 35) N (%) | Moderate (Total N = 11) N (%) | Severe (Total N = 3) N (%) | p-Value | |
| Virulent, Moxifloxacin-sensitive | 11 (52.4) | 12 (42.8) | 0.65 | 17 (48.6) | 6 (54.5) | 0 (0) | 0.3 |
| Virulent, Moxifloxacin-resistant | 5 (23.8) | 11 (39.3) | 11 (31.4) | 2 (18.2) | 3 (100) | ||
| Hypervirulent, Moxifloxacin-resistant (078, 126) | 1 (4.8) | 1 (3.6) | 2 (5.7) | 0 (0) | 0 (0) | ||
| Hypervirulent ribotype027 | 0 (0) | 1 (3.6) | 1 (2.9) | 0 (0) | 0 (0) | ||
| Unclear strain | 4 (19) | 3 (10.7) | 4 (11.4) | 3 (27.3) | 0 (0) | ||
| Etest Result | |||
|---|---|---|---|
| Sensitive | Resistant | ||
| GenoType CDiff | Resistant (gyrA MUT1A) | 1 (2.1%) | 19 (38.7%) |
| Sensitive (gyrA WT) | 23 (47%) | 6 (12.2%) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mizrahi, S.; Hamo, Z.; Azrad, M.; Peretz, A. Molecular Characterization and Moxifloxacin Susceptibility of Clostridium difficile. Antibiotics 2019, 8, 118. https://doi.org/10.3390/antibiotics8030118
Mizrahi S, Hamo Z, Azrad M, Peretz A. Molecular Characterization and Moxifloxacin Susceptibility of Clostridium difficile. Antibiotics. 2019; 8(3):118. https://doi.org/10.3390/antibiotics8030118
Chicago/Turabian StyleMizrahi, Sarah, Zohar Hamo, Maya Azrad, and Avi Peretz. 2019. "Molecular Characterization and Moxifloxacin Susceptibility of Clostridium difficile" Antibiotics 8, no. 3: 118. https://doi.org/10.3390/antibiotics8030118
APA StyleMizrahi, S., Hamo, Z., Azrad, M., & Peretz, A. (2019). Molecular Characterization and Moxifloxacin Susceptibility of Clostridium difficile. Antibiotics, 8(3), 118. https://doi.org/10.3390/antibiotics8030118







