Abstract
Stress and starvation causes bacterial cells to activate the stringent response. This results in down-regulation of energy-requiring processes related to growth, as well as an upregulation of genes associated with survival and stress responses. Guanosine tetra- and pentaphosphates (collectively referred to as (p)ppGpp) are critical for this process. In Gram-positive bacteria, a main function of (p)ppGpp is to limit cellular levels of GTP, one consequence of which is reduced transcription of genes that require GTP as the initiating nucleotide, such as rRNA genes. In Streptomycetes, the stringent response is also linked to complex morphological differentiation and to production of secondary metabolites, including antibiotics. These processes are also influenced by the second messenger c-di-GMP. Since GTP is a substrate for both (p)ppGpp and c-di-GMP, a finely tuned regulation of cellular GTP levels is required to ensure adequate synthesis of these guanosine derivatives. Here, we discuss mechanisms that operate to control guanosine metabolism and how they impinge on the production of antibiotics in Streptomyces species.
1. Introduction
Bacteria experience constant challenges, either in the environment or when infecting a host. They utilize various mechanisms to survive such stresses, which may include changes in temperature, pH, or oxygen content as well as limited access to carbon or nitrogen sources. A stress response that is mobilized to deal with events such as nutrient limitation is the stringent response. The purpose of this response is to focus cellular resources on survival, as opposed to growth [1]. To achieve this outcome, bacteria synthesize the hyperphosphorylated nucleotides guanosine pentaphosphate and guanosine tetraphosphate, collectively called (p)ppGpp, from GTP/GDP and ATP. Based on the critical role of (p)ppGpp, it is also called an alarmone, as it responds to starvation or stress and conveys such distressing circumstances to the cellular machineries in order to conserve resources. Exposure to challenging environmental conditions may also induce formation of biofilms, protective bacterial communities that confer a number of fitness advantages, including resistance to nutrient deprivation or antimicrobial agents. The second messenger cyclic-di-GMP (c-di-GMP), which is synthesized from two molecules of GTP, is intimately involved in the switch between a motile planktonic lifestyle and a sessile biofilm community in unicellular bacteria [2]. In multicellular Actinomycetes, such as Streptomyces spp., c-di-GMP is involved in controlling developmental processes [3]. Collectively, these signaling molecules require the presence of purines to sustain their synthesis under conditions of stress and starvation, circumstances under which the bacterial cell must conserve essential nutrients. Purine salvage pathways are, therefore, expected to be important for the generation of substrates for (p)ppGpp and c-di-GMP synthesis. Additionally, members of the genus Streptomyces respond to changes in environmental conditions by a morphological differentiation and multicellular development that may be closely tied to their production of secondary metabolites, including a wide range of antibiotics. Indeed, two thirds of clinically relevant antibiotics such as chloramphenicol, streptomycin, and tetracycline are synthesized by Streptomyces spp. [4]. In this review, we discuss how purine metabolism affects antibiotic production, with an emphasis on the stringent response.
2. Stringent Response and Morphological Differentiation
Nutrient limitation or starvation induces the stringent response in the vast majority of bacteria. This response was first described in Escherichia coli and shown to involve reduced accumulation of stable RNAs (rRNA and tRNA); this stringent response was ‘relaxed’ by mutation in the gene accordingly named relA. Stringent response depends on a transient increase in the level of (p)ppGpp, also known as the magic spot [5]. Members of the RSH (RelA/SpoT Homolog) protein superfamily are responsible for (p)ppGpp synthesis from ATP and either GTP or GDP and for the degradation of (p)ppGpp to GTP or GDP and pyrophosphate, with some members possessing both enzymatic functions. The activity of different RSH family proteins is controlled by distinct mechanisms, including interaction with idling ribosomes in many cases, allowing responses to various stresses [1]. The stringent response may be initiated by limitation of nutrients such as amino acids, nitrogen, phosphorous, and carbon sources. Once (p)ppGpp accumulates, it exerts global changes designed to downregulate transcription of genes linked to growth (e.g., ribosome biogenesis) and upregulation of genes required for survival (e.g., nutrient acquisition and stress responses), and it also directly binds a number of proteins to regulate their activity [6]. While specific binding targets may differ between Gram-negative and Gram-positive species, (p)ppGpp has been shown to regulate replication, transcription, translation and GTP biosynthesis by binding to proteins that participate in these processes, including DNA primase, RNA polymerase, small GTPases, enzymes involved in purine biosynthesis, and transcriptional regulators [7,8,9,10,11].
Considerable differences exist between bacterial species with regard to both the mode of action and metabolism of (p)ppGpp. In Gram-negative species such as E. coli, ribosomes sense the uncharged tRNAs at the ribosomal A site during amino acid starvation, causing protein synthesis to stall and resulting in the activation of RelA [12]. This leads to synthesis of (p)ppGpp, which acts as an allosteric regulator of RNA polymerase. The (p)ppGpp binds together with the regulatory protein DksA; this leads to inhibition of transcription from stable RNA promoters and promoters driving expression of genes encoding, for example, ribosomal proteins, and it mediates an activation of genes critical for cell survival [13,14]. This mechanism is likely to be conserved among other proteobacteria [14].
