Abstract
Antimicrobial agents are used in both veterinary and human medicine. The intensive use of antimicrobials in animals may promote the fixation of antimicrobial resistance genes in bacteria, which may be zoonotic or capable to transfer these genes to human-adapted pathogens or to human gut microbiota via direct contact, food or the environment. This review summarizes the current knowledge of the use of antimicrobial agents in animal health and explores the role of bacteria from animals as a pool of antimicrobial resistance genes for human bacteria. This review focused in relevant examples within the ESC(K)APE (Enterococcus faecium, Staphylococcus aureus, Clostridium difficile (Klebsiella pneumoniae), Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae) group of bacterial pathogens that are the leading cause of nosocomial infections throughout the world.
1. Introduction
The discovery of antimicrobial agents in the mid-twentieth century revolutionized the management and therapy of bacterial infections. Infections that would normally have been fatal became curable. Ever since then, the antimicrobial agents have saved the lives of millions of people. However, these gains are now seriously jeopardized by the rapid emergence and spread of antimicrobial-resistant bacteria []. Antimicrobial resistance (AMR) is a major health problem rapidly spreading across the world. The Review on Antimicrobial Resistance report [] estimates that at least 700,000 annual deaths are due to infections by drug-resistant strains of common bacterial infections, human immunodeficiency virus (HIV), tuberculosis and malaria. Numbers suggested that up to 50,000 lives are lost each year due to antibiotic-resistant infections in Europe and the US alone []. The inappropriate use of antibiotics in food animals, as well as in the medical practice has potentiated the risk of untreatable infections. Due to the free movement of people and goods between countries, and the intensive international transport of livestock, the problem of AMR is becoming by nature a global problem. Moreover, the AMR emergence is accompanied with a decline in the discovery of new antimicrobial agents. It has been estimated that most of the antibiotics used presently for common human and animal infections will be useless within five to ten years, turning back the clock to the pre-antibiotic era [].
Antimicrobial agents are principally used for therapy and prevention of human and animal diseases, but they are still used in some countries for growth-promotion in food animal productions []. Their indiscriminate use has contributed to the emergence of bacterial resistance, in hospitals, community and livestock settings. AMR may spread from animals to humans and vice versa; directly by the spread of the resistant bacteria or indirectly by the spread of resistance genes from animal bacteria to human bacteria. In this manuscript, we overview the current knowledge about the use of antimicrobial agents of critical importance in veterinary medicine, and investigate the potential of bacteria from animals as an AMR-gene reservoir. We have also underlined some resistance genes that were firstly described in bacteria from animals and later were found in human bacteria. This review focused in relevant examples within the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) or ESCAPE (E. faecium, S. aureus, Clostridium difficile, A. baumannii, P. aeruginosa, and Enterobacteriaceae) bacterial pathogens that are the leading cause of nosocomial infections throughout the world [,].
2. Use of Antimicrobials in Animal Health
Antimicrobial agents play a key role in the treatment of bacterial infections in human and veterinary medicine. In fact, AMR has been considered the quintessential One Health issue []. This One Health approach recognizes that the human health is connected to the animal health and the environment [].
The use of antimicrobials in veterinary medicine creates a selective pressure for the emergence of antimicrobial resistant bacteria, including animal pathogens, human pathogens that have animal reservoirs and commensal bacteria from animals [] The bacteria selected by this pressure can spread to humans either by direct contact with animals or food products, or indirectly via environmental pathways and/or non-food producing animals [] (Figure 1).
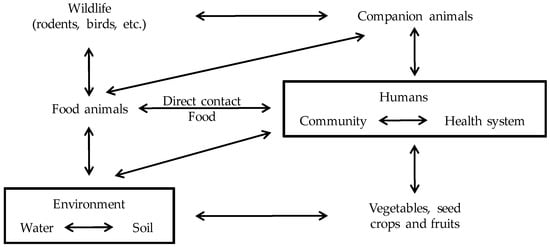
Figure 1.
Interactions between groups. Antimicrobial-resistant bacteria can spread to humans either by the food supply, direct contact with food or companion animals or, more indirectly, through environmental pathways, including waterways, soils and vegetables contaminated with human or animals waste, and vectors such as rodents, insects, and birds. Based on da Costa et al. [] and McEwen et al. [] with modifications.
