Genetic Profiling and Comparison of Human and Animal Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Serbia
Abstract
1. Introduction
2. Results
2.1. Antibiotic Susceptibility Testing
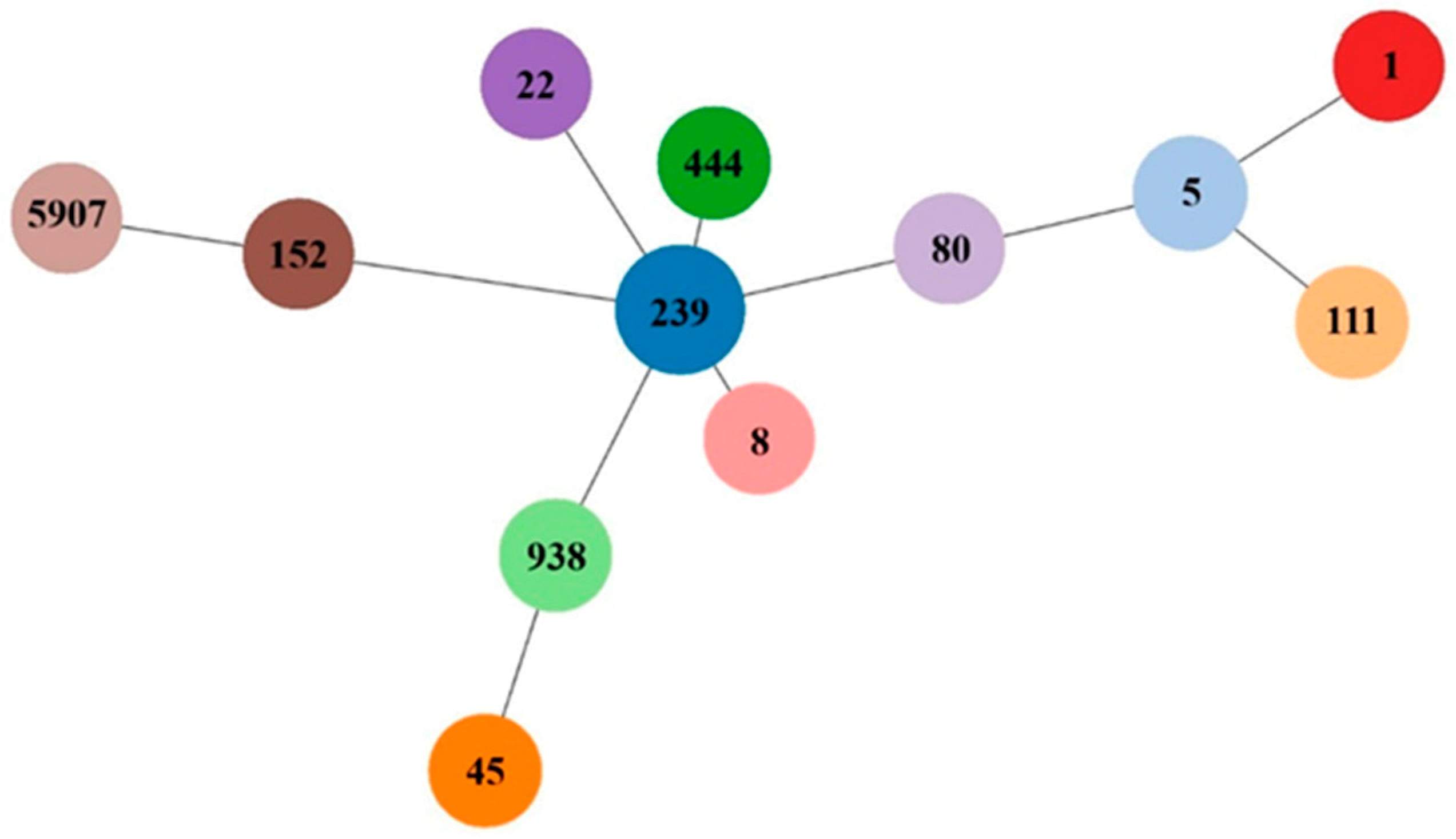
2.2. Molecular Characterization
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Identification of Methicillin-Resistant Staphylococcus aureus
4.3. Antibiotic Susceptibility Testing
4.4. Genotyping of MRSA
4.5. Detection of Virulence and Other Determinants
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morgan, M. Methicillin-resistant Staphylococcus aureus and animals: Zoonosis or humanosis? J. Antimicrob. Chemother. 2008, 62, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for Reporting Novel SCCmec Elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Slickers, P.; Gawlik, D.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; de Jäckel, S.C.; Feßler, A.T.; Frank, M.; Hotzel, H.; Kadlec, K.; et al. Variability of SCCmec elements in livestock-associated CC398 MRSA. Vet. Microbiol. 2018, 217, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Baig, S.; Johannesen, T.B.; Overballe-Petersen, S.; Larsen, J.; Larsen, A.R.; Stegger, M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2018, 61, 74–76. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Köck, R.; Becker, K.; Cookson, B.; van Gemert-Pijnen, J.E.; Harbarth, S.; Kluytmans, J.; Mielke, M.; Peters, G.; Skov, R.L.; Struelens, M.J.; et al. Methicillin-resistant Staphylococcus aureus (MRSA): Burden of disease and control challenges in Europe. Eur. Surveill. 2010, 15, 19688. [Google Scholar] [CrossRef]
- Malachowa, N.; DeLeo, F. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A Field Guide to Pandemic, Epidemic and Sporadic Clones of Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef]
- McDougal, L.K.; Fosheim, G.E.; Nicholson, A.; Bulens, S.N.; Limbago, B.M.; Shearer, J.E.S.; Summers, A.O.; Patel, J.B. Emergence of Resistance among USA300 Methicillin-Resistant Staphylococcus aureus Isolates Causing Invasive Disease in the United States. Antimicrob. Agents Chemother. 2010, 54, 3804–3811. [Google Scholar] [CrossRef]
- Uhlemann, A.C.; Otto, M.; Lowy, F.D.; DeLeo, F.R. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2013, 21, 563–574. [Google Scholar] [CrossRef]
- Morris, D.O.; Loeffler, A.; Davis, M.F.; Guardabassi, L.; Weese, J.S. Recommendations for approaches to meticillin-resistant staphylococcal infections of small animals: Diagnosis, therapeutic considerations and preventative measures. Clinical Consensus Guidelines of the World Association for Veterinary Dermatology. Vet. Dermatol. 2017, 28, 304.e69. [Google Scholar] [CrossRef]
- De Backer, S.; Xavier, B.B.; Vanjari, L.; Coppens, J.; Lammens, C.; Vemu, L.; Carevic, B.; Hryniewicz, W.; Jorens, P.; Kumar-Singh, S.; et al. Remarkable geographical variations between India and Europe in carriage of the staphylococcal surface protein-encoding sasX/sesI and in the population structure of methicillin-resistant Staphylococcus aureus belonging to clonal complex 8. Clin. Microbiol. Infect. 2018. [Google Scholar] [CrossRef]
