Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics
Abstract
1. Introduction
2. Results and Discussion
2.1. Origin of Phages B1 and JA1
2.2. Morphology and Host Range of Phages K, B1 and JA1
2.3. Phage Adsorption on Phage Resistant Isolates
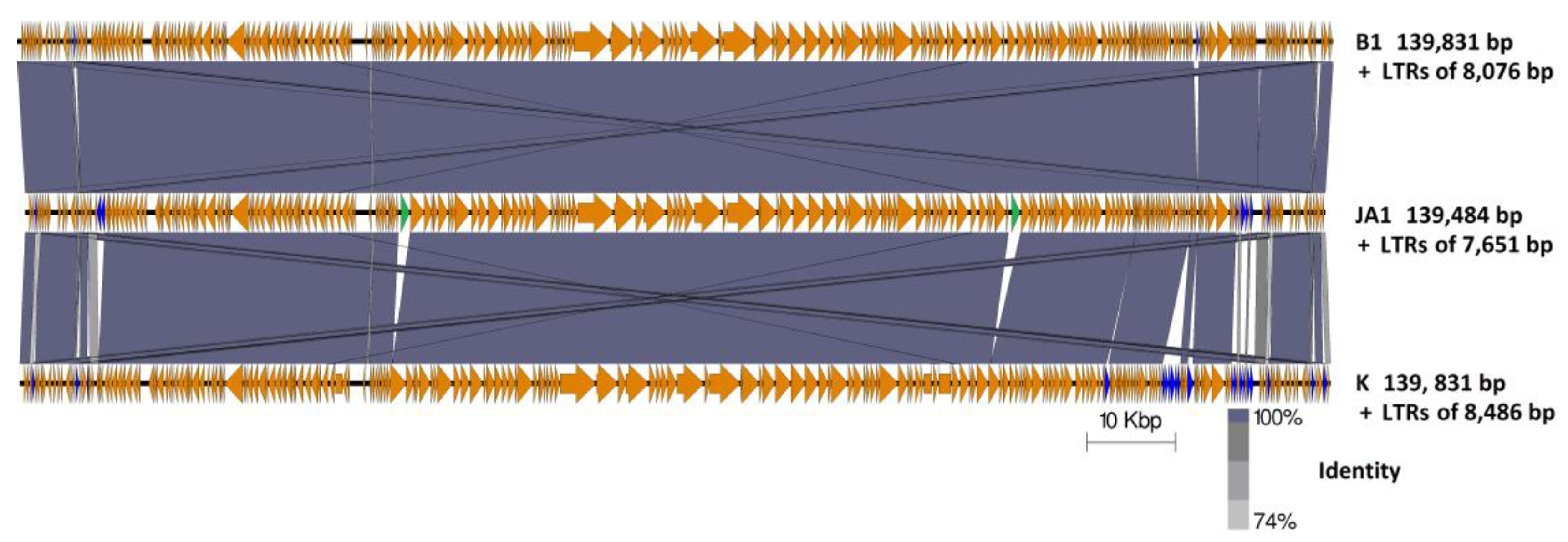
2.4. Genome Comparison between Phages B1, JA1, and K
2.4.1. Characteristic Features of Phage K ORFs Absent in Both JA1 and B1
2.4.2. Characteristic Features of Phages B1 and JA1 ORFs Absent in Phage K
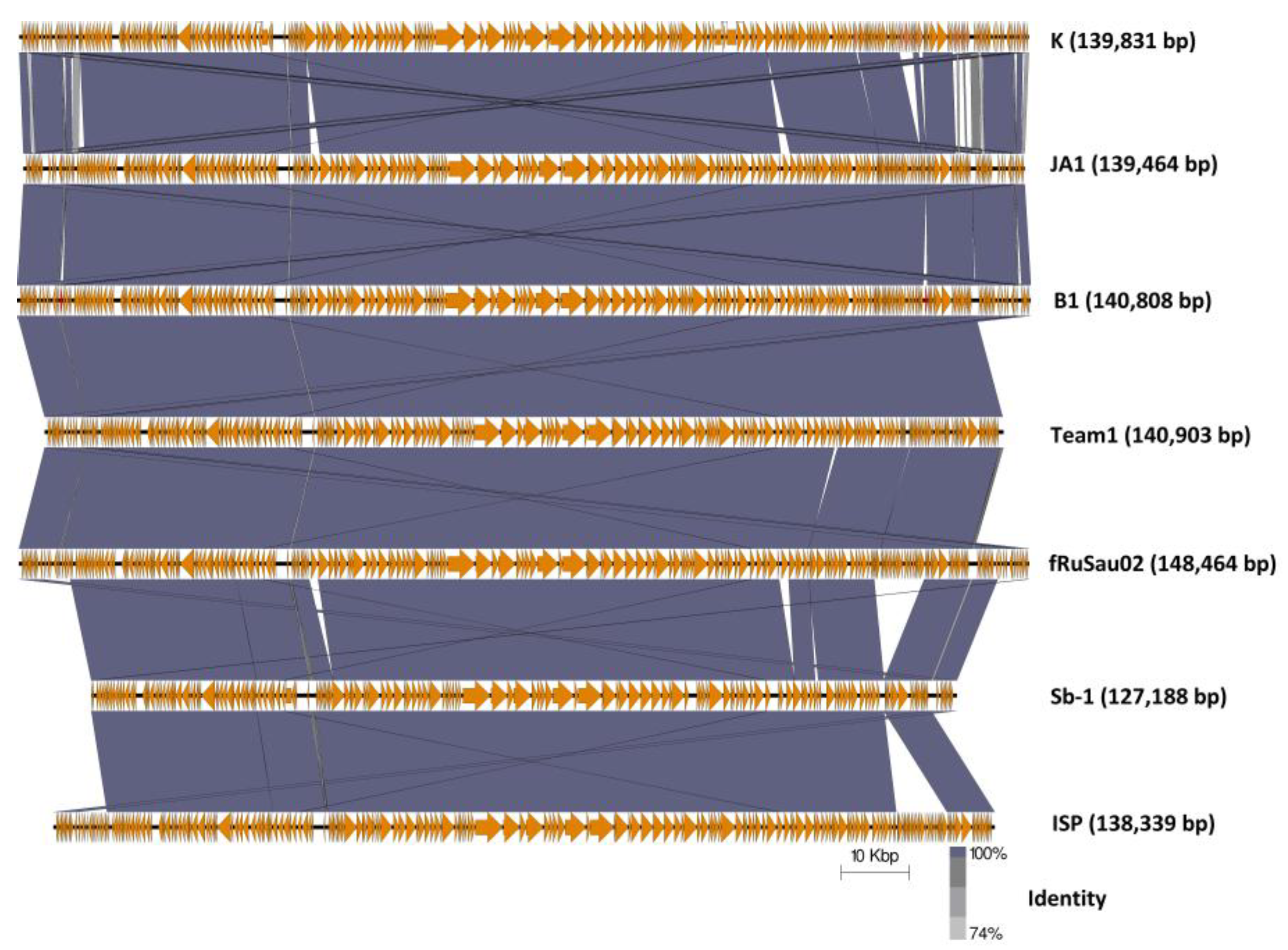
2.4.3. Comparison of Phages K, B1, and JA1 with other Similar Therapeutic Phages (Team1, vB_SauM-fRuSau02, Sb-1 and ISP)
3. Materials and Methods
3.1. Bacterial Strains, Phage and Growth Requirement
3.2. CsCl Gradient Purification
3.3. Phage Host Range and Adsorption Study
3.4. Transmission Electron Microscopy
3.5. Phage DNA Isolation
3.6. Phage DNA Sequencing
3.7. Bioinformatic Analysis
3.8. Nucleotide Sequence Accession Number
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interests
References
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.C.; Rossney, A.S.; O’Connell, B.; Herra, C.M.; Sullivan, D.J.; Humphreys, H.; Coleman, D.C. Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA. Antimicrob. Agents Chemother. 2008, 52, 4407–4419. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Cui, L.; Kuroda, M.; Ito, T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001, 9, 486–493. [Google Scholar] [CrossRef]
- Jevons, M.P. “Celbenin”—Resistant Staphylococci. BMJ 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Klein, E.; Smith, D.L.; Laxminarayan, R. Hospitalizations and Deaths Caused by Methicillin-Resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 2007, 13, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Ross, R.P.; Coffey, A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 2009, 33, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Loessner, M.J. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014, 4, e28137. [Google Scholar] [CrossRef] [PubMed]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Bacteriophage Host Range and Bacterial Resistance. In Advances in Applied Microbiology, 1st ed.; Laskin, A.I., Sarislani, S., Gadd, G.M., Eds.; Elsevier Inc.: Cambridge, MA, USA, 2010; Volume 70, pp. 217–248. ISBN 9780123809919. [Google Scholar]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Scanlan, P.D.; Buckling, A. Bacteria-Phage Coevolution and the Emergence of Generalist Pathogens. Am. Nat. 2011, 177, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.P.J.; Harrison, E.; Brockhurst, M.A. Viral host-adaptation: Insights from evolution experiments with phages. Curr. Opin. Virol. 2013, 3, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, J.; Larsen, M.V.; Kilstrup, M.; Nielsen, M. Metagenomic analysis of therapeutic PYO phage cocktails from 1997 to 2014. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Zschach, H.; Joensen, K.G.; Lindhard, B.; Lund, O.; Goderdzishvili, M.; Chkonia, I.; Jgenti, G.; Kvatadze, N.; Alavidze, Z.; Kutter, E.M.; et al. What can we learn from a metagenomic analysis of a georgian bacteriophage cocktail? Viruses 2015, 7, 6570–6589. [Google Scholar] [CrossRef] [PubMed]
- Kvachadze, L.; Balarjishvili, N.; Meskhi, T.; Tevdoradze, E.; Skhirtladze, N.; Pataridze, T.; Adamia, R.; Topuria, T.; Kutter, E.; Rohde, C.; et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb. Biotechnol. 2011, 4, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Vandersteegen, K.; Mattheus, W.; Ceyssens, P.J.; Bilocq, F.; de Vos, D.; Pirnay, J.P.; Noben, J.P.; Merabishvili, M.; Lipinska, U.; Hermans, K.; et al. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS ONE 2011, 6, e24418. [Google Scholar] [CrossRef] [PubMed]
- Leskinen, K.; Tuomala, H.; Wicklund, A.; Horsma-Heikkinen, J.; Kuusela, P.; Skurnik, M.; Kiljunen, S. Characterization of vB_SauM-fRuSau02, a Twort-Like Bacteriophage Isolated from a Therapeutic Phage Cocktail. Viruses 2017, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- El Haddad, L.; Abdallah, N.B.; Plante, P.L.; Dumaresq, J.; Katsarava, R.; Labrie, S.; Corbeil, J.; St-Gelais, D.; Moineau, S. Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS ONE 2014, 9, e102600. [Google Scholar] [CrossRef] [PubMed]