The antimicrobial use in animals selects for AMR in commensal and zoonotic bacteria []. Soil treated with manure represents a “hot spot” of bacteria carrying AMR-genes []. However, soil itself is also a natural reservoir for antimicrobial-resistant bacteria []. The fecal wastes from animals contaminate groundwater, streams and other waterways, contributing to the spread of bacteria carrying AMR-genes []. Human wastes from homes, hospitals and offices also contribute to contaminate rivers and waterways with antimicrobial-resistant bacteria []. In fact, treated wastewater and lake water have been shown to contain AMR-genes and antimicrobial-resistant bacteria []. Soils and irrigation water are contamination sources for vegetables and fruits, in which resistant bacteria have been detected []. Antimicrobial-resistant bacteria may also spread between farms via infected carrier animals, companion animals or wildlife vectors []. Finally, there is a flow of patients and bacteria between community and hospital environments (Figure 1).
These complex transmission routes within farm animals, between farm animals and humans and the transfer of AMR-genes among bacteria, make it challenging to prove whether a reservoir of AMR-genes in livestock poses a risk for animal or human health []. However, some antimicrobial-resistant bacteria are zoonotic agents or can colonize and/or infect several hosts. In this sense, the current approach to evaluate the reservoir of AMR-genes in farm animals is to study the AMR-level of commensal bacteria and zoonotic agents in healthy farm animals and slaughter []. Reports from the European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC) monitoring AMR in animals are increasing. Yet, there are some limitations in the current data, and Thanner et al. [] have recently suggested a voluntary monitoring program by researchers.
In order to underline the importance of the current available antimicrobials classes, the World Health Organization (WHO) started categorizing the most important antimicrobials in human medicine. The last revision of the list was done in 2016 []. The importance of each antimicrobial group is based on two criteria: C1, “the antimicrobial class is the sole, or one of limited available therapies, to treat serious bacterial infections in people”; and C2, “the antimicrobial class is used to treat infections in people caused by either: (i) bacteria that may be transmitted to humans from nonhuman sources, or (ii) bacteria that may acquire resistance genes from non-human sources”. Antimicrobials that meet both criteria are considered “critically important” in human medicine, antimicrobials that meet one of the criteria are considered “highly important”, and antimicrobials that meet none of the two criteria are considered “important” [].
Similarly to the WHO, the World Organization for Animal Health (also named Organisation mondiale de la santé animale, OIE) [] has developed a list of the antimicrobial agents of veterinary importance []. Since in veterinary medicine, many different species have to be treated the criteria to classify the antimicrobials were different than for the human medicine. The OIE criteria were based on a questionnaire prepared by the ad hoc group, which was sent to the OIE delegates of all member countries and international organizations which had signed a co-operation agreement with the OIE. The responses were analyzed by the ad hoc group and discussed in some international committees. The criterion C1 was based on the response rate to the questionnaire: “This criterion was met when a majority of the respondents (more than 50%) identified the importance of the antimicrobial class in their response to the questionnaire”. The criterion C2 was based on the treatment of each serious animal disease and the availability of alternative antimicrobial agents: “This criterion was met when compounds within the class were identified as essential against specific infections and there was a lack of sufficient therapeutic alternatives”. Similarly to the WHO list, antimicrobials that meet both criteria are considered “critically important” in veterinary medicine, antimicrobials that meet one of the criteria are considered “highly important”, and antimicrobials that meet none of the two criteria are considered “important”.
After the ban of antimicrobial growth promoters, antimicrobial agents are still allowed with veterinary prescription []. Around 37% of the antimicrobials (mainly ionophores) used in food animal production do not have equivalent drugs used for human therapeutic purposes []. Similarly, tetracyclines, that are not considered a first-line antimicrobial therapy in human medicine, make up another 44% of total antimicrobial used in animal agriculture []. While not all antimicrobial agents used in animal health are used in human medicine, most antimicrobials used in food animals are analogs to those used in human medicine []. In both WHO and OIE lists, substances belonging to certain groups (aminoglycosides, cephalosporins of third generation, macrolides, penicillins, and quinolones) were considered critical important antimicrobial groups [,]. In fact, some specific antibiotics are critically and/or highly important in both human and veterinary medicine (Table 1). Interestingly, the antibiotic streptomycin is also used in plant agriculture in the prevention of fire blight disease in apple and pear tree caused by the phytopathogenic Erwinia amylovora [].

Table 1.
Antimicrobials used in both human and veterinary medicine.