- Cirkovic, I.; Stepanovic, S.; Skov, R.; Trajkovic, J.; Grgurevic, A.; Larsen, A.R. Carriage and genetic diversity of methicillin-resistant Staphylococcus aureus among patients and healthcare workers in a Serbian University Hospital. PLoS ONE 2015, 10, e0127347. [Google Scholar] [CrossRef]
- Cirkovic, I.; Djukic, S.; Carevic, B.; Mazic, N.; Mioljevic, V.; Stepanovic, S. Methicillin-resistant Staphylococcus aureus nasal carriage among hospitalized patients and healthcare workers in the Clinical Centre of Serbia. Arch. Biol Sci. 2014, 66, 87–92. [Google Scholar] [CrossRef]
- Cirkovic, I.; Sørum, M.; Radenkovic, D.; Vlahovic, M.S.; Larsen, A.R. National surveillance reveals findings of Panton-Valentine leukocidin positive meticillin-resistant Staphylococcus aureus in Serbia. J. Med. Microbiol. 2013, 62, 342–344. [Google Scholar] [CrossRef]
- Zutic, M.; Cirkovic, I.; Pavlovic, Lj.; Zutic, J.; Asanin, J.; Radanovic, O.; Pavlovic, N. Occurrence of methicillin-resistant Staphylococcus aureus in milk samples from Serbian cows with subclinical mastitis. Afr. J. Microbiol. Res. 2012, 6, 5887–5889. [Google Scholar] [CrossRef]
- Zutic, M.; Cirkovic, I.; Pavlovic, Lj.; Asanin, J.; Jovanovic, S.; Zutic, J.; Asanin, R. First isolation of methicillin-resistant Staphylococcus aureus from pigs’clinical samples in Serbia. Acta Vet. Brno. 2012, 81, 225–227. [Google Scholar] [CrossRef]
- Velebit, B.; Fetsch, A.; Mirilovic, M.; Teodorovic, V.; Jovanovic, M. MRSA in pigs in Serbia. Vet. Rec. 2010, 167, 183–184. [Google Scholar] [CrossRef]
- Sweeney, M.; Lubbers, B.; Schwarz, S.; Watts, J. Applying definitions for multidrug resistance extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef]
- Van Belkum, A.; Melles, D.C.; Nouwen, J.; van Leeuwen, W.B.; van Wamel, W.; Vos, M.C.; Wertheim, H.F.L.; Verbrugh, H.A. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 2009, 9, 32–47. [Google Scholar] [CrossRef]
- Bierowiec, K.; Płoneczka-Janeczko, K.; Rypuła, K. Is the Colonisation of Staphylococcus aureus in Pets Associated with Their Close Contact with Owners? PLoS ONE 2016, 11, e0156052. [Google Scholar] [CrossRef]
- Guardabassi, L.; Larsen, J.; Weese, J.S.; Butaye, P.; Battisti, A.; Kluytmans, J.; Lloyd, D.H.; Skov, R.L. Public health impact and antimicrobial selection of meticillin-resistant staphylococci in animals. J. Glob. Antimicrob. Resist. 2013, 1, 55–62. [Google Scholar] [CrossRef]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet animals as reservoirs of antimicrobial-resistant bacteria: Review. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef]
- Loncaric, I.; Brunthaler, R.; Spergser, J. Suspected goat-to-human transmission of methicillin-resistant Staphylococcus aureus sequence type 398. J. Clin. Microbiol. 2013, 51, 1625–1626. [Google Scholar] [CrossRef]
- Loncaric, I.; Künzel, F.; Klang, A.; Wagner, R.; Licka, T.; Grunert, T.; Feßler, A.T.; Geier-Dömling, D.; Rosengarten, R.; Müller, E.; et al. Carriage of meticillin-resistant staphylococci between humans and animals on a small farm. Vet. Dermatol. 2016, 27, 191-e48. [Google Scholar] [CrossRef]
- Baldan, R.; Rancoita, P.M.V.; Di Serio, C.; Mazzotti, M.; Cichero, P.; Ossi, C.; Biancardi, A.; Nizzero, P.; Saracco, A.; Scarpellini, P.; et al. Epidemic MRSA clone ST22-IV is more resistant to multiple host- and environment-related stresses compared with ST228-I. J. Antimicrob. Chemother. 2015, 70, 757–765. [Google Scholar] [CrossRef]
