Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis
Abstract
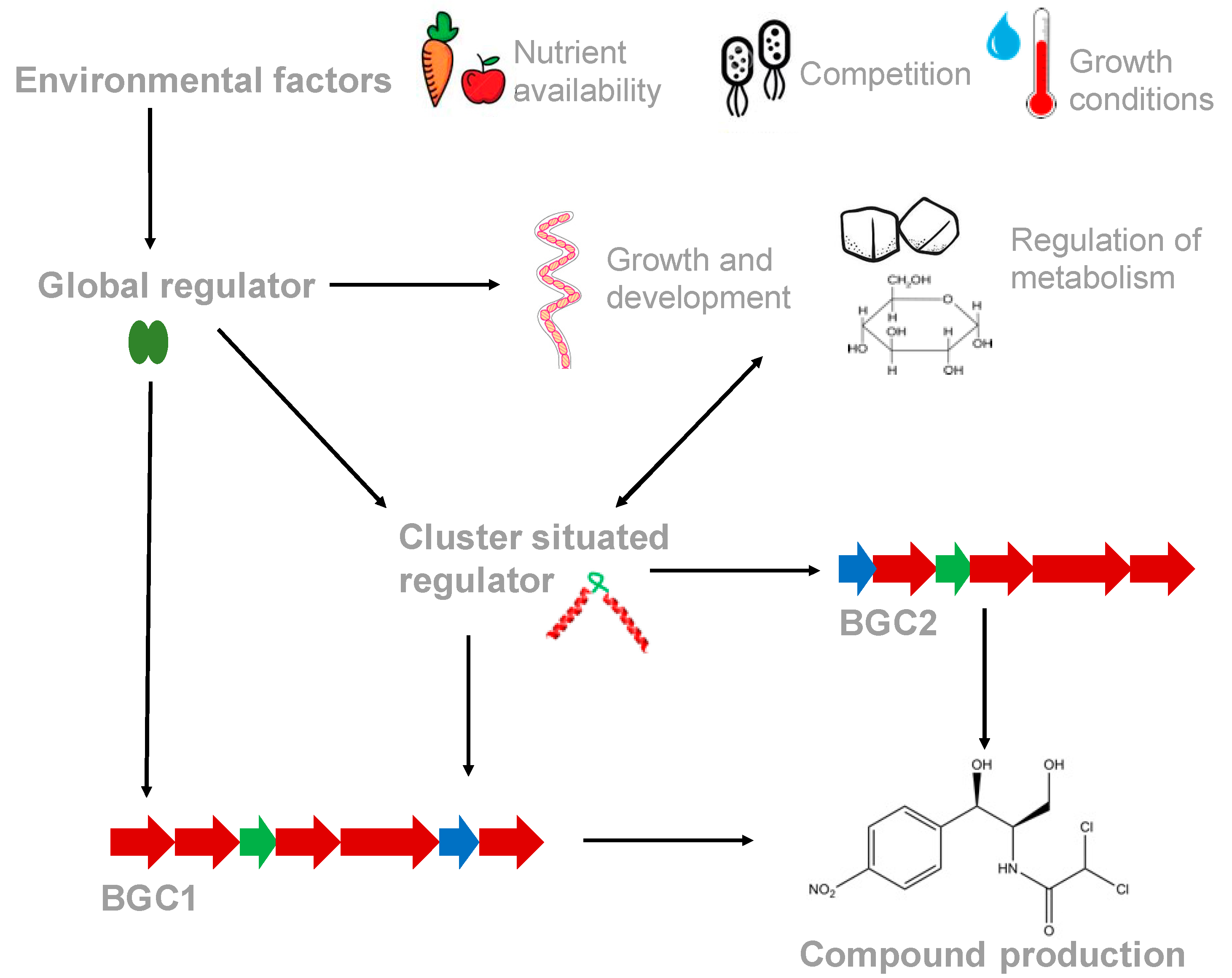
1. Introduction
1.1. Cluster Situated Regulators (CSRs) Control the Expression of Biosynthetic Genes within their Pathways
1.2. Antibiotic Biosynthesis is Controlled by Both High- and Low-Level Regulatory Systems
2. Coordinate Regulation of BGCs
2.1. FscRI—Antimycins and Candicidin
2.2. CcaR—Cephamycin C and Clavulanic Acid
2.3. GdmRIII
2.4. JadR1
2.5. Phloroglucinol/PltM/PltR
2.6. Crp
2.7. MtrA
2.8. Studying Cross-Regulation of Secondary Metabolite BGCs Enables the Discovery of Novel Natural Products
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Devine, R.; Hutchings, M.; Holmes, N. Future directions for the discovery of antibiotics from actinomycete bacteria. Emerg. Top. Life Sci. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Ishikawa, J.; Hara, H.; Suzuki, H.; Ikenoya, M.; Ikeda, H.; Yamashita, A.; Hattori, M.; Horinouchi, S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008, 190, 4050–4060. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.; Challis, G. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Corre, C.; Challis, G. Heavy Tools for Genome Mining. Chem. Biol. 2007, 14, 7–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bibb, M. Regulation of secondary metabolism in Streptomyces. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Chater, K. Recent advances in understanding Streptomyces. F1000Research 2016, 5, 2795–2811. [Google Scholar] [CrossRef] [PubMed]
- Aigle, B.; Corre, C. Waking up Streptomyces Secondary Metabolism by Constitutive Expression of Activators or Genetic Disruption of Repressors. In Methods in Enzymologyv; Academic Press: Cambridge, MA, USA, 2012; Volume 517, pp. 343–366. [Google Scholar]
- Romero-Rodríguez, A.; Robledo-Casados, I.; Sánchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 1017–1039. [Google Scholar] [CrossRef] [PubMed]
- Busby, S.J.W. Transcription activation in bacteria: Ancient and modern. Microbiology 2019, 165, 386–395. [Google Scholar] [CrossRef]
- Okamoto, S.; Taguchi, T.; Ochi, K.; Ichinose, K. Biosynthesis of Actinorhodin and Related Antibiotics: Discovery of Alternative Routes for Quinone Formation Encoded in the act Gene Cluster. Chem. Biol. 2009, 16, 226–236. [Google Scholar] [CrossRef]
- Wei, J.; He, L.; Niu, G. Regulation of antibiotic biosynthesis in actinomycetes: Perspectives and challenges. Synth. Syst. Biotechnol. 2018, 3, 229–235. [Google Scholar] [CrossRef]