The use of antimicrobial agents in human medicine is restricted to therapy and prophylaxis. However, the antimicrobials in farm animals have therapeutic, prophylactic, metaphylactic and sub-therapeutic uses [,]. Therapeutic treatments are planned for individual animals that are diseased, but in food animal productions it is often more efficient to treat entire groups by medicating feed or water [,]. This metaphylactic use is particularly common and it implies the use of antimicrobials in the whole herd or flock for disease prophylaxis and/or therapy in case of presence of clinical illness in one individual of the group [,]. Moreover, for some animals (poultry and fish) this mass medication is the only feasible means of treatment []. This metaphylactic use results in the frequent exposition of entire groups of animals, healthy and diseased, to antimicrobial agents []. Prophylactic antimicrobial treatments are typically used during high-risk periods for infectious disease such as after weaning or transport []. Antimicrobials may also be administered in relatively low (sub-therapeutic) concentrations to food animals to promote growth and to enhance feed efficiency [,]. A ban on the use of growth promoters was implemented in Europe in 2006, but it has not led to any consistent decrease in antimicrobials consumption since this ban has been compensated by metaphylactic and prophylactic uses [].
In addition to antimicrobials, metals compounds, such as zinc and copper, are also used to supplement animal (mainly in pigs) feed for the prevention of post-weaning diarrhea and the stimulation of growth. Resistance to these compounds is often associated to resistance to antimicrobial drugs such as methicillin in staphylococci, or macrolides and glycopeptides in enterococci []. It has been shown that these metal resistance genes are frequent in animal-associated bacteria [,,,]. The use of antimicrobials, biocides and metal compounds in animal productions in sub-therapeutic doses and with long exposure periods, may promote that bacteria fix genes that confer AMR [,]. These resistance genes can subsequently be transmitted to human-adapted pathogens or to human gut microbiota via people, contaminated food or the environment [,].
The multiple pathways involved in AMR-genes dissemination and exchange within the agriculture, the environment and the food processing industry (Figure 1) make difficult to track the movement of these genes in vivo []. In this regard, Thanner et al. [] underlined some gaps regarding our current knowledge about AMR in plant and animal agriculture, and proposed a worldwide surveillance program of soil, plants, animals, water and wastewater treatment plants using the same methods that for AMR monitoring of human hospitals isolates. As for AMR in human bacteria, a better knowledge of the AMR in animal bacteria will help to achieve an effective and controlled use of antimicrobials in animals, thereby avoiding the dissemination of known and novel AMR-genes [].
3. Presence of AMR-Genes in Animals: The Metagenomics Evidence
Although some bacterial species (such as Mycobacterium tuberculosis and Streptococcus pneumoniae) are specialist human pathogens, a larger number of species are opportunistic pathogens (such as Escherichia coli) causing disease in humans and other hosts including livestock and wildlife species, and are also present in the wider environment []. The interplay of these ecologies is important, since animals and the environment represent a major AMR reservoir. Through evolution, microorganisms have synthetized antibiotics and/or develop resistance methods for microbial competition in the environment []. In fact, AMR-genes have been found in soils not exposed to antimicrobials [,]. However, the human activity is increasing and changing this environmental resistome []. Indeed, animal microbiomes have acquired genes over years of exposure to antimicrobial agents and heavy metals compounds used as therapeutics, metaphylactics, prophylactics and growth promoters [,].
Current studies based on metagenomics and/or real-time polymerase chain reaction (PCR) approaches have given diverse results regarding the human, animal and environmental resistome. These novel technologies offer the possibility to elucidate the presence of AMR-genes in human, animal and environmental microbiomes and to identify the factors causing their persistence, selection and spread [].
Some studies have suggested that human and animal microbiomes are different []. Agga et al. [] compared environments related to animal (cattle and swine) and municipal (human) waste and saw that antimicrobial-resistant bacteria populations associated with animal agriculture were distinct from those associated to human activity. However, regarding the gene content, 25 out of 61 unique AMR-genes identified were common between municipal waste and animal samples. The half of the AMR-genes detected in another study [], were only found in external environments. These genes from external environment microbiomes were mainly related to biocide and metal resistance []. Human microbiota had the highest abundance and diversity of AMR-genes, and the lowest taxonomic diversity []. Nevertheless, it was seen that tetracycline resistance genes dominated in both human and animal microbiomes []. Moreover, 20.5% of the AMR-genes detected were found in human, animal and environmental samples []. A recent study based on the comparison of published data on metagenomics made similar conclusions []. This study showed that the environment is a reservoir of the basic forms of resistance genes (such as blaTEM), while both the human and mammalian gut microbiomes contained the widest diversity of clinically relevant resistance genes [].