The stringent response in Gram-positive bacteria is not mediated by direct binding of (p)ppGpp to RNA polymerase [15,16]. In Bacillus subtilis, (p)ppGpp regulates intracellular GTP levels, even during normal growth, and dysregulated GTP homeostasis leads to cell death [17,18]. Two mechanisms contribute to the observed reduction in GTP levels during the stringent response, namely, consumption of GTP for the purpose of (p)ppGpp synthesis and inhibition of GTP biosynthesis. GTP can be synthesized by both expensive de novo and less resource-demanding salvage pathways, and (p)ppGpp binds directly to several proteins that participate in these pathways to inhibit their activity [10,18]. Figure 1 summarizes purine metabolism, identifying key regulated enzymes participating in synthesis of GTP; de novo synthesis initiated by GuaB-catalyzed conversion of inosine monophosphate (IMP) to XMP, or formation of XMP by purine salvage, which requires xanthine dehydrogenase (Xdh). As a consequence of reduced cellular GTP levels, transcription from genes whose initiating nucleotide is GTP is decreased, thereby accounting for the reduced accumulation of stable RNAs [15]. Expression of other genes is affected indirectly by the reduced levels of GTP, for example, by favoring transcription of genes whose initiating nucleotide is ATP [19].
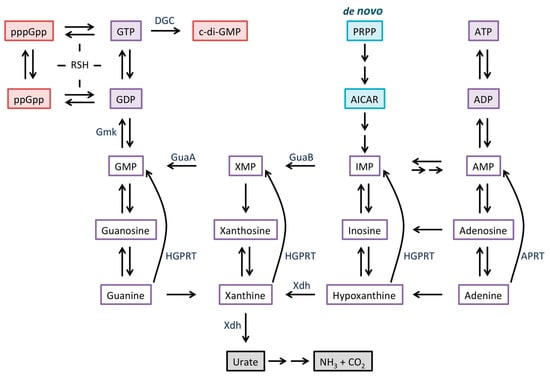
Figure 1.
Overview of purine metabolism. The de novo biosynthesis (turquoise) starts with phosphoribosyl pyrophosphate (PRPP) and ends with inosine monophosphate (IMP); the intermediate AICAR (5-aminoimidazole-4-carboxamide ribonucleotide) accumulates in presence of dihydrofolate reductase inhibitors [33]. Intermediates in salvage pathways are outlined in purple. In some species, guanine, xanthine, and hypoxanthine are converted to the corresponding mononucleotides by different enzymes. Xdh can initiate degradation of purines (gray). RelA/SpoT Homolog (RSH) family enzymes are responsible for (p)ppGpp synthesis and hydrolysis, while diguanylate cyclase (DGC) synthesizes c-di-GMP (red).
Two proteins, RelA and RSH, participate in (p)ppGpp metabolism in Streptomyces spp., likely with a division of labor in which RelA and RSH respond to amino acid/glucose starvation and phosphate starvation, respectively [20,21,22]. Starvation of Streptomyces spp. results in elevated levels of (p)ppGpp as well as a reduced GTP pool, as also seen in B. subtilis, accompanied by reduced transcription of rRNA genes [23,24,25,26]. For example, S. clavuligerus in which relA is inactivated fails to synthesize (p)ppGpp and maintains steady levels of GTP during growth, whereas wild-type cells exhibit an approximately three-fold decrease in GTP levels as (p)ppGpp accumulates [27]. A similar (p)ppGpp-dependent decrease in cellular GTP levels was previously demonstrated in S. griseus during nutritional shift-down, and it was attributed to ppGpp-mediated inhibition of IMP dehydrogenase (GuaB; Figure 1) [28]. Starvation also elicits a complex morphological differentiation in Streptomycetes, a process that involves formation of aerial hyphae and unicellular spores for dispersal, and this differentiation may be accompanied by production of bioactive secondary metabolites. Erection of aerial hyphae requires the bld (bald) gene products, while differentiation of aerial hyphae into spores requires the whi (white) gene products [29]. The transcriptional regulator BldD controls a large number of genes, many of which are involved in differentiation and antibiotic production [3].
The secondary messenger c-di-GMP, which is synthesized from two molecules of GTP by diguanylate cyclases and degraded by phosphodiesterases, also participates in differentiation. The two S. coelicolor proteins RmdA and RmdB possess phosphodiesterase activity and the rmdA-rmdB double mutant accumulates three-fold higher levels of c-di-GMP and is completely deficient in aerial hyphae formation [30]. Deficiency in generation of aerial mycelium is also a characteristic of S. coelicolor deleted for cdgB, which encodes a diguanylate cyclase, and of cells over-expressing cdgB; cdgB over-expression also inhibits formation of the antibiotic actinorhodin [31]. Expression of cdgB is controlled by BldD; notably, DNA binding by BldD is activated by direct binding of c-di-GMP, leading to repression of the BldD-controlled sporulation genes during vegetative growth [32].
3. Secondary Metabolism in Streptomyces
Members of this genus produce numerous antibiotics or secondary metabolites that are commercially or clinically relevant. These natural products possess, for example, antibacterial, antiviral, anti-tumor, and anti-fungal properties [4]. Secondary metabolites are produced in response to environmental conditions, including starvation or growth of other nearby organisms that may belong to the same or different bacterial genera, or to different species such as plants or fungi; such compounds are therefore thought to furnish the producer with a competitive advantage [34]. Under laboratory conditions, many of the gene clusters that encode proteins involved in generation of natural products are silent. Efforts to elicit their expression by co-culturing Streptomyces spp. with various other microbes or by addition of natural and synthetic chemicals have been implemented, and this has resulted in the identification of numerous antibiotics [35]. Combining drug resistant mutations also resulted in activation of antibiotic gene clusters [36,37]. Inducers of secondary metabolism are likely to act indirectly through intracellular signaling, including the generation of alarmones. Since many gene clusters have resisted efforts to elicit their expression during laboratory conditions, Streptomyces spp. (and other bacterial species known for production of bioactive secondary metabolites, such as Burkholderia spp. [38]) are predicted to constitute a valuable resource for discovery of novel biologically active compounds.