- Wilson, D.J.; Xue, Y.; Reynolds, K.A.; Sherman, D.H. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 2001, 183, 3468–3475. [Google Scholar] [CrossRef] [PubMed]
- Kuscer, E.; Coates, N.; Challis, I.; Gregory, M.; Wilkinson, B.; Sheridan, R.; Petković, H. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 2007, 189, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Nodwell, J.R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 2013, 77, 440–475. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Grove, A. Regulation of Metabolic Pathways by MarR Family Transcription Factors. Comput. Struct. Biotechnol. J. 2017, 15, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, X.; Lu, X.; Liu, W.; Chen, Z.; Li, J.; Deng, L.; Wen, Y. SAV4189, a MarR-Family Regulator in Streptomyces avermitilis, Activates Avermectin Biosynthesis. Front. Microbiol. 2018, 9, 1358. [Google Scholar] [CrossRef]
- Huang, J.; Shi, J.; Molle, V.; Sohlberg, B.; Weaver, D.; Bibb, M.J.; Karoonuthaisiri, N.; Lih, C.J.; Kao, C.M.; Buttner, M.J.; et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 2005, 58, 1276–1287. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Martínez-Burgo, Y.; Álvarez-Álvarez, R.; Rodríguez-García, A.; Liras, P. The Pathway-Specific Regulator ClaR of Streptomyces clavuligerus has a Global Effect on the Expression of Genes for Secondary Metabolism and Differentiation. Appl. Environ. Microbiol. 2015, 81, 6637–6648. [Google Scholar] [CrossRef]
- Hunt, A.C.; Servín-González, L.; Kelemen, G.H.; Buttner, M.J. The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family. J. Bacteriol. 2005, 187, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.J.; Tschowri, N.; Schlimpert, S.; Flärdh, K.; Buttner, M.J. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 2015, 13, 749. [Google Scholar] [CrossRef] [PubMed]
- Den Hengst, C.D.; Tran, N.T.; Bibb, M.J.; Chandra, G.; Leskiw, B.K.; Buttner, M.J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010, 78, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Chng, C.; Lum, A.M.; Vroom, J.A.; Kao, C.M. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc. Natl. Acad. Sci. USA 2008, 105, 11346–11351. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, I.; Korostelev, Y.; Gelfand, M. GntR Family of Bacterial Transcription Factors and Their DNA Binding Motifs: Structure, Positioning and Co-Evolution. PLoS ONE 2015, 10, e0132618. [Google Scholar] [CrossRef] [PubMed]