- Markoishvili, K.; Tsitlanadze, G.; Katsarava, R.; Morris, J.G.; Sulakvelidze, A. A novel sustained-release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int. J. Dermatol. 2002, 41, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Jikia, D.; Chkhaidze, N.; Imedashvili, E.; Mgaloblishvili, I.; Tsitlanadze, G.; Katsarava, R.; Morris, J.G.; Sulakvelidze, A. The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multidrug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90. Clin. Exp. Dermatol. 2005, 30, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Clokie, C.M.R.; Sullivan, M.B.; Gillis, A.; Jens Kuhn, B.H.; Kropinski, A.M. Taxonomy of prokaryotic viruses: 2016 update from the ICTV bacterial and archaeal viruses subcommittee. Arch. Virol. 2017, 162, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Ross, R.P.; Meaney, W.; Fitzgerald, G.F.; Elbreki, M.F.; Coffey, A. Potential of the Polyvalent Anti-Staphylococcus Bacteriophage K for Control of Antibiotic-Resistant Staphylococci from Hospitals. Appl. Environ. Microbiol. 2005, 71, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Rountree, P.M. The serological differentiation of staphylococcal bacteriophages. J. Gen. Microbiol. 1949, 3, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 2001, 146, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Rossney, A.S.; Lawrence, M.J.; Morgan, P.M.; Fitzgibbon, M.M.; Shore, A.; Coleman, D.C.; Keane, C.T.; O’Connell, B. Epidemiological typing of MRSA isolates from blood cultures taken in Irish hospitals participating in the European Antimicrobial Resistance Surveillance System (1999–2003). Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Feng, T.; Gu, F.; Li, Q.; Dong, K.; Zhang, Y.; Zhu, Y.; Han, L.; Qin, J.; Guo, X. Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai, China. Virol. J. 2017, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J. Revised Genome Sequence of Staphylococcus aureus Bacteriophage K. Genome Announc. 2014, 2, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Hendrix, H.; Oliveira, H.; Casey, A.; Neve, H.; McAuliffe, O.; Paul Ross, R.; Hill, C.; Noben, J.P.; O’Mahony, J.; et al. Things are getting hairy: Enterobacteria bacteriophage vB_PcaM_CBB. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fan, H.; An, X.; Fan, H.; Jiang, H.; Chen, Y.; Tong, Y. Scrutinizing virus genome termini by high-throughput sequencing. PLoS ONE 2014, 9, e85806. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E.; Klumpp, J.; Bishop-Lilly, K.A.; Rajavel, M.; Willner, K.M.; Butani, A.; Henry, M.; Biswas, B.; Li, M.; Albert, M.J.; et al. Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol. J. 2013, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.S.; Dąbrowski, K.; Gozdek, A.; Kosakowski, J.; Witkowska, M.; Ulatowska, M.I.; Weber-Dąbrowska, B.; Kwiatek, M.; Parasion, S.; et al. Genomics of staphylococcal Twort-like phages—Potential therapeutics of the post-antibiotic era. Adv. Virus Res. 2012, 83, 143–216. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Coffey, A.; Edwards, R.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. Genome of Staphylococcal Phage K: A New Lineage of Myoviridae Infecting Gram-Positive Bacteria with a Low G+C Content. J. Bacteriol. 2004, 186, 2862–2871. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Rodney Brister, J. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Matange, N.; Podobnik, M.; Visweswariah, S.S. Metallophosphoesterases: structural fidelity with functional promiscuity. Biochem. J. 2015, 467, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, J.P.; Hilario, E. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 2006, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Vandersteegen, K.; Kropinski, A.M.; Nash, J.H.E.; Noben, J.-P.; Hermans, K.; Lavigne, R. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J. Virol. 2013, 87, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Stewart, C.R. A Cytotoxic Early Gene of Bacillus subtilis Bacteriophage SPOl. J. Bacteriol. 1993, 175, 7887–7900. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.R.; Gaslightwala, I.; Hinata, K.; Krolikowski, K.A.; Needleman, D.S.; Peng, A.S.Y.; Peterman, M.A.; Tobias, A.; Wei, P. Genes and regulatory sites of the “host-takeover module” in the terminal redundancy of Bacillus subtilis bacteriophage SPO1. Virology 1998, 246, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Seed, K.D. Battling Phages: How Bacteria Defend against Viral Attack. PLoS Pathog. 2015, 11, e1004847. [Google Scholar] [CrossRef] [PubMed]