Genes linked to biosynthesis of secondary metabolites generally form very large clusters, often comprising multiple operons, and analysis of genome sequences suggests that the majority are species specific [39]. For instance, S. coelicolor strain A3(2) produces several compounds, including the red-pigmented undecylprodigiosin (RED, a member of the prodiginine family of bacterial alkaloids), the deep blue polyketide actinorhodin (ACT), and methylenomycin (Figure 2; reviewed in [40]), while S. griseus produces streptomycin, S. virginia produces virginiamycin, S. natalensis produces pimaricin, and S. avermitilis produces avermectin [40]. The antibiotic biosynthetic gene clusters often include regulatory genes referred to as cluster-situated regulators (CSR) [40,41]. The CSRs may directly regulate the transcription of the corresponding antibiotic gene cluster or they may control antibiotic production indirectly through other mechanisms [40]. Activation of antibiotic synthesis is complex and may involve multiple regulators, including factors that affect both antibiotic production and differentiation such as BldD, and it may require a variety of signals, including accumulation of (p)ppGpp and c-di-GMP.
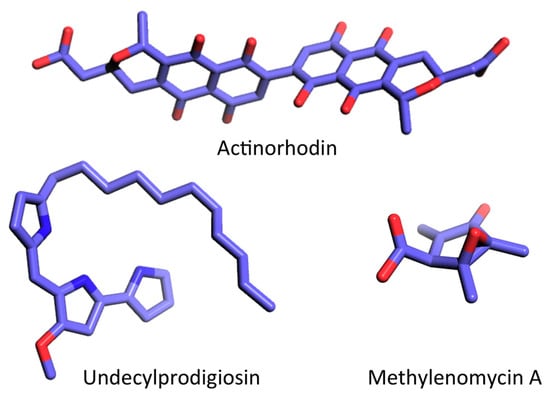
Figure 2.
Examples of secondary metabolites produced by S. coelicolor. Structures were obtained from the PubChem database and rendered using PyMol (C, light blue; N, dark blue; O, red). Actinorhodin, CID 441143; undecylprodigiosin, CID 135515151; methylenomycin A, CID 122733.
4. Link Between Purine Salvage and Production of Guanosine Nucleotide Second Messengers
While (p)ppGpp was first identified under conditions of amino acid starvation, it has since been associated with a number of other stress conditions, most notably conditions faced by bacterial pathogens when they infect a host [42]. Since guanosine nucleotide second messengers are synthesized from GDP or GTP (and at the expense of ATP hydrolysis in case of (p)ppGpp), the enzymes required for purine de novo synthesis or salvage are critical for the generation of these second messengers. Direct links between activity of purine metabolic enzymes and (p)ppGpp accumulation have been documented in several bacterial species, including the effects on virulence in the case of pathogens.
4.1. Cellular Levels of GTP and (p)ppGpp are Inter-Dependent
De novo purine biosynthesis involves the assembly of the purine ring on phosphoribosyl-pyrophosphate (PRPP) to generate inosine monophosphate (IMP), which is the precursor to both AMP and GMP (Figure 1) [43,44]. Purine bases or nucleotides, including those acquired from exogenous sources, may be inter-converted using salvage pathways. Activity of a number of enzymes in these pathways has been reported to affect the generation of (p)ppGpp, including hypoxanthine guanine phosphoribosyl transferase (HGPRT), which salvages guanine, xanthine, and hypoxanthine nucleobases by converting them to the corresponding mononucleotides, IMP dehydrogenase (GuaB), which catalyzes the committed step in de novo GMP biosynthesis, GMP synthetase (GuaA), and xanthine dehydrogenase (Xdh). Xdh converts hypoxanthine to xanthine, thus favoring the conversion of adenine to guanine, and it can divert purines away from salvage pathways and towards degradation by catalyzing the oxidation of xanthine to urate [45]. For example, guaA and guaB mutants of the Lyme disease pathogen Borrelia burgdorferi fail to infect a mouse model, illustrating that purine metabolic enzymes play an important role in virulence [46]. Lacking enzymes involved in de novo purine biosynthesis, this bacterium in addition depends on transport proteins to salvage purines from the host [47]. Both (p)ppGpp and c-di-GMP play a vital role during starvation and survival in the tick host, and failure to synthesize these messengers reduces persistence [48,49]. In Sinorhizhobium meliloti, (p)ppGpp synthesis is increased upon carbon or nitrogen starvation, concomitant with an increase in the transcript levels of xdh. This indicates that the xdh gene product is linked to synthesis of (p)ppGpp during starvation [50]. Consistent with this interpretation, xdh expression is also increased in stationary phase in S. coelicolor, whereas inhibition of Xdh with allopurinol attenuates (p)ppGpp production [51]. In Listeria monocytogenes, both hgprt and relA mutants are incapable of synthesizing (p)ppGpp and fail to cause infection in mice [52]. Evidently, the enzymes of purine metabolism are critical for maintaining the requisite levels of guanosine nucleotide second messengers.
A feedback response in which (p)ppGpp negatively controls activity of purine metabolic enzymes also exists. As noted above, S. griseus GuaB is inhibited by ppGpp, which accounts for reduced cellular levels of GTP during stringent response (Figure 3) [28]. In B. subtilis, (p)ppGpp inhibits several GTP biosynthesis enzymes, but primarily HGPRT and Gmk, which converts GMP to GDP, in order to regulate the GTP levels in the cell [9]; in absence of (p)ppGpp, excess GTP leads to cell death [18]. Staphylococcus aureus HGPRT and Gmk are also direct targets for (p)ppGpp, suggesting a conserved regulatory mechanism [10]. By contrast, Gmk is not specifically inhibited by (p)ppGpp in Enterococcus faecalis, whereas HGPRT is [53]. In addition, HPRT/GPRT were recently confirmed as physiologically relevant targets of (p)ppGpp in E. coli as well, suggesting that direct regulation of purine metabolic enzymes by (p)ppGpp is not unique to Gram-positive species [8]. That (p)ppGpp targets different enzymes in various bacterial species speaks to distinct mechanisms for control of GTP homeostasis.