Some studies have identified AMR-genes in animal samples regardless of the antibiotic exposure. Sequence-based metagenomics analysis of conventionally raised cattle without therapeutic antibiotics exposure revealed that 3.7% of the sequences encoded resistance to antibiotics and toxic compounds, and nearly half of these genes encoded multidrug resistance efflux pumps []. A similar metagenomics study in chicken ceca, revealed that around 2% of the sequences encoded resistance to antibiotics and toxic compounds []. At least one quarter of these genes were related to tetracycline and fluoroquinolones resistance []. Studies in swine fecal samples revealed the existence of at least 149 AMR-genes in non-medicated animals [,].
Although non-exposed animals may already carry bacteria with resistance genes, some studies have underlined that their resistome can change after antibiotic exposure. Diverse studies have shown that antibiotic treatment increased diversity of antibiotic resistance genes [,]. Moreover, some enriched genes, such as an aminoglycoside O-phosphotransferase, confer resistance to antibiotics that were not administered, demonstrating the potential for indirect selection of resistance to classes of antibiotics not fed []. The effects of administering sub-therapeutic concentrations of antimicrobials to beef cattle were investigated in a recent study, showing that the antimicrobial treatment differentially affected the abundance of certain resistance genes in fecal deposits, but not their persistence []. In other study, the administration of the third generation cephalosporin ceftiofur in dairy cows increased the β-lactam and multidrug resistance genes in feces []. Nevertheless, another study identified approximately the same number (21–26) of unique AMR-genes in manure samples of four dairy cows despite different prior exposure to antibiotics [].
Some metagenomics studies have underlined the dominance of tetracycline resistance genes in animal microbiomes [,]. These results may be partially explained by the historical and current exposure to tetracyclines in the animal husbandry. Yet, a metagenomics study in pigs reared in an antibiotic-free environment revealed also the presence of diverse tetracycline resistance genes including novel genes, as well as a gene [tet(40)] not previously observed outside the human gut microbiome [].
Although the metagenomics studies seem promising, they have some limitations. Since, some annotated genes (ex. efflux pumps) that confer AMR, perform other basic functions, more research on gene expression and functional analysis is needed to determine whether these genes can confer phenotypical resistance to antimicrobials []. Moreover, the sequence-based metagenomics approach does not provide information about the genetic context []. Nevertheless, some promising plasmid metagenomics studies have showed a broad dissemination of plasmid carrying AMR-genes in pig and bovine samples [,]. The functional genomics study about the tetracycline resistome of the pig gut has shown that most of the genes resided on putative mobile genetic elements (MGEs), which may contribute to the maintenance and dissemination of antibiotic resistance in antibiotic-free environments []. Nevertheless, other studies have probed that the presence of MGEs is also affected by antibiotic exposure [,]. It has been seen that the antimicrobial exposure increased the abundance of phage integrase-encoding genes (that may carry virulence or AMR-genes) in the viromes of swine, demonstrating the induction of prophages with antibiotic treatment []. In the study based on the administration of ceftiofur in dairy cows, an increase in gene sequences associated with phages, prophages, transposable elements and plasmids was observed [].
Some metagenomics studies have proven that the animal microbiome is different of the human microbiome, but, interestingly, their shared a part of their resistome. Acquired resistance genes seem disseminated in the absence of selective pressure, but their abundance is affected by antibiotic exposition. These findings confirm that continue antimicrobial selective pressure in both humans and animals may benefit the dissemination of acquired resistance genes []. As other authors have concluded, a prudent use of antibiotics in human and veterinary health is needed to slow down the AMR spread and prevent the emergence of novel AMR-genes [].
4. AMR-Genes in Gram-Positive Bacteria from Animals
Three Gram-positive species are considered members of the ESC(K)APE group: C. difficile, E. faecium, and S. aureus. This section focuses on the current knowledge about these pathogens regarding their AMR in animals, particularly on the genes common within these species (Table 2).

Table 2.
Examples of common AMR-genes found in Clostridium, Enterococci and Staphylococci.
4.1. Clostridium difficile
C. difficile is an ubiquitous environmental organism widespread in rivers, lakes and soils. It is also found in the hospital environment where it is difficult to eradicate, as well as in meat products and animals including diverse species (calves, ostriches, chickens, elephants, dogs, horses and pigs) []. Although there are some current genomic updates of the clostridial phylogeny [,], in this review we use the classical nomenclature of Clostridium difficile.