- Roberts, G.A.; Houston, P.J.; White, J.H.; Chen, K.; Stephanou, A.S.; Cooper, L.P.; Dryden, D.T.F.; Lindsay, J.A. Impact of target site distribution for Type i restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res. 2013, 41, 7472–7484. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Gao, C.H.; Zhu, J.; Zhao, L.; Wu, Q.; Li, M.; Sun, B. Identification and functional study of type III—A CRISPR-Cas systems in clinical isolates of Staphylococcus aureus. Int. J. Med. Microbiol. 2016, 306, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Purification of bacteriophage lamda particles by isopycnic centrifugation through CsCl gradients. In Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1, p. 247. ISBN 0879695773. [Google Scholar]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two Phages, phiIPLA-RODI and phiIPLA-C1C, Lyse Mono- and Dual-Species Staphylococcal Biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Koç, C.; Kühner, P.; Stierhof, Y.-D.; Krismer, B.; Enright, M.C.; Penadés, J.R.; Wolz, C.; Stehle, T.; Cambillau, C.; et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 2016, 6, 26455. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A web tool for the prediction of rho-independent transcription terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.-A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
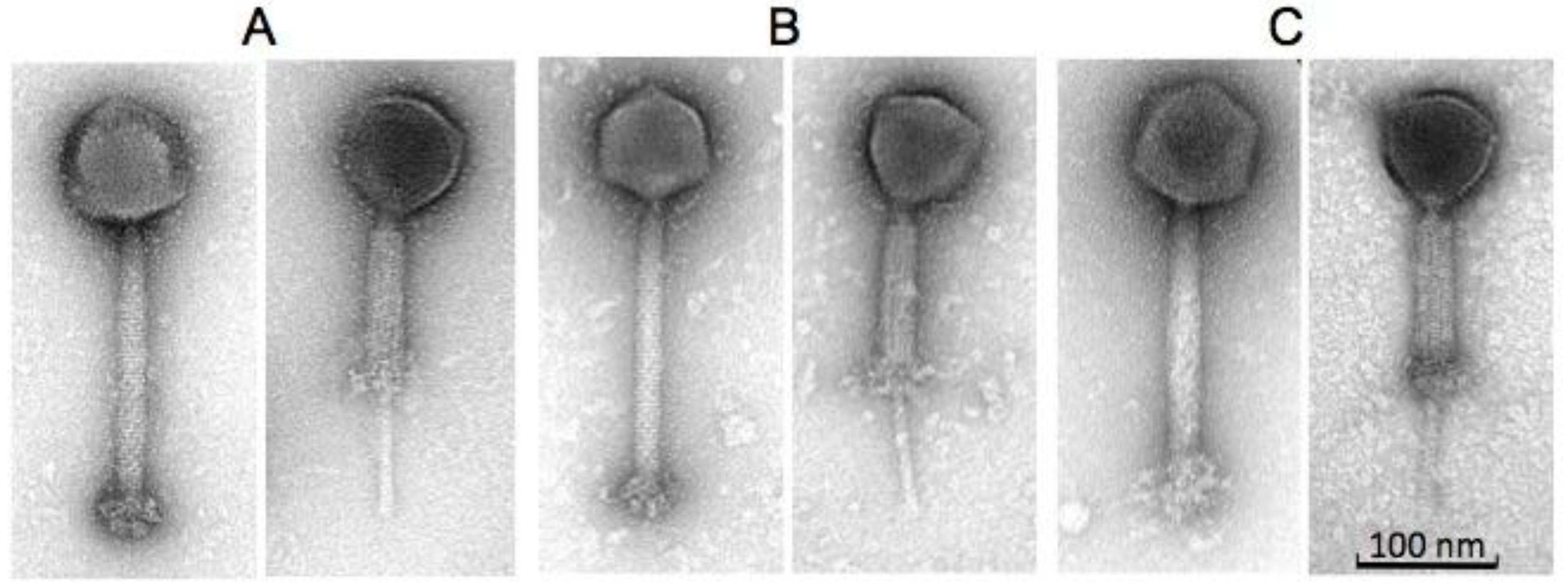
| Phages | Head (nm) | Tail Length (nm) (incl. “knob”) | Tail Width (nm) | Baseplate “knob” Length (nm) | Baseplate “knob” Width (nm) |
|---|---|---|---|---|---|
| B1 | 92.9 ± 4.0 (n = 11) | 233.0 ± 4.4 (n = 12) | 23.4 ± 1.2 (n = 12) | 30.1 ± 1.8 (n = 12) | 47.2 ± 3.7 (n = 10) |
| JA1 | 87.0 ± 2.1 (n = 9) | 231.5 ± 4.7 (n = 9) | 22.7 ± 0.9 (n = 9) | 32.5 ± 7.9 (n = 9) | 45.8 ± 1.4 (n = 9) |
| K | 92.9 ± 3.8 (n = 16) | 227.5 ± 5.5 (n = 16) | 23.8 ± 1.0 (n = 16) | 36.6 ± 5.1 (n = 16) | 41.7 ± 2.6 (n = 16) |
| S. aureus Strain | Phage K | Phage B1 | Phage JA1 |
|---|---|---|---|
| DPC5246* | 1.00 ± 0.0 | 1.00 ± 0.0 | 8.98 × 10−1 ± 0.8 |
| CIT281189* | No infection | No infection | 1.00 ± 0.0 |