- Asadollahi, P.; Farahani, N.N.; Mirzaii, M.; Khoramrooz, S.S.; van Belkum, A.; Asadollahi, K.; Dadashi, M.; Darban-Sarokhalil, D. Distribution of the Most Prevalent Spa Types among Clinical Isolates of Methicillin-Resistant and –Susceptible Staphylococcus aureus around the World: A Review. Front. Microbiol. 2018, 9, 163. [Google Scholar] [CrossRef]
- Smyth, D.S.; McDougal, L.K.; Gran, F.W.; Manoharan, A.; Enright, M.C.; Song, J.-H.; de Lencastre, H.; Robinson, D.A. Population Structure of a Hybrid Clonal Group of Methicillin-Resistant Staphylococcus aureus, ST239-MRSA-III. PLoS ONE 2010, 5, e8582. [Google Scholar] [CrossRef]
- Harris, S.R.; Feil, E.J.; Holden, M.T.G.; Quail, M.A.; Nickerson, E.K.; Chantratita, N.; Gardete, S.; Tavares, A.; Day, N.; Lindsay, J.A.; et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010, 327, 469–474. [Google Scholar] [CrossRef]
- Monecke, S.; Slickers, P.; Gawlik, D.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; Akpaka, P.E.; Bandt, D.; Bes, M.; Boswihi, S.S.; et al. Molecular Typing of ST239-MRSA-III from Diverse Geographic Locations and the Evolution of the SCCmec III Element during Its Intercontinental Spread. Front. Microbiol. 2018, 9, 1436. [Google Scholar] [CrossRef]
- Stegger, M.; Wirth, T.; Andersen, P.S.; Skov, R.L.; De Grassi, A.; Simões, P.M.; Tristan, A.; Petersen, A.; Aziz, M.; Kiil, K.; et al. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. MBio 2014, 5, e01044-14. [Google Scholar] [CrossRef]
- Larsen, A.R.; Böcher, S.; Stegger, M.; Goering, R.; Pallesen, L.V.; Skov, R. Epidemiology of European Community-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 80 Type IV Strains Isolated in Denmark from 1993 to 2004. J. Clin. Microbiol. 2008, 46, 62–68. [Google Scholar] [CrossRef]
- Budimir, A.; Deurenberg, R.H.; Bosnjak, Z.; Stobberingh, E.E.; Cetkovic, H.; Kalenic, S. A variant of the Southern German clone of methicillin-resistant Staphylococcus aureus is predominant in Croatia. Clin. Microbiol. Infect. 2010, 16, 1077–1083. [Google Scholar] [CrossRef]
- Petersson, A.C.; Olsson-Liljequist, B.; Miorner, H.; Hæggman, S. Evaluating the usefulness of spa typing, in comparison with pulsed-field gel electrophoresis, for epidemiological typing of methicillin-resistant Staphylococcus aureus in a low-prevalence region in Sweden 2000–2004. Clin. Microbiol. Infect. 2010, 16, 456–462. [Google Scholar] [CrossRef]
- Strauß, L.; Stegger, M.; Akpaka, P.E.; Alabi, A.; Breurec, S.; Coombs, G.; Egyir, B.; Larsen, A.R.; Laurent, F.; Monecke, S.; et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. PNAS 2017, 114, E10596–E10604. [Google Scholar] [CrossRef]
- Garofalo, A.; Giai, C.; Lattar, S.; Gardella, N.; Mollerach, M.; Kahl, B.C.; Becker, K.; Prince, A.S.; Sordelli, D.O.; Gómez, M.I. The Length of the Staphylococcus aureus Protein A Polymorphic Region Regulates Inflammation: Impact on Acute and Chronic Infection. J. Infect. Dis. 2012, 206, 81–90. [Google Scholar] [CrossRef]