- Hillerich, B.; Westpheling, J. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 2006, 188, 7477–7487. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; Sola-Landa, A.; Santos-Beneit, F.; Fernández-Martínez, L.T.; Prieto, C.; Rodríguez-García, A. Minireview Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 2010, 4, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-H.; Xu, Y.; Rigali, S.; Ye, B.-C. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2015, 99, 10215–10224. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhu, J.; Chen, Z.; Li, J.; Wen, Y. SAV742, a Novel AraC-Family Regulator from Streptomyces avermitilis, Controls Avermectin Biosynthesis, Cell Growth and Development. Sci. Rep. 2016, 6, 36915. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Yamazaki, H.; Kato, J.; Tomono, A.; Horinouchi, S. AdpA, a Central Transcriptional Regulator in the A-Factor Regulatory Cascade That Leads to Morphological Development and Secondary Metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 2005, 69, 431–439. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Hoskisson, P.A.; Chandra, G.; Buttner, M.J. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3 (2). Microbiology 2004, 150, 2795–2806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Rourke, S.; Widdick, D.; Bibb, M. A novel mechanism of immunity controls the onset of cinnamycin biosynthesis in Streptomyces cinnamoneus DSM 40646. J. Ind. Microbiol. Biotechnol. 2017, 44, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Hutchings, M.I. MtrAB-LpqB: A conserved three-component system in actinobacteria? Trends Microbiol. 2006, 14, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Som, N.F.; Heine, D.; Holmes, N.A.; Munnoch, J.T.; Chandra, G.; Seipke, R.F.; Hoskisson, P.A.; Wilkinson, B.; Hutchings, M.I. The Conserved Actinobacterial Two-Component System MtrAB Coordinates Chloramphenicol Production with Sporulation in Streptomyces venezuelae NRRL B-65442. Front. Microbiol. 2017, 8, 1145. [Google Scholar] [CrossRef] [PubMed]
- McLean, T.C.; Lo, R.; Tschowri, N.; Hoskisson, P.A.; Al-Bassam, M.M.; Hutchings, M.I.; Som, N.F. Sensing and responding to diverse extracellular signals: An updated analysis of the sensor kinases and response regulators of Streptomyces spp. Microbiology. in press.