| 0.0066 (IIIV) ST239 | No infection | No infection | 2.59 ± 2.5 |
| 0.1206 (IV) ST250 | No infection | 3.89 × 10−1 ± 0.3 | 1.35 ± 1.2 |
| 0.1239 (III) ST239 | No infection | 1.46 × 10−1 ± 0.1 | 4.17 × 10−2 ± 0.0 |
| 0.1345 (II) ST5 | No infection | No infection | 2.08 × 10−1 ± 0.1 |
| 0073 (III) ST239 | No infection | 3.21 × 10−1 ± 0.2 | No infection |
| 0104 (III) ST239 | No infection | 3.95 × 10−1 ± 0.2 | 1.82 ± 1.6 |
| 0220 (II) ST5 | 3.03 × 10−1 ± 0.1 | 2.17 × 10−1 ± 0.2 | 2.38 × 10−1 ± 0.2 |
| 0242 (IV) ST30 | 4.43 × 10−1 ± 0.1 | 5.23 × 10−1 ± 0.5 | 4.90 × 10−1 ± 0.3 |
| 0308 (IA) ST247 | 1.40 ± 0.2 | 1.36 ± 1.3 | 1.71 ± 1.6 |
| 3045 (IIV) ST8 | No infection | 4.93 × 10−2 ± 0.0 | 1.69 ± 0.7 |
| 3144 (IIV) ST8 | No infection | 1.21 ± 1.0 | 2.17 ± 1.2 |
| 3488 (VV) ST8 | No infection | No infection | No infection |
| 3581 (IA) ST247 | No infection | No infection | 9.26 × 10−1 ± 0.7 |
| 3594 (II) ST36 | 4.38 × 10−1 ± 0.1 | 8.67 × 10−1 ± 0.4 | 1.06 ± 0.7 |
| 3596 (IIV) ST8 | 2.49 × 10−4 ± 0.0 | 1.29 ± 0.9 | 3.59 ± 2.7 |
| E1038 (IIV) ST8 | 1.27 × 10−4 ± 0.0 | 2.02 × 10−1 ± 0.2 | 1.89 ± 1.4 |
| E1139 (IV) ST45 | No infection | 3.88 × 10−6 ± 0.0 | No infection |
| E1174 (IV) ST22 | 7.03 × 10−1 ± 0.7 | 3.11 × 10−1 ± 0.2 | No infection |
| E1185 (IV) ST12 | 1.16 × 10−6 ± 0.0 | No infection | No infection |
| E1202 (II) ST496 | No infection | 4.79 × 10−1 ± 0.2 | 9.49 × 10−1 ± 0.8 |
| M03/0073 (III) ST239 | 1.76 ± 0.5 | 1.51 ± 0.8 | 2.30 ± 0.7 |
| S. aureus Strain | Phage K | Phage B1 | Phage JA1 |
|---|---|---|---|
| DPC5246 | 2 mm | 1 mm with halo to 2 mm | 1 mm with halo to 2 mm |
| CIT281189 | No plaques | No plaques | 1.5 mm |
| 0.0066 (IIV) ST239 | No plaques | No plaques | 1 mm |
| 0.1206 (IV) ST250 | No plaques | 2 mm | 0.5 mm with halo to 1 mm |
| 0.1239 (III) ST239 | No plaques | 0.5 mm, faint plaques | 1 mm |
| 0.1345 (II) ST5 | No plaques | No plaques | 1 mm |
| 0073 (III) ST239 | No plaques | 0.5 mm | No plaques |
| 0104 (III) ST239 | No plaques | 0.5 mm | 1 mm |
| 0220 (II) ST5 | 0.5 mm | 1 mm | 1 mm |
| 0242 (IV) ST30 | 1 mm | 1.5 mm | 1.5 mm |
| 0308 (IA) ST247 | 1 mm | 1 mm | 0.5 mm, faint plaques |