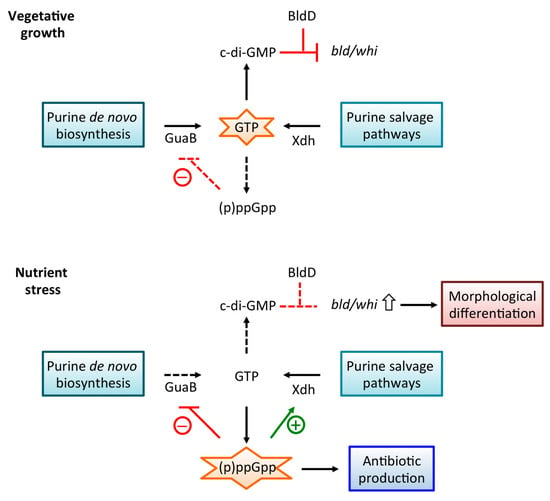
Figure 3.
Summary of link between guanosine metabolism and antibiotic production. During vegetative growth, cellular concentrations of GTP are high, but maintained by (p)ppGpp-mediated inhibition of GTP biosynthesis, most likely direct inhibition of GuaB. Under these conditions, c-di-GMP is produced, and it binds BldD to form a complex, which represses bld and whi genes. Under stress conditions, (p)ppGpp accumulates, resulting in efficient inhibition of GTP biosynthesis. However, high levels of (p)ppGpp also mediate an increase in Xdh production, averting excessive depletion of GTP. Reduced production of c-di-GMP results in derepression of bld and whi genes, leading to morphological differentiation. The accumulation of (p)ppGpp generally leads to production of secondary metabolites, including antibiotics. The role of BldD in antibiotic production remains to be fully elucidated.
4.2. Production of Xdh As a Mechanism to Promote Purine salvage
Control of cellular GTP levels is thought to be a primary function of (p)ppGpp in Gram-positives, guarding against excess GTP accumulation during balanced growth and effecting a decrease during the stringent response [9,18,53]. However, excessive depletion of GTP is also deleterious, and mechanisms exist that favor synthesis of guanosine nucleotides. In S. coelicolor, xanthine dehydrogenase regulator (XdhR) represses expression of the xdh operon; XdhR binds ppGpp (and with lower affinity to GTP), a result of which is attenuated DNA binding and increased expression of the xdhABC gene cluster during the stringent response [51]. This mode of regulation would bias purine salvage pathways towards production of guanosine nucleotides, particularly during stringent response when de novo pathways are downregulated and the bacteria are dependent upon salvage pathways. The ~0.2 mM affinity of ppGpp for XdhR suggests that xdh expression (and hence GTP synthesis) will be induced when (p)ppGpp accumulates to mM concentrations; under these conditions, (p)ppGpp-mediated repression of target enzymes in the purine biosynthesis pathways would be efficient, and production of Xdh may avert excessive depletion of GTP. If (p)ppGpp preferentially inhibits GuaB, as reported in S. griseus [28], then GTP production would depend on synthesis of XMP via xanthine (Figure 1), rationalizing the need for (p)ppGpp-dependent control of xdh expression (Figure 3).
The xdhR-xdhABC gene locus is not conserved in B. subtilis, where instead expression of xdh is induced in the presence of hypoxanthine [54]. By contrast, hypoxanthine is not a ligand for S. coelicolor XdhR [51]. This may reflect that B. subtilis Xdh has a primary role in purine catabolism, whereas S. coelicolor Xdh activity is more important for GTP homeostasis. Consistent with this interpretation, the catabolism of xanthine to urate appears to be disfavored in Agrobacterium fabrum during the stringent response when (p)ppGpp accumulates and xdh expression is increased; A. fabrum encodes a locus that is homologous to the xdhR-xdhABC gene cluster found in Streptomyces spp. [55]. Furthermore, an S. coelicolor xdhR mutant, which overproduces Xdh, is defective in forming aerial mycelium and spores and it overproduces the antibiotic ACT [56,57,58]; the ΔxdhR strain does not express whi genes, which are important for sporulation, likely on account of increased (p)ppGpp synthesis and dysregulated GTP homeostasis.
A. fabrum XdhR binds c-di-GMP with even higher affinity than ppGpp [51]. As noted above, c-di-GMP is also involved in morphological differentiation and antibiotic production in Streptomyces spp., and it activates DNA binding by the key regulator BldD. S. coelicolor XdhR likewise responds to c-di-GMP by attenuated DNA binding (Sivapragasam and Grove, unpublished), which may favor synthesis of GTP to sustain production of secondary messengers that derive from GTP. Taken together, we propose that regulation of purine metabolism and GTP homeostasis in Streptomycetes is distinct from that reported in B. subtilis and that these processes need to be carefully managed to maintain the optimal synthesis of guanosine nucleotide second messengers.
Starvation of nutrients, stationary phase, and slow growth leads to degradation of stable RNAs, rRNA and tRNA. These RNAs, which are typically not degraded during balanced growth, account for the vast majority of total RNA, and they represent a valuable source of nutrients [59]. For instance, extensive degradation (>90%) of the 23S rRNA and 16S rRNA occurs in Salmonella strains when they reach stationary phase [60]. A similar degradation of rRNA is observed in a relA mutant of E. coli following nutrient starvation [61]. Even during slow growth, almost 70% of the rRNA that is newly synthesized is degraded. This degradation is considered to be a vital survival strategy as it furnishes the bacteria with a source of nucleotides for salvage pathways during stress or starvation.