- McLean, T.C.; Hoskisson, P.A.; Seipke, R.F. Coordinate Regulation of Antimycin and Candicidin Biosynthesis. mSphere 2016, 1, e00305-16. [Google Scholar] [CrossRef]
- Pérez-Llarena, F.J.; Liras, P.; Rodríguez-García, A.; Martín, J.F. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: Amplification results in overproduction of both beta-lactam compounds. J. Bacteriol. 1997, 179, 2053–2059. [Google Scholar] [CrossRef][Green Version]
- Jiang, M.; Yin, M.; Wu, S.H.; Han, X.L.; Ji, K.Y.; Wen, M.L.; Lu, T. GdmRIII, a TetR Family Transcriptional Regulator, Controls Geldanamycin and Elaiophylin Biosynthesis in Streptomyces autolyticus CGMCC0516. Sci. Rep. 2017, 7, 4803. [Google Scholar] [CrossRef]
- Sekurova, O.N.; Zhang, J.; Kristiansen, K.A.; Zotchev, S.B. Activation of chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712 by ethanol shock: Insights from the promoter fusion studies. Microb. Cell Fact. 2016, 15, 85. [Google Scholar] [CrossRef]
- Yan, Q.; Philmus, B.; Chang, J.H.; Loper, J.E. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. Elife 2017, 6, e22835. [Google Scholar] [CrossRef]
- Gao, C.; Mulder, D.; Yin, C.; Elliot, M.A. Crp is a Global Regulator of Antibiotic Production in Streptomyces. MBio 2012, 3, e00407-12. [Google Scholar] [CrossRef]
- Som, N.F.; Heine, D.; Holmes, N.; Knowles, F.; Chandra, G.; Seipke, R.F.; Hoskisson, P.A.; Wilkinson, B.; Hutchings, M.I. The MtrAB two-component system controls antibiotic production in Streptomyces coelicolor A3(2). Microbiology 2017, 163, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Campelo-Diez, A. Candicidin biosynthesis in Streptomyces griseus. Appl. Microbiol. Biotechnol. 2003, 60, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pinto-Tomás, A.A.; Rong, X.; Cheng, K.; Liu, M.; Huang, Y. Population Genomics Insights into Adaptive Evolution and Ecological Differentiation in Streptomycetes. Appl. Environ. Microbiol. 2019, 85, e02555-18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, Z.; Li, H.; Chen, X.L.; Deng, Z.; Bai, L.; Pang, X. Production of the antibiotic FR-008/candicidin in Streptomyces sp. FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 2015, 161, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Antón, N.; Santos-Aberturas, J.; Mendes, M.V.; Guerra, S.M.; Martín, J.F.; Aparicio, J.F. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 2007, 153, 3174–3183. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Patrick, E.; Hutchings, M.I. Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factor σAntA. PeerJ 2014, 2, e253. [Google Scholar] [CrossRef]
- Schoenian, I.; Spiteller, M.; Ghaste, M.; Wirth, R.; Herz, H.; Spiteller, D. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc. Natl. Acad. Sci. USA 2011, 108, 1955–1960. [Google Scholar] [CrossRef]