- Rijnders, M.I.A.; Deurenberg, R.H.; Boumans, M.L.L.; Hoogkamp-Korstanje, J.A.A.; Beisser, P.S.; Stobberingh, E.E. The Antibiotic Resistance Surveillance Group. Population Structure of Staphylococcus aureus Strains Isolated from Intensive Care Unit Patients in The Netherlands over an 11-Year Period (1996 to 2006). J. Clin. Microbiol. 2009, 47, 4090–4095. [Google Scholar] [CrossRef]
- Cuny, C.; Layer, F.; Werner, G.; Harmsen, D.; Daniels-Haardt, I.; Jurke, A.; Mellmann, A.; Witte, W.; Köck, R. State-wide surveillance of antibiotic resistance patterns and spa types of methicillin-resistant Staphylococcus aureus from blood cultures in North Rhine-Westphalia, 2011–2013. Clin. Microbiol. Infect. 2015, 21, 750–757. [Google Scholar] [CrossRef]
- Aqel, A.A.; Alzoubi, H.M.; Vickers, A.; Pichon, B.; Kearns, A.M. Molecular epidemiology of nasal isolates of methicillin-resistant Staphylococcus aureus from Jordan. J. Infect. Public Health 2015, 8, 90–97. [Google Scholar] [CrossRef]
- Udo, E.E.; Boswihi, S.S.; Al-Sweih, N. High prevalence of toxic shock syndrome toxin–producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infect. 2016, 12, 24–30. [Google Scholar] [CrossRef]
- Franco, A.; Hasman, H.; Iurescia, M.; Lorenzetti, R.; Stegger, M.; Pantosti, A.; Feltrin, F.; Ianzano, A.; Porrero, M.C.; Liapi, M.; et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J. Antimicrob. Chemother. 2011, 66, 1231–1235. [Google Scholar] [CrossRef]
- Hopman, J.; Peraza, G.T.; Espinosa, F.; Klaassen, C.; Velázquez, D.; Meis, J.; Voss, A. Methicillin-resistant Staphylococcus aureus without borders: USA300 in Cuba. BMC Proc. 2011, 5 (Suppl. 6), 172. [Google Scholar] [CrossRef]
- Wendlandt, S.; Kadlec, K.; Feßler, A.T.; Monecke, S.; Ehricht, R.; van de Giessen, A.W.; Hengeveld, P.D.; Huijsdens, X.; Schwarz, S.; van Duijkeren, E. Resistance phenotypes and genotypes of methicillin-resistant Staphylococcus aureus isolates from broiler chickens at slaughter and abattoir workers. J. Antimicrob. Chemother. 2013, 68, 2458–2463. [Google Scholar] [CrossRef]
- Butin, M.; Rasigade, J.P.; Martins-Simões, P.; Meugnier, H.; Lemriss, H.; Goering, R.V.; Kearns, A.; Deighton, M.A.; Denis, O.; Ibrahimi, A.; et al. Wide geographical dissemination of the multiresistant Staphylococcus capitis NRCS-A clone in neonatal intensive-care units. Clin. Microbiol. Infect. 2016, 22, 46–52. [Google Scholar] [CrossRef]
- Bartels, M.D.; Boye, K.; Oliveira, D.C.; Worning, P.; Goering, R.; Westh, H. Associations between dru Types and SCCmec Cassettes. PLoS ONE 2013, 8, e61860. [Google Scholar] [CrossRef]
- Goering, R.V.; Morrison, D.; Al-Doori, Z.; Edwards, G.F.S.; Gemmell, C.G. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 2008, 14, 964–969. [Google Scholar] [CrossRef]
- Lim, K.T.; Yeo, C.C.; Suhaili, Z.; Thong, K.L. Comparison of Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Strains Isolated from a Tertiary Hospital in Terengganu, Malaysia. Jpn. J. Infect. Dis. 2012, 65, 502–509. [Google Scholar] [CrossRef]
- Shore, A.C.; Rossney, A.S.; Kinnevey, P.M.; Brennan, O.M.; Creamer, E.; Sherlock, O.; Dolan, A.; Cunney, R.; Sullivan, D.J.; Goering, R.V.; et al. Enhanced Discrimination of Highly Clonal ST22-Methicillin-Resistant Staphylococcus aureus IV Isolates Achieved by Combining spa, dru, and Pulsed-Field Gel Electrophoresis Typing Data. J. Clin. Microbiol. 2010, 48, 1839–1852. [Google Scholar] [CrossRef]