5. Guanosine Metabolism Impinges on Antibiotic Production
Analysis of annotated Streptomyces genomes suggests the presence of >20 species-specific gene clusters implicated in synthesis of secondary metabolites per genome, many of them antibiotics [39]. Production of many such natural products is coordinated with morphological differentiation, however, regulatory mechanisms are complex, and (p)ppGpp has been reported to serve as both negative and positive regulator.
5.1. Production of Actinorhodin (ACT) and Undecylprodigiosin (RED)
Production of antibiotics such as ACT and RED (Figure 2) in S. coelicolor generally takes place in stationary phase when (p)ppGpp accumulates (Figure 3) and genes encoding the biosynthetic enzymes are activated by cluster-situated regulators ActII-ORF4 and RedD, respectively [62,63]. Inactivation of relA in S. coelicolor A2(3) abolishes (p)ppGpp synthesis and production of both ACT and RED under nitrogen-limiting conditions, and this correlates with the reduced accumulation of both actII-ORF4 and redD transcripts (Table 1) [64]. Under conditions of nutrient sufficiency, induced production of (p)ppGpp results in activation of actII-ORF4, but not redD, indicating that accumulation of (p)ppGpp per se does not induce RED production [21,65]. While inactivation of either relA or rshA in S. coelicolor M600 leads to a complete lack of ACT production in batch culture, RED production is higher than in wild-type cells [20]. Based on the analysis of (p)ppGpp levels and antibiotic production in a strain carrying a mutation in the principal sigma factor (a mutation that was speculated to reduce levels of substrates for (p)ppGpp synthesis), it was inferred that (p)ppGpp-mediated induction of antibiotic production is extremely sensitive to differences in cellular (p)ppGpp pools [66]. In S. coelicolor and S. griseus, the cyclic AMP (cAMP)-binding protein EshA is essential for ensuring adequate (p)ppGpp synthesis by mechanisms that remain unclear and this, in turn, is required for (p)ppGpp-mediated control of GTP levels. EshA is therefore important for morphological differentiation and ACT/streptomycin production [67]. This analysis also concluded that finely tuned (p)ppGpp accumulation is essential for ensuring antibiotic production.

Table 1.
Examples of factors involved in antibiotic production in Streptomyces spp. that are linked to stress response or GTP synthesis.
Transcription of actII-ORF4 is under control of numerous regulators, whereas redD is activated by RedZ; redZ expression, however, is controlled by some of the same factors that regulate actII-ORF4 expression (reviewed in [40]). The transcriptional regulator XdhR, which was previously shown to control expression of the xdh gene cluster by mediating a (p)ppGpp-dependent increase in xdh expression during nutrient limitation and stationary phase [51,56], also binds the promoter regions of actII-ORF4 and actII-ORF1. The gene actII-ORF1 encodes another cluster-situated regulator of ACT biosynthesis. The binding of XdhR leads to the repression of genes encoding ACT biosynthetic enzymes and hence production of ACT in S. coelicolor M145 [58]. Since (p)ppGpp is a ligand for XdhR, entry into stationary phase or stringent response would be expected to lead to dissociation of XdhR from the actII-ORF4 and actII-ORF1 promoters, thus facilitating ACT biosynthesis. Accordingly, XdhR directly links regulation of purine metabolism with (p)ppGpp–mediated control of antibiotic production.
5.2. Production of Other Antibiotics
(p)ppGpp has also been implicated in controlling the antibiotic production in other Streptomyces spp. For example, an S. antibioticus relA mutant is deficient in (p)ppGpp synthesis and actinomycin production, a deficiency that is not alleviated by growth under phosphate-limiting conditions [69]. In S. griseus, inhibition of GMP synthetase/GuaA (and hence GTP and (p)ppGpp production) by addition of decoyinine leads to formation of aerial mycelium, but a decrease in the production of streptomycin [23]. A rel mutant (tentatively identified as a relC mutant) fails to accumulate (p)ppGpp in response to nutrient limitation, and it is deficient in both formation of aerial mycelium and submerged spores and in production of enzymes required for streptomycin biosynthesis. In this rel mutant, the addition of decoyinine is accompanied by a marked reduction in cellular GTP levels and it restores the generation of submerged spores; this indicates that morphological differentiation is initiated when GTP levels decrease, but that streptomycin biosynthesis requires (p)ppGpp.
In S. clavuligerus, inactivation of relA leads to the expected failure to accumulate (p)ppGpp and an inability to sporulate. However, production of clavulanic acid and cephamycin C and transcription of biosynthetic genes is increased in the rel mutant, indicating that antibiotic production is negatively regulated by (p)ppGpp [27]. Curiously, the production of these antibiotics in a relA mutant strain also appears to depend on strain background and/or growth media as reflected in a separate study in which an S. clavuligerus relA disruptant strain was deficient in antibiotic production [71]. The general role of (p)ppGpp in eliciting an upregulation of antibiotic production in a number of bacterial species has also motivated efforts towards metabolic engineering by expressing a (p)ppGpp synthetase. In the actinomycete Saccharopolyspora erythraea, this approach yields increased production of erythromycin, but only in a wild-type strain and not in an industrial strain, which already exhibits elevated (p)ppGpp levels [72].