- Nagarajan, R.; Boeck, L.D.; Gorman, M.; Hamill, R.L.; Higgens, C.E.; Hoehn, M.M.; Stark, W.M.; Whitney, J.G. beta.-Lactam antibiotics from Streptomyces. J. Am. Chem. Soc. 1971, 93, 2308–2310. [Google Scholar] [CrossRef]
- Reading, C.; Cole, M. Clavulanic acid: A beta-lactamase-inhibiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 1977, 11, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, J.E.; Ward, J.M. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS Microbiol. Lett. 1993, 110, 239–242. [Google Scholar]
- De la Fuente, A.; Lorenzana, L.M.; Martín, J.F.; Liras, P. Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: Possible cross-regulation of two unrelated secondary metabolic pathways. J. Bacteriol. 2002, 184, 6559–6565. [Google Scholar] [CrossRef] [PubMed]
- Santamarta, I.; López-García, M.T.; Kurt, A.; Nárdiz, N.; Alvarez-Álvarez, R.; Pérez-Redondo, R.; Martín, J.F.; Liras, P. Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Mol. Microbiol. 2011, 81, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Easton, J.; Noble, S.; Perry, C.M. Amoxicillin/Clavulanic Acid. Drugs 2003, 63, 311–340. [Google Scholar] [CrossRef] [PubMed]
- Todd, P.A.; Benfield, P. Amoxicillin/Clavulanic Acid. Drugs 1990, 39, 264–307. [Google Scholar] [CrossRef] [PubMed]
- Finlay, J. A review of the antimicrobial activity of clavulanate. J. Antimicrob. Chemother. 2003, 52, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Arcamone, F.M.; Bertazzoli, C.; Ghione, M.; Scotti, T. Melanosporin and Elaiophylin, New Antibiotics from Streptomyces melanosporus (sive melonsporofaciens) n. sp. G. Microbiol. 1959, 7, 207–216. [Google Scholar]
- Zhao, X.; Fang, Y.; Yang, Y.; Qin, Y.; Wu, P.; Wang, T.; Lai, H.; Meng, L.; Wang, D.; Zheng, Z.; et al. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy 2015, 11, 1849–1863. [Google Scholar] [CrossRef]
- Stebbins, C.E.; Russo, A.A.; Schneider, C.; Rosen, N.; Hartl, F.U.; Pavletich, N.P. Crystal Structure of an Hsp90–Geldanamycin Complex: Targeting of a Protein Chaperone by an Antitumor Agent. Cell 1997, 89, 239–250. [Google Scholar] [CrossRef]
- Fernández-Martínez, L.T.; Borsetto, C.; Gomez-Escribano, J.P.; Bibb, M.J.; Al-Bassam, M.M.; Chandra, G.; Bibb, M.J. New insights into chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712. Antimicrob. Agents Chemother. 2014, 58, 7441–7450. [Google Scholar] [CrossRef] [PubMed]
- Forget, S.M.; McVey, J.; Vining, L.C.; Jakeman, D.L. Streptomyces venezuelae ISP5230 Maintains Excretion of Jadomycin upon Disruption of the MFS Transporter JadL Located within the Natural Product Biosynthetic Gene Cluster. Front. Microbiol. 2017, 8, 432. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, J.; Wang, L.; Tian, X.; Yang, H.; Fan, K.; Yang, K.; Tan, H. ‘Pseudo’ gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 2010, 285, 27440–27448. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tian, X.; Wang, J.; Yang, H.; Fan, K.; Xu, G.; Yang, K.; Tan, H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. USA 2009, 106, 8617–8622. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.N.; Loper, J.E. Soilborne Plant Diseases Caused by Pythium spp.: Ecology, Epidemiology, and Prospects for Biological Control. Crit. Rev. Plant Sci. 1999, 18, 111–181. [Google Scholar] [CrossRef]