- Worthing, K.A.; Abraham, S.; Pang, S.; Coombs, G.W.; Saputra, S.; Jordan, D.; Wong, H.S.; Abraham, R.J.; Trott, D.J.; Norris, J.M. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Isolated from Australian Animals and Veterinarians. Microb. Drug. Resist. 2018, 24, 203–212. [Google Scholar] [CrossRef]
- Feßler, A.; Scott, C.; Kadlec, K.; Ehricht, R.; Monecke, S.; Schwarz, S. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 2010, 65, 619–625. [Google Scholar] [CrossRef]
- Ionescu, R.; Mediavilla, J.R.; Chen, L.; Grigorescu, D.O.; Idomir, M.; Kreiswirth, B.N.; Roberts, R.B. Molecular Characterization and Antibiotic Susceptibility of Staphylococcus aureus from a Multidisciplinary Hospital in Romania. Microb. Drug. Resist. 2010, 16, 263–272. [Google Scholar] [CrossRef]
- Chen, L.; Mediavilla, J.R.; Smyth, D.S.; Chavda, K.D.; Ionescu, R.; Roberts, R.B.; Robinson, D.A.; Kreiswirth, B.N. Identification of a Novel Transposon (Tn6072) and a Truncated Staphylococcal Cassette Chromosome mec Element in Methicillin-Resistant Staphylococcus aureus ST239. Antimicrob. Agents Chemother. 2010, 54, 3347–3354. [Google Scholar] [CrossRef]
- Strommenger, B.; Layer, F.; Werner, G. Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in workers in the food industry. In Staphylococcus aureus, 1st ed.; Fetsch, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 163–188. [Google Scholar]
- Hennekinne, J.A. Staphylococcus aureus as a leading cause of foodborne outbreaks worldwide. In Staphylococcus aureus, 1st ed.; Fetsch, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 129–146. [Google Scholar]
- Smyth, D.S.; Hartigan, P.J.; Meaney, W.J.; Fitzgerald, J.R.; Deobald, C.F.; Bohach, G.A.; Smyth, C.J. Superantigen genes encoded by the egc cluster and SaPIbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J. Med. Microbiol. 2005, 54, 401–411. [Google Scholar] [CrossRef]
- Couto, N.; Belas, A.; Kadlec, K.; Schwarz, S.; Pomba, C. Clonal diversity, virulence patterns and antimicrobial and biocide susceptibility among human, animal and environmental MRSA in Portugal. J. Antimicrob. Chemother. 2015, 70, 2483–2487. [Google Scholar] [CrossRef]
- Zarazaga, M.; Gómez, P.; Ceballos, S.; Torres, C. Molecular epidemiology of Staphylococcus aureus lineages in the animal-human interface. In Staphylococcus aureus, 1st ed.; Fetsch, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 189–214. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI supplement M100S; CLSI: Wayne, PA, USA, 2016; pp. 74–80. [Google Scholar]
- Loncaric, I.; Kübber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 2013, 68, 2222–2225. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; AgersŁ, Y.; Ahrens, P.; JŁrgensen, J.C.; Madsen, M.; Jensen, L.B. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 2000, 74, 353–364. [Google Scholar] [CrossRef]
- Martineau, F.; Picard, F.J.; Lansac, N.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus Antimicrob. Agents Chemother. 2000, 44, 231–238. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef]
- Schnellmann, C.; Gerber, V.; Rossano, A.; Jaquier, V.; Panchaud, Y.; Doherr, M.G.; Thomann, A.; Straub, R.; Perreten, V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 2006, 44, 4444–4454. [Google Scholar] [CrossRef]