| 3045 (IIV) ST8 | No plaques | 1 mm | 1 mm |
| 3144 (IIV) ST8 | No plaques | 1.5 mm, faint plaques | 1 mm |
| 3488 (VV) ST8 | No plaques | 0.5 mm, faint plaques | 0.5 mm with halo to 1 mm |
| 3581 (IA) ST247 | No plaques | No plaques | 1 mm |
| 3594 (II) ST36 | 1.5 mm | 1 mm | 1.5 mm |
| 3596 (IIV) ST8 | 0.5 mm | 0.5 mm with halo to 1.5 mm | 0.5 mm with halo to 1.5 mm |
| E1038 (IIV) ST8 | 0.5 mm, faint plaques | 0.5 mm, faint plaques | 1.5 mm |
| E1139 (IV) ST45 | No plaques | 0.5 mm, faint plaques | No plaques |
| E1174 (IV) ST22 | 0.5 mm, faint plaques | 0.5 mm | No plaques |
| E1185 (IV) ST12 | 0.5 mm, faint plaques | No plaques | No plaques |
| E1202 (II) ST496 | No plaques | 1 mm | 0.5 mm |
| M03/0073 (III) ST239 | 2 mm | 0.5 mm with halo to 1.5 mm | 0.5 mm with halo to 1.5 mm |
| ORFs | Amino Acid Numbers | Protein Size (kDa) | Predicted Function |
|---|---|---|---|
| PhageB1_009 | 112 | 13.5 | Terminal repeat encoded protein |
| PhageB1_016 | 107 | 12.4 | Terminal repeat encoded protein |
| PhageB1_202 | 32 | 3.5 | Unknown |
| PhageB1_203 | 104 | 11.6 | Unknown |
| ORFs | Amino Acid Number | Protein Size (kDa) | Predicted Function |
|---|---|---|---|
| PhageK_004 | 108 | 12.7 | Unknown |
| PhageK_016* | 107 | 12.4 | Unknown |
| PhageK_019 | 57 | 4.7 | Unknown |
| PhageK_020 | 89 | 10.2 | Unknown |
| PhageK_168 | 185 | 21.7 | Predicted to contain a transmembrane region based on InterProScan |
| PhageK_187 | 101 | 11.7 | Unknown |
| PhageK_188 | 123 | 13.8 | Predicted to contain a transmembrane region based on InterProScan |
| PhageK_189 | 78 | 9.2 | Unknown |
| PhageK_190 | 175 | 20.6 | Predicted as a putative metallophoshatase |
| PhageK_191 | 106 | 12.9 | Unknown |
| PhageK_192 | 76 | 8.9 | Predicted to contain a transmembrane region based on InterProScan |
| PhageK_196 | 226 | 25.8 | Unknown |
| PhageK_205 | 83 | 9.7 | Unknown |
| PhageK_206 | 98 | 11.2 | Unknown |
| PhageK_208 | 99 | 11.6 | Unknown |
| PhageK_209 | 75 | 8.9 | Unknown |
| PhageK_211 | 117 | 13.9 | Predicted to possess a transmembrane region based on InterProScan |
| PhageK_212 | 128 | 15.6 | Unknown |
| ORFs | Amino Acid Number | Protein Size (kDa) | Predicted Function |
|---|---|---|---|