- Ohmori, T.; Hagiwara, S.; Ueda, A.; Minoda, Y.; Yamada, K. Production of Pyoluteorin and Its Derivatives from n-Paraffin by Pseudomonas aeruginosa S10B2. Agric. Biol. Chem. 1978, 42, 2031–2036. [Google Scholar] [CrossRef]
- Howell, C.R.; Stipanovic, R.D. Suppression of Pythium ultium-Induced Damping-Off of Cotton Seedlings by Pseudomanas fluorescens and its Antibiotic, Pyoluteorin. Phytopathology 1980, 70, 712–715. [Google Scholar] [CrossRef]
- Harrison, L.A.; Letendre, L.; Kovacevich, P.; Pierson, E.; Weller, D. Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol. Biochem. 1993, 25, 215–221. [Google Scholar] [CrossRef]
- Bangera, M.G.; Thomashow, L.S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999, 181, 3155–3163. [Google Scholar]
- Heel, C.; Schnider, U.; Maurhofer, M.; Voisard, C.; Laville, J.; Burger, U.; Wirthner, P.; Haas, D.; Défago, G. Suppression of Root Diseases by Pseudomonas fluorescens CHA0: Importance of the Bacterial Secondary Metabolite 2,4-Diacetylphloroglucinol. Mol. Plant-Microbe Interact 1992, 5, 4–13. [Google Scholar]
- Paulsen, I.T.; Press, C.M.; Ravel, J.; Kobayashi, D.Y.; Myers, G.S.; Mavrodi, D.V.; DeBoy, R.T.; Seshadri, R.; Ren, Q.; Madupu, R.; et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 2005, 23, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Brodhagen, M.; Henkels, M.D.; Loper, J.E. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 2004, 70, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Kidarsa, T.A.; Goebel, N.C.; Zabriskie, T.M.; Loper, J.E. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 2011, 81, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Clifford, J.C.; Buchanan, A.; Vining, O.; Kidarsa, T.A.; Chang, J.H.; McPhail, K.L.; Loper, J.E. Phloroglucinol functions as an intracellular and intercellular chemical messenger influencing gene expression in Pseudomonas protegens. Environ. Microbiol. 2016, 18, 3296–3308. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Bonkowski, M. The model predator Acanthamoeba castellanii induces the production of 2,4, DAPG by the biocontrol strain Pseudomonas fluorescens Q2-87. Soil Biol. Biochem. 2010, 42, 1647–1649. [Google Scholar] [CrossRef]