- Hauschild, T.; Vukovic, D.; Dakić, I.; Ježek, P.; Djukić, S.; Dimitrijević, V.; Stepanović, S.; Schwarz, S. Aminoglycoside Resistance in Members of the Staphylococcus sciuri Group. Microb. Drug Resist. 2007, 13, 77–84. [Google Scholar] [CrossRef]
- Argudín, M.A.; Tenhagen, B.A.; Fetsch, A.; Sachsenröder, J.; Käsbohrer, A.; Schroeter, A.; Hammerl, J.A.; Hertwig, S.; Helmuth, R.; Bräunig, J.; et al. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 2011, 77, 3052–3060. [Google Scholar] [CrossRef]
- Shittu, A.O.; Okon, K.; Adesida, S.; Oyedara, O.; Witte, W.; Strommenger, B.; Layer, F.; Nübel, U. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011, 11, 92. [Google Scholar] [CrossRef]
- Loncaric, I.; Künzel, F.; Licka, T.; Simhofer, H.; Spergser, J.; Rosengarten, R. Identification and characterization of methicillin-resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Vet. Microbiol. 2014, 168, 381–387. [Google Scholar] [CrossRef]
- Schauer, B.; Krametter-Frötscher, R.; Knauer, F.; Ehricht, R.; Monecke, S.; Feßler, A.T.; Schwarz, S.; Grunert, T.; Spergser, J.; Loncaric, I. Diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from Austrian ruminants and New World camelids. Vet. Microbiol. 2018, 215, 77–82. [Google Scholar] [CrossRef]
- Feßler, A.T.; Li, J.; Kadlec, K.; Wang, Y.; Schwarz, S. Antimicrobial resistance properties of Staphylococcus aureus. In Staphylococcus aureus, 1st ed.; Fetsch, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 57–85. [Google Scholar]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef]
- Mehrotra, M.; Wang, G.; Johnson, W.M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 2000, 38, 1032–1035. [Google Scholar]
- Van Wamel, W.J.; Rooijakkers, S.H.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef]
- Peacock, S.J.; Moore, C.E.; Justice, A.; Kantzanou, M.; Story, L.; Mackie, K.; O’Neill, G.; Day, N.P. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 2002, 70, 4987–4996. [Google Scholar] [CrossRef]
| Antimicrobial Resistance | Virulence Factors | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Host | Clinical Site | spa | dru | SCCmec | MLVA * | ST | CC | Phenotype ** | Genes Detected | IEC *** | Miscellaneous Genes **** | |
| S264 | Cat | Skin swab | t127 | dt7o | NT | 15 | ST1 | CC1 | β-Lactams, CIP, GEN, RIF | mecA, aacA-aphD, ant(6’)-la | seg | sak, sea, scn, hlb | fnbA, clfA, clfB, icaA |
| NN | Human | Wound swab | t223 | nt | IV | 18 | ST22 | CC22 | β-Lactams | mecA | seg, sei, tsst-1, PVL | chp, sak, scn, hlb | fnbA, clfB |
| S164 | Human | Sputum | t037 | dt10a | NT | 9 | ST152 | CC152 | β-Lactams, CIP, GEN, TET, CHL, ERY, CLI | mecA,aacA-aphD, ant(6′)-Ia, erm(C), tet(K), tet(M), catpC221 | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, icaA |