| PhageJA1_003 (PhageB1_003) | 96 | 11.3 | Unknown |
| PhageJA1_020 (PhageB1_022) | 161 | 19.1 | Unknown |
| PhageJA1_021 (PhageB1_023) | 135 | 16.5 | Unknown |
| PhageJA1_084 (PhageB1_087) | 323 | 39.6 | Predicted as a putative endonuclease interrupting the terminase large subunit [PhageJA1_083 (PhageB1_086) and PhageJA1_085 (PhageB1_088)] |
| PhageJA1_152 (PhageB1_155) | 322 | 38.3 | Predicted as a putative endonuclease containing a LAGLIDADG-like domain and an Intein splicing domain and interrupts the DNA repair protein [PhageJA1_151 (PhageB1_154) and PhageJA1_153 (PhageB1_156)] |
| PhageJA1_206 (PhageB1_212) | 73 | 8.9 | Unknown |
| PhageJA1_208 (PhageB1_214) | 169 | 20.3 | HHpred indicates homology to cell wall hydrolases |
| PhageJA1_209 (PhageB1_215) | 109 | 12.6 | Unknown |
| PhageJA1_211 (PhageB1_217) | 104 | 12.0 | Unknown |
| PhageJA1_212 (PhageB1_218) | 55 | 6.5 | Unknown |
| PhageJA1_213 (PhageB1_219) | 33 | 3.7 | Predicted to possess a transmembrane region based on InterProScan |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajuebor, J.; Buttimer, C.; Arroyo-Moreno, S.; Chanishvili, N.; Gabriel, E.M.; O’Mahony, J.; McAuliffe, O.; Neve, H.; Franz, C.; Coffey, A. Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics. Antibiotics 2018, 7, 37. https://doi.org/10.3390/antibiotics7020037
Ajuebor J, Buttimer C, Arroyo-Moreno S, Chanishvili N, Gabriel EM, O’Mahony J, McAuliffe O, Neve H, Franz C, Coffey A. Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics. Antibiotics. 2018; 7(2):37. https://doi.org/10.3390/antibiotics7020037
Chicago/Turabian StyleAjuebor, Jude, Colin Buttimer, Sara Arroyo-Moreno, Nina Chanishvili, Emma M. Gabriel, Jim O’Mahony, Olivia McAuliffe, Horst Neve, Charles Franz, and Aidan Coffey. 2018. "Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics" Antibiotics 7, no. 2: 37. https://doi.org/10.3390/antibiotics7020037
APA StyleAjuebor, J., Buttimer, C., Arroyo-Moreno, S., Chanishvili, N., Gabriel, E. M., O’Mahony, J., McAuliffe, O., Neve, H., Franz, C., & Coffey, A. (2018). Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics. Antibiotics, 7(2), 37. https://doi.org/10.3390/antibiotics7020037