5.3. Regulation of Antibiotic Production by c-di-GMP
Both (p)ppGpp and c-di-GMP affect morphological differentiation and regulation of antibiotic production. As noted above, the master regulator BldD represses genes involved in formation of aerial hyphae (the bld genes) and sporulation (the whi genes) during vegetative growth, and this occurs when BldD binds c-di-GMP [29,32]. Accordingly, c-di-GMP is required for repression of the BldD regulon, and overexpression of the diguanylate cyclases CdgA or CdgB blocks formation of aerial hyphae [3,31]. BldD directly controls expression of cdgA and cdgB, a regulatory mechanism that may prevent excessive accumulation of c-di-GMP. A bldD mutant likewise exhibits a bald phenotype as it bypasses formation of aerial hyphae and sporulates prematurely [32]. The production of antibiotics correlates with these morphological phenotypes as both overexpression of diguanylate cyclases and inactivation of bldD leads to a defect in ACT production [73]. BldD controls activity of the adpA gene (bldH in S. coelicolor), which encodes an AraC-family activator of actII-ORF4 and redD expression, and it binds directly to nsdA, which encodes a repressor of ACT production [3,74]. Accordingly, either absence of BldD or hyper-activation of its DNA binding by excess c-di-GMP is likely to result in failure to control expression of cluster-situated activators of ACT and RED. Since XdhR also binds actII-ORF4, and since DNA binding by XdhR is attenuated by c-di-GMP (Sivapragasam and Grove, unpublished), we speculate that XdhR may also participate in fine-tuning of actII-ORF expression in response to cellular levels of c-di-GMP.
6. Conclusions
Antibiotic production in Streptomyces spp. is linked to the stringent response and morphological differentiation (Figure 3). During vegetative growth, cellular GTP levels are high, conditions under which synthesis of c-di-GMP would be favored, and BldD–c-di-GMP represses bld and whi genes. As (p)ppGpp levels increase, the attendant decrease in cellular GTP levels is associated with de-repression of bld and whi genes and erection of aerial hyphae and sporulation. By contrast, antibiotic production generally occurs as a direct consequence of (p)ppGpp production, with BldD–c-di-GMP playing a poorly understood role in controlling expression of several regulators of antibiotic biosynthetic genes. As both signaling molecules are synthesized from GTP, purine metabolism affects their accumulation, yet GTP homeostasis is likely controlled by distinct mechanisms in different bacterial species, as evidenced for example by the specific purine metabolic enzymes that are direct targets for (p)ppGpp. Notably, recent evidence suggests that the transcription factor XdhR furnishes a direct link between purine salvage and ACT production. Accumulation of (p)ppGpp is associated with increased production of Xdh, as (p)ppGpp attenuates binding of XdhR to the xdh promoter; this would circumvent the (p)ppGpp-mediated inhibition of GuaB and allow GTP production to proceed via xanthine and XMP. Under these conditions, XdhR-mediated repression of actII-ORF4 would also be relieved by (p)ppGpp. Evidently, finely tuned mechanisms operate to control guanosine metabolism, in turn impinging on production of antibiotics.
Author Contributions
S.S. and A.G; writing—original draft preparation, review and editing, A.G.; visualization.
Funding
Research in the authors’ lab was supported by National Science Foundation award MCB-1714219 (to Anne Grove).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed]
- Den Hengst, C.D.; Tran, N.T.; Bibb, M.J.; Chandra, G.; Leskiw, B.K.; Buttner, M.J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010, 78, 361–379. [Google Scholar] [CrossRef]
- Procopio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araujo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Cashel, M.; Gallant, J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef]
- Gaca, A.O.; Colomer-Winter, C.; Lemos, J.A. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 2015, 197, 1146–1156. [Google Scholar] [CrossRef]
- Zhang, Y.; Zbornikova, E.; Rejman, D.; Gerdes, K. Novel (p)ppGpp Binding and Metabolizing Proteins of Escherichia coli. mBio 2018, 9. [Google Scholar] [CrossRef]
- Kriel, A.; Bittner, A.N.; Kim, S.H.; Liu, K.; Tehranchi, A.K.; Zou, W.Y.; Rendon, S.; Chen, R.; Tu, B.P.; Wang, J.D. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol. Cell 2012, 48, 231–241. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Bellows, L.E.; Wood, A.; Grundling, A. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2016, 113, 1710–1719. [Google Scholar] [CrossRef]
- Wang, B.; Dai, P.; Ding, D.; Del Rosario, A.; Grant, R.A.; Pentelute, B.L.; Laub, M.T. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 2019, 15, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Agirrezabala, X.; Fernandez, I.S.; Kelley, A.C.; Carton, D.G.; Ramakrishnan, V.; Valle, M. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013, 14, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.J.; Barker, M.M.; Ross, W.; Schneider, D.A.; Webb, C.; Foster, J.W.; Gourse, R.L. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 2004, 118, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.; Vrentas, C.E.; Sanchez-Vazquez, P.; Gaal, T.; Gourse, R.L. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 2013, 50, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Gourse, R.L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004, 23, 4473–4483. [Google Scholar] [CrossRef] [PubMed]