| S239 | Human | Nipple discharge | t037 | dt11c | III | 5 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S241 | Human | Wound swab | t037 | dt11c | III + SCCmercury | 6 | ST239 | CC239 | β-Lactams, CIP, GEN, TET, ERY, CHL, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(K), tet(M), fexA, catpC221 | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S245 | Human | Wound swab | t037 | dt11c | III + SCCmercury | 7 | ST239 | CC239 | β-Lactams, CIP, GEN, AMK, TET, CHL | mecA, aacA-aphD, ant(6′)-Ia, tet(K), tet(M), fexA, catpC221 | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S246 | Human | Wound swab | t037 | dt11c | III | 8 | ne | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S255 | Human | Wound swab | t037 | dt11c | III | 8 | ne | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S257 | Human | Sputum | t037 | dt11c | III | 8 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S399 | Human | Wound swab | t037 | dr11c | NT | 10 | ST239 | CC239 | β-Lactams, CIP, GEN | mecA, aacA-aphD, ant(6′)-Ia, tet(M) | sea, sei | sak, sep, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S401 | Human | Wound swab | t037 | dr11c | NT | 11 | ne | CC239 | β-Lactams, CIP, GEN | mecA, aacA-aphD, ant(6′)-Ia, tet(M) | sea | sak, sep, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S473 | Human | Wound swab | t037 | dt11c | III | 11 | ne | CC239 | β-Lactams, CIP, GEN | mecA, aacA-aphD, ant(6′)-Ia, tet(M) | sea | sak, sea, scn, hlb | fnbA, clfB, cna |
| S474 | Human | Wound swab | t038 | dt11c | III | 11 | ne | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea, sei | sak, sea, scn, hlb | fnbA, clfB, cna |
| S475 | Human | Wound swab | t039 | dt11c | III | 12 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia erm(C), tet(M) | - | sak, scn, hlb | fnbA, clfB, cna |
| S476 | Human | Wound swab | t040 | dt11c | III | 11 | ne | CC239 | β-Lactams, CIP, GEN | mecA, aacA-aphD, ant(6′)-Ia, tet(M) | sea | sak, sea, scn, hlb | fnbA, clfB, cna |
| S478 | Human | Wound swab | t041 | dt11c | III | 11 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfB, cna |
| S386 | Dog | Eye swab | t2029 | dt11c | III | 17 | ST239 | CC239 | β-Lactams, CIP, TET, ERY, CLI (inducible) | mecA, ant(6′)-Ia, erm(C), tet(K), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S400 | Human | Wound swab | t4789 | dr11c | NT | 22 | ne | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea, sei | sak, sep, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S402 | Human | Wound swab | t4789 | dr11c | III | 22 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sep, scn | fnbA, clfA, clfB, cna, icaA |
| S403 | Human | Wound swab | t4789 | dr11c | III | 23 | ST239 | CC239 | β-Lactams, CIP, GEN | mecA, aacA-aphD, ant(6′)-Ia, tet(M) | sea | sak, sep, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S480 | Human | Wound swab | t4789 | dt11c | NT | 22 | ne | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, scn, hlb | clfB, cna |
| S479 | Human | Wound swab | t487 | dt11c | III | 25 | ST239 | CC239 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfB, cna |
| S244a | Human | Wound swab | t685 | dt10a | NT | 26 | ST938 | CC30 | β-Lactams, ERY, CLI | mecA, ant(6′)-Ia, erm(A), erm(B) | seg, sei, tsst-1 | sak, scn, hlb | fnbA, clfA, clfB, icaA |
| S244b | Human | Wound swab | t685 | dt10a | IV | 27 | ST938 | CC30 | β-Lactams, ERY, CLI | mecA, erm(A), erm(B) | seg, sei, tsst-1 | sak, scn, hlb | fnbA, clfA, clfB, icaA |
| S398 | Dog | Skin swab | t487 | dt10g | NT | 24 | ST45 | CC45 | β-Lactams | mecA, ant(6′)-Ia | seg, sei | chp, sak, scn | fnbA, clfA, clfB, icaA |