- Baehler, E.; Bottiglieri, M.; Péchy-Tarr, M.; Maurhofer, M.; Keel, C. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 2005, 99, 24–38. [Google Scholar] [CrossRef]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Derouaux, A.; Halici, S.; Nothaft, H.; Neutelings, T.; Moutzourelis, G.; Dusart, J.; Titgemeyer, F.; Rigali, S. Deletion of a Cyclic AMP Receptor Protein Homologue Diminishes Germination and Affects Morphological Development of Streptomyces coelicolor. J. Bacteriol. 2004, 186, 1893–1897. [Google Scholar] [CrossRef]
- Piette, A.; Derouaux, A.; Gerkens, P.; Noens, E.E.; Mazzucchelli, G.; Vion, S.; Koerten, H.K.; Titgemeyer, F.; De Pauw, E.; Leprince, P.; et al. From Dormant to Germinating Spores of Streptomyces coelicolor A3(2): New Perspectives from the crp Null Mutant. J. Proteome Res. 2005, 4, 1699–1708. [Google Scholar] [CrossRef]
- Zahrt, T.C.; Deretic, V. An Essential Two-Component Signal Transduction System in Mycobacterium tuberculosis. J. Bacteriol. 2000, 182, 3832–3838. [Google Scholar] [CrossRef]
- Möker, N.; Brocker, M.; Schaffer, S.; Krämer, R.; Morbach, S.; Bott, M. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 2004, 54, 420–438. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Tong, Y.; Han, Y.-J.; Ye, B.-C. Two amino acids missing of MtrA resulted in increased erythromycin level and altered phenotypes in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2019, 103, 4539–4548. [Google Scholar] [CrossRef] [PubMed]
- Purushotham, G.; Sarva, K.B.; Blaszczyk, E.; Rajagopalan, M.; Madiraju, M.V. Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol. Microbiol. 2015, 98, 586–604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, L.; Zhu, Y.; Liu, M.; Wang, Y.; Cao, G.; Chen, X.L.; Tao, M.; Pang, X. Deletion of MtrA Inhibits Cellular Development of Streptomyces coelicolor and Alters Expression of Developmental Regulatory Genes. Front. Microbiol. 2017, 8, 2013. [Google Scholar] [CrossRef] [PubMed]
- Munnoch, J.T.; Martinez, M.T.; Svistunenko, D.A.; Crack, J.C.; Le Brun, N.E.; Hutchings, M.I. Characterization of a putative NsrR homologue in Streptomyces venezuelae reveals a new member of the Rrf2 superfamily. Sci. Rep. 2016, 6, 31597. [Google Scholar] [CrossRef] [PubMed]
- Alberti, F.; Leng, D.J.; Wilkening, I.; Song, L.; Tosinb, M.; Corre, C. Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chem. Sci. 2019, 10, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Funk, A.N.; Scherlach, K.; Schroeckh, V.; Shelest, E.; Horn, U.; Hertweck, C.; Brakhage, A.A. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol. 2010, 76, 8143–8149. [Google Scholar] [CrossRef]
- Lo, H.-C.; Entwistle, R.; Guo, C.J.; Ahuja, M.; Szewczyk, E.; Hung, J.H.; Chiang, Y.M.; Oakley, B.R.; Wang, C.C. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 2012, 134, 4709–4720. [Google Scholar] [CrossRef]
- Tong, Y.; Weber, T.; Lee, S. CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 2018. [Google Scholar] [CrossRef]