- Vrentas, C.E.; Gaal, T.; Berkmen, M.B.; Rutherford, S.T.; Haugen, S.P.; Vassylyev, D.G.; Ross, W.; Gourse, R.L. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008, 377, 551–564. [Google Scholar] [CrossRef]
- Geiger, T.; Wolz, C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 2014, 304, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Kriel, A.; Brinsmade, S.R.; Tse, J.L.; Tehranchi, A.K.; Bittner, A.N.; Sonenshein, A.L.; Wang, J.D. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J. Bacteriol. 2014, 196, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Tiserova, H.; Jonak, J.; Rejman, D.; Sanderova, H. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus Subtilis. Mol. Microbiol. 2008, 69, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.G.; Kim, E.S.; Kim, D.W.; Kim, S.K.; Lee, K.J. Differential stringent responses of Streptomyces coelicolor M600 to starvation of specific nutrients. J. Microbiol. Biotechnol. 2007, 17, 305–312. [Google Scholar]
- Hesketh, A.; Sun, J.; Bibb, M. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 2001, 39, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.G.; Jin, W.; Kim, J.Y.; Kim, J.Y.; Lee, S.H.; Lee, K.J. Stringent factor regulates antibiotics production and morphological differentiation of Streptomyces clavuligerus. J. Microbiol. Biotechnol. 2004, 14, 1170–1175. [Google Scholar]
- Ochi, K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: Significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 1987, 169, 3608–3616. [Google Scholar] [CrossRef] [PubMed]
- Strauch, E.; Takano, E.; Baylis, H.A.; Bibb, M.J. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 1991, 5, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Riesenberg, D.; Bergter, F.; Kari, C. Effect of serine hydroxamate and methyl alpha-D-glucopyranoside treatment on nucleoside polyphosphate pools, RNA and protein accumulation in Streptomyces hygroscopicus. J. Gen. Microbiol. 1984, 130, 2549–2558. [Google Scholar] [PubMed]
- Bascaran, V.; Sanchez, L.; Hardisson, C.; Brana, A.F. Stringent response and initiation of secondary metabolism in Streptomyces clavuligerus. J. Gen. Microbiol. 1991, 137, 1625–1634. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Martin, J.F.; Hesketh, A.; Bibb, M.J.; Liras, P. Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: Negative regulation of secondary metabolism by (p)ppGpp. Microbiology 2008, 154, 744–755. [Google Scholar] [CrossRef]
- Ochi, K. Changes in Nucleotide Pools during Sporulation of Streptomyces-Griseus in Submerged Culture. J. Gen. Microbiol. 1987, 133, 2787–2795. [Google Scholar] [CrossRef][Green Version]
- McCormick, J.R.; Flardh, K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 2012, 36, 206–231. [Google Scholar] [CrossRef]
- Hull, T.D.; Ryu, M.H.; Sullivan, M.J.; Johnson, R.C.; Klena, N.T.; Geiger, R.M.; Gomelsky, M.; Bennett, J.A. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces Coelicolor. J. Bacteriol. 2012, 194, 4642–4651. [Google Scholar] [CrossRef]
- Tran, N.T.; Den Hengst, C.D.; Gomez-Escribano, J.P.; Buttner, M.J. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 2011, 193, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Tschowri, N.; Schumacher, M.A.; Schlimpert, S.; Chinnam, N.B.; Findlay, K.C.; Brennan, R.G.; Buttner, M.J. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 2014, 158, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.K.; Higgins, M.B.; Rabinowitz, J.D. Antifolate-Induced Depletion of Intracellular Glycine and Purines Inhibits Thymineless Death in E. coli. ACS Chem. Biol. 2010, 5, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Bosso, J.A.; Mauldin, P.D.; Salgado, C.D. The association between antibiotic use and resistance: The role of secondary antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ochi, K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Env. Microbiol. 2001, 67, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Shentu, X.P.; Cao, Z.Y.; Xiao, Y.; Tang, G.; Ochi, K.; Yu, X.P. Substantial improvement of toyocamycin production in Streptomyces diastatochromogenes by cumulative drug-resistance mutations. PloS ONE 2018, 13, 0203006. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bedre, R.; Thapa, S.S.; Sabrin, A.; Wang, G.; Dassanayake, M.; Grove, A. Global Awakening of Cryptic Biosynthetic Gene Clusters in Burkholderia thailandensis. ACS Chem. Biol. 2017, 12, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Huang, J.; Shi, J.; Molle, V.; Sohlberg, B.; Weaver, D.; Bibb, M.J.; Karoonuthaisiri, N.; Lih, C.J.; Kao, C.M.; Buttner, M.J.; et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 2005, 58, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp conjures bacterial virulence. Microbiol. Mol. Biol Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Mantsala, P.; Zalkin, H. Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP. J. Bacteriol. 1992, 174, 1883–1890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hedstrom, L.; Liechti, G.; Goldberg, J.B.; Gollapalli, D.R. The antibiotic potential of prokaryotic IMP dehydrogenase inhibitors. Curr. Med. Chem. 2011, 18, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Schneider, B.L.; Reitzer, L. Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J. Bacteriol. 2000, 182, 5332–5341. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.W.; Lawrence, K.A.; Bestor, A.; Byram, R.; Gherardini, F.; Rosa, P.A. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 2009, 191, 6231–6241. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Sutchu, S.; Rosa, P.A.; Byram, R.; Jewett, M.W. Borrelia burgdorferi harbors a transport system essential for purine salvage and mammalian infection. Infect. Immun. 2012, 80, 3086–3093. [Google Scholar] [CrossRef] [PubMed]