| S395 | Human | Wound swab | nt | nt | NT | 1 | ST111 | CC5 | β-Lactams, CIP, GEN, ERY, CLI, RIF | mecA, aacA-aphD, ant(6’)-la, erm(A), erm(B) | sea | sak, scn, hlb | fnbA, clfB, icaA |
| S258 | Human | Wound swab | t041 | dt8h | I | 13 | ST111 | CC5 | β-Lactams, CIP, GEN, ERY, CLI | mecA, aacA-aphD, ant(6′)-Ia, erm(A), erm(B), tet(M) | sea, seg, sei | sak, sea, scn, hlb | fnbA, clfB, icaA |
| S396 | Human | Nose swab | t12886 | dt10a | IV | 16 | ST5 | CC5 | β-Lactams, ERY, CLI (inducible) | erm(A), erm(B) | sea, sed, seg, sei | chp, sak, scn, hlb | fnbA, clfA, clfB, icaA |
| MRS1 | Dog | Wound swab | t242 | dt10a | NT | 19 | CC5 | β-Lactams, GEN | aacA-aphD | seg, sei | chp, sak, sea, scn, hlb | fnbA, clfA, clfB, icaA | |
| MRS2 | Dog | Wound swab | t242 | dt10a | NT | 19 | ST5 | CC5 | β-Lactams, GEN | aacA-aphD | seg, sei | chp, sak, scn, hlb | fnbA, clfA, clfB, icaA |
| MRS3 | Dog | Ear swab | t242 | dt10a | V | 20 | ST5 | CC5 | β-Lactams | aacA-aphD | seg, sei | chp, sak, scn, hlb | fnbA, clfB |
| S422 | Human | Ear swab | t024 | dt7f | IV | 2 | ST8 | CC8 | β-Lactams, GEN, ERY, CLI (inducible) | aacA-aphD, ant(6′)-Ia, erm(C) | sej | sak, scn, hlb | fnbA, clfA, clfB |
| S423 | Human | Nose swab | t044 | dt10 | IV | 14 | ST80 | CC80 | β-Lactams, TET | aacA-aphD, ant(6′)-Ia, tet(K) | PVL | sak, scn, hlb | fnbA, clfA, clfB, icaA |
| S256 | Human | Wound swab | t030 | dt8a | III + SCCmercury | 3 | ST444 | _ | β-Lactams, CIP, GEN, TET, ERY, CLI, RIF | aacA-aphD, ant(6′)-Ia, erm(A), erm(B), erm(C), tet(M) | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA, qacAB |
| S195 | Human | Skin swab | t030 | dt8a | III | 4 | ST444 | _ | β-Lactams, CIP, GEN, TET, CHL, ERY, CLI, SXT | aacA-aphD, erm(A), erm(B), erm(C), tet(M), catpC221 | sea | sak, sea, scn, hlb | fnbA, clfA, clfB, cna, icaA |
| S394 | Human | Wound swab | t4272 | dt10q | NT | 21 | ST5907 | _ | β-Lactams, GEN, ERY, CLI | aacA-aphD, erm(C) | _ | sak, sea, scn, hlb | fnbA, clfA, clfB, icaA, arsA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asanin, J.; Misic, D.; Aksentijevic, K.; Tambur, Z.; Rakonjac, B.; Kovacevic, I.; Spergser, J.; Loncaric, I. Genetic Profiling and Comparison of Human and Animal Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Serbia. Antibiotics 2019, 8, 26. https://doi.org/10.3390/antibiotics8010026
Asanin J, Misic D, Aksentijevic K, Tambur Z, Rakonjac B, Kovacevic I, Spergser J, Loncaric I. Genetic Profiling and Comparison of Human and Animal Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Serbia. Antibiotics. 2019; 8(1):26. https://doi.org/10.3390/antibiotics8010026
Chicago/Turabian StyleAsanin, Jelena, Dusan Misic, Ksenija Aksentijevic, Zoran Tambur, Bojan Rakonjac, Ivana Kovacevic, Joachim Spergser, and Igor Loncaric. 2019. "Genetic Profiling and Comparison of Human and Animal Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Serbia" Antibiotics 8, no. 1: 26. https://doi.org/10.3390/antibiotics8010026
APA StyleAsanin, J., Misic, D., Aksentijevic, K., Tambur, Z., Rakonjac, B., Kovacevic, I., Spergser, J., & Loncaric, I. (2019). Genetic Profiling and Comparison of Human and Animal Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates from Serbia. Antibiotics, 8(1), 26. https://doi.org/10.3390/antibiotics8010026