- Alberti, F.; Corre, C. Editing streptomycete genomes in the CRISPR/Cas9 age. Nat. Prod. Rep. 2019. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, D.; Yan, Y.; Pan, G.; Xiang, W.; Shen, B. Overproduction of lactimidomycin by cross-overexpression of genes encoding Streptomyces antibiotic regulatory proteins. Appl. Microbiol. Biotechnol. 2016, 100, 2267–2277. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santos-Aberturas, J.; Payero, T.D.; Vicente, C.M.; Guerra, S.M.; Cañibano, C.; Martín, J.F.; Aparicio, J.F. Functional conservation of PAS–LuxR transcriptional regulators in polyene macrolide biosynthesis. Metab. Eng. 2011, 13, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Colombo, V.; Fernández-de-Heredia, M.; Malpartida, F. A polyketide biosynthetic gene cluster from Streptomyces antibioticus includes a LysR-type transcriptional regulator. Microbiology 2001, 147, 3083–3092. [Google Scholar] [CrossRef] [PubMed]
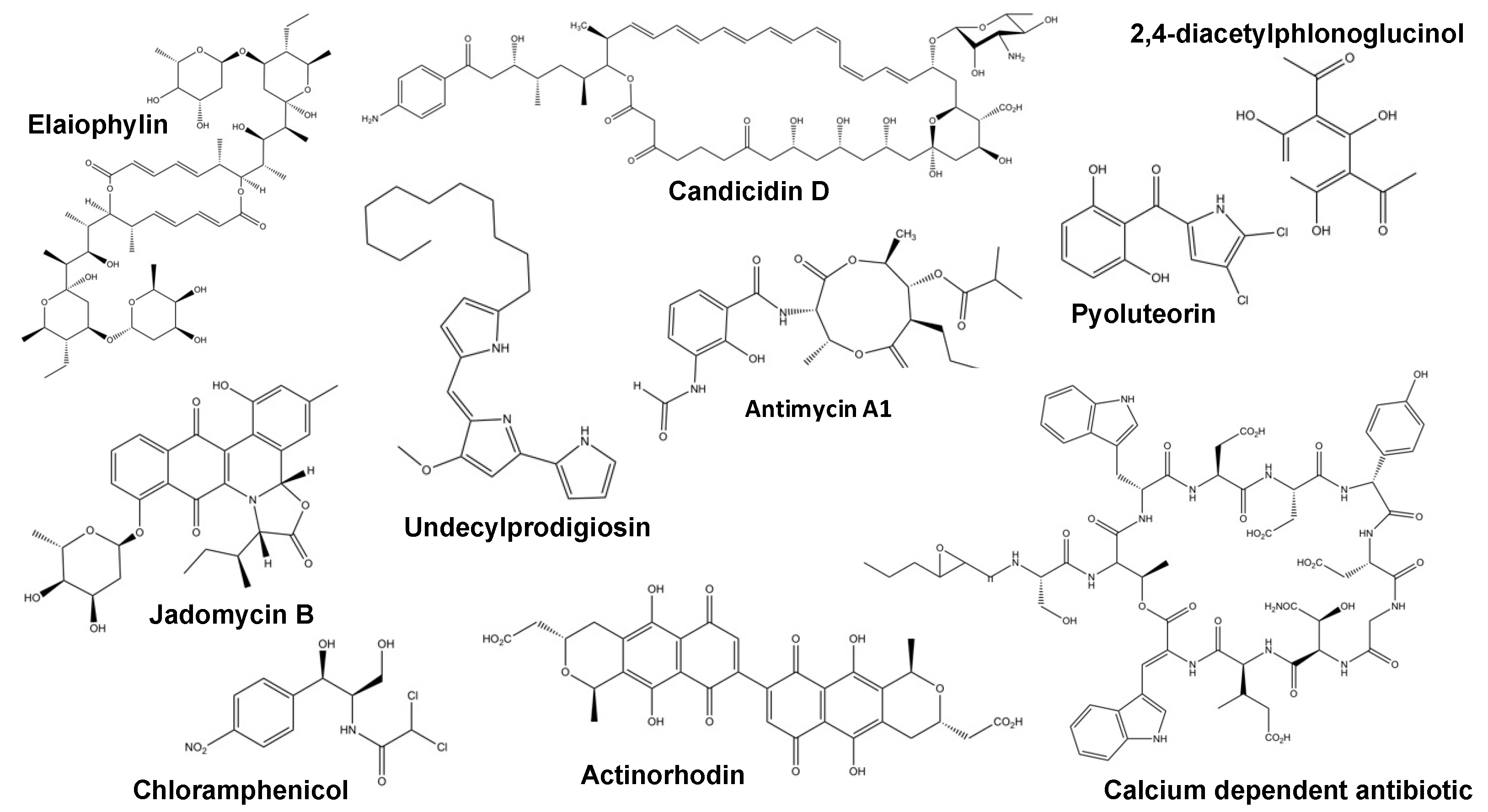
| Regulator Name. | Organism | Regulator Type | Antibiotics Regulated | Type of Regulation | Ref. |
|---|---|---|---|---|---|
| FscRI | Streptomyces albus | PAS-LuxR family | Candicidin | Positive direct | [37] |
| Antimycins | |||||
| CcaR | Streptomyces clavuligerus | SARP-type | Cephamycin C | Positive direct | [38] |
| Clavulanic acid | |||||
| GdmRIII | Streptomyces autolyticus | TetR-family | Geldanamycin | Antagonistic direct | [39] |
| Elaiophylin | |||||
| JadR1 | Streptomyces venezuelae | Atypical OmpR-family | Jadomycin | Antagonistic direct | [40] |
| Chloramphenicol | |||||
| PG-Cl2/PG-Cl | Pseudomonas protegens | Chemical | 2,4-diacetylphloroglucinol | Dose dependant | [41] |
| Pyoluteorin | |||||
| Crp | Streptomyces coelicolor | Crp-Fnr family | Actinorhodin | Global | [42] |
| Undecylprodiogisin | |||||
| Calcium-dependant antibiotic | |||||
| yellow-pigmented polyketide | |||||
| MtrA | Streptomyces coelicolor | OmpR-family | Actinorhodin | Global | [35,43] |
| Undecylprodiogisin | |||||
| Streptomyces venezuelae | Chloramphenicol | ||||
| Jadomycin |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McLean, T.C.; Wilkinson, B.; Hutchings, M.I.; Devine, R. Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis. Antibiotics 2019, 8, 83. https://doi.org/10.3390/antibiotics8020083
McLean TC, Wilkinson B, Hutchings MI, Devine R. Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis. Antibiotics. 2019; 8(2):83. https://doi.org/10.3390/antibiotics8020083
Chicago/Turabian StyleMcLean, Thomas C., Barrie Wilkinson, Matthew I. Hutchings, and Rebecca Devine. 2019. "Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis" Antibiotics 8, no. 2: 83. https://doi.org/10.3390/antibiotics8020083
APA StyleMcLean, T. C., Wilkinson, B., Hutchings, M. I., & Devine, R. (2019). Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis. Antibiotics, 8(2), 83. https://doi.org/10.3390/antibiotics8020083