- Drecktrah, D.; Lybecker, M.; Popitsch, N.; Rescheneder, P.; Hall, L.S.; Samuels, D.S. The Borrelia burgdorferi RelA/SpoT Homolog and Stringent Response Regulate Survival in the Tick Vector and Global Gene Expression during Starvation. PLoS Pathog. 2015, 11, 1005160. [Google Scholar] [CrossRef] [PubMed]
- Novak, E.A.; Sultan, S.Z.; Motaleb, M.A. The cyclic-di-GMP signaling pathway in the Lyme disease spirochete, Borrelia burgdorferi. Front. Cell Infect. Microbiol. 2014, 4, 56. [Google Scholar] [CrossRef]
- Krol, E.; Becker, A. ppGpp in Sinorhizobium meliloti: Biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol. Microbiol. 2011, 81, 1233–1254. [Google Scholar] [CrossRef]
- Sivapragasam, S.; Grove, A. Streptomyces coelicolor XdhR is a direct target of (p)ppGpp that controls expression of genes encoding xanthine dehydrogenase to promote purine salvage. Mol. Microbiol. 2016, 100, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; Beresford, M.; Epton, H.A.; Sigee, D.C.; Shama, G.; Andrew, P.W.; Roberts, I.S. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol 2002, 184, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Gaca, A.O.; Kajfasz, J.K.; Miller, J.H.; Liu, K.Q.; Wang, J.D.; Abranches, J.; Lemos, J.A. Basal Levels of (p)ppGpp in Enterococcus faecalis: The Magic beyond the Stringent Response. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, L.C.; Schou, S.; Nygaard, P.; Saxild, H.H. Xanthine metabolism in Bacillus subtilis: Characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 1997, 179, 2540–2550. [Google Scholar] [CrossRef] [PubMed]
- Sivapragasam, S.; Deochand, D.K.; Meariman, J.K.; Grove, A. The Stringent Response Induced by Phosphate Limitation Promotes Purine Salvage in Agrobacterium fabrum. Biochemistry 2017, 56, 5831–5843. [Google Scholar] [CrossRef]
- Hillerich, B.; Westpheling, J. A new TetR family transcriptional regulator required for morphogenesis in Streptomyces coelicolor. J. Bacteriol. 2008, 190, 61–67. [Google Scholar] [CrossRef]
- Sprusansky, O.; Zhou, L.; Jordan, S.; White, J.; Westpheling, J. Identification of three new genes involved in morphogenesis and antibiotic production in Streptomyces coelicolor. J. Bacteriol. 2003, 185, 6147–6157. [Google Scholar] [CrossRef]
- Fu, J.; Zong, G.; Zhang, P.; Zhao, Z.; Ma, J.; Pang, X.; Cao, G. XdhR negatively regulates actinorhodin biosynthesis in Streptomyces coelicolor M145. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Deutscher, M.P. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 2009, 85, 369–391. [Google Scholar]
- Hsu, D.; Shih, L.M.; Zee, Y.C. Degradation of rRNA in Salmonella strains: A novel mechanism to regulate the concentrations of rRNA and ribosomes. J. Bacteriol. 1994, 176, 4761–4765. [Google Scholar] [CrossRef][Green Version]
- Molin, S.; Von Meyenburg, K.; Maaloe, O.; Hansen, M.T.; Pato, M.L. Control of ribosome synthesis in Escherichia coli: Analysis of an energy source shift-down. J. Bacteriol. 1977, 131, 7–17. [Google Scholar]
- Gramajo, H.C.; Takano, E.; Bibb, M.J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 1993, 7, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Takano, E.; Gramajo, H.C.; Strauch, E.; Andres, N.; White, J.; Bibb, M.J. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 1992, 6, 2797–2804. [Google Scholar] [CrossRef] [PubMed]
- Chakraburtty, R.; Bibb, M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1997, 179, 5854–5861. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, A.; Chen, W.J.; Ryding, J.; Chang, S.; Bibb, M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 2007, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tanaka, Y.; Ochi, K. The G243D mutation (afsB mutation) in the principal sigma factor sigmaHrdB alters intracellular ppGpp level and antibiotic production in Streptomyces coelicolor A3(2). Microbiology 2010, 156, 2384–2392. [Google Scholar] [CrossRef][Green Version]
- Saito, N.; Xu, J.; Hosaka, T.; Okamoto, S.; Aoki, H.; Bibb, M.J.; Ochi, K. EshA accentuates ppGpp accumulation and is conditionally required for antibiotic production in Streptomyces coelicolor A3(2). J. Bacteriol. 2006, 188, 4952–4961. [Google Scholar] [CrossRef]
- Ochi, K.; Zhang, D.; Kawamoto, S.; Hesketh, A. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 1997, 256, 488–498. [Google Scholar] [CrossRef]
- Hoyt, S.; Jones, G.H. relA is required for actinomycin production in Streptomyces antibioticus. J. Bacteriol. 1999, 181, 3824–3829. [Google Scholar]
- Kelly, K.S.; Ochi, K.; Jones, G.H. Pleiotropic effects of a relC mutation in Streptomyces antibioticus. J. Bacteriol. 1991, 173, 2297–2300. [Google Scholar] [CrossRef]
- Jin, W.; Ryu, Y.G.; Kang, S.G.; Kim, S.K.; Saito, N.; Ochi, K.; Lee, S.H.; Lee, K.J. Two relAlspoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces Clavuligerus. Microbiol. 2004, 150, 1485–1493. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, Z.; You, D.; Tang, L.Y.; Zhou, Y.; Ye, B.C. Metabolic Engineering Strategies Based on Secondary Messengers (p)ppGpp and C-di-GMP To Increase Erythromycin Yield in Saccharopolyspora erythraea. ACS Synth. Biol. 2019, 8, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Elliot, M.; Damji, F.; Passantino, R.; Chater, K.; Leskiw, B. The bldD gene of Streptomyces coelicolor A3(2): A regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 1998, 180, 1549–1555. [Google Scholar] [PubMed]
- Park, S.S.; Yang, Y.H.; Song, E.; Kim, E.J.; Kim, W.S.; Sohng, J.K.; Lee, H.C.; Liou, K.K.; Kim, B.G. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J. Indust. Microbiol. Biotechnol. 2009, 36, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).