C. difficile is recognized as the major cause of healthcare antibiotic-associated diarrhea []. Over the last decade, an alarming increase in incidence of C. difficile infection was observed across the USA, Canada and Europe, and it has been associated with the emergence of the highly virulent (hypervirulent) clone BI/NAP1/027 [named according to its restriction enzyme analysis (REA), pulsed-field gel electrophoresis (PFGE) and PCR Ribotype (RT)] [,]. This dramatic increase in C. difficile infections was associated to the fluoroquinolone resistance of this clone []. Fluoroquinolones are broad-spectrum antimicrobials highly effective for the treatment of bacterial infections in animals and humans. Most RT027 isolates harbored mutations in the quinolone-resistance determining region (QRDR) of the DNA gyrase subunit gene, gyrA, that confer resistance to fluoroquinolones []. Nevertheless, clinical C. difficile strains acquire fluoroquinolone resistance due to alterations in the QRDR of either GyrA or GyrB DNA gyrase subunits [].
C. difficile infection has also emerged as a cause of diarrhea in the community, especially in populations previously considered at low risk, such as young people, antibiotic-naive patients or people without healthcare exposure []. Studies in the community indicated that up to 13% healthy human subjects are asymptomatically colonized with C. difficile []. In addition to RT027, a number of emergent hypervirulent RTs have recently been identified, notably the hypervirulent RT078 recognized as infection cause in hospitals, the community and animals []. The use of fluoroquinolones in the pork industry may have also contributed to the emergence of the multidrug-resistant RT078 []. In a study by Keessen et al. [], most human and porcine isolates were resistant to ciprofloxacin (96%), and some were also resistant to moxifloxacin (16% for both human and porcine isolates). Resistance to moxifloxacin in this study was associated with a gyrA mutation []. Moreover, the use of fluoroquinolones [ciprofloxacin in humans, and enrofloxacin in pigs] was significantly associated with isolation of moxifloxacin-resistant isolates in both populations []. The authors proposed that the increased fluoroquinolone use could have contributed to the spread of C. difficile RT078 [].
RT078 is commonly isolated from swine and other food animals []. It is likely that RT078 is an important pathogen of piglet diarrhea worldwide [,]. Molecular genotyping has suggested that RT078 isolates of human and swine origin are highly related and may therefore represent a potential zoonotic transmission []. Several studies have suggested that transmission from swine to humans may occur in farms or in large integrated swine operations, and moreover, the aerial dissemination of C. difficile from pig farms has been shown [,]. Due to the increased incidence of C. difficile infection outside the hospital environment and the presence of the same genetic lineage in food animals and its products, some authors have suggested that C. difficile may be considered as a foodborne pathogen []. Although RT078 is predominant in studies on swine, other RTs have been described in animals []. For example, RT046 has been found in both piglets and humans in Sweden [].
AMR in C. difficile has been less intensively investigated than in other Gram-positive pathogens (such as S. aureus). In fact, few AMR-genes [such as erm(A), erm(B), tet(M), tet(44), tet(W), ant(6)-Ib, catD and cfr(B)] have been characterized in C. difficile isolates []. Resistance to cephalosporins is still uncharacterized, although most clinical C. difficile strains are resistant to these antibiotics. Similarly, some C. difficile human and animal isolates with reduced susceptibility to metronidazole have been found, although the resistance mechanism is not completely understood [].
The most widespread mechanism of resistance to the antibiotics of the macrolide-lincosamide-streptogramin B (MLSB) group in C. difficile is ribosomal methylation due to the erythromycin ribosomal methylases (erm) genes of class B []. It is to note, that erm(B) is the erm gene with the widest bacteria host range, and it has been found in both Gram-positive and Gram-negative bacteria, aerobic, and anaerobic genera and in most ecosystems []. However, erm-negative C. difficile strains resistant to both erythromycin and clindamycin or only to erythromycin have been also described []. Alterations in the 23S rRNA or ribosomal proteins (L4 or L22) have been found in some of these strains, but the presence of these changes in susceptible isolates had excluded their role in resistance []. Interestingly, a multidrug resistant cfr-like gene, cfr(B), that modifies the 23S rRNA has been recently found in clinical C. difficile isolates [,] (see Section 4.3.2).
The tet(M) gene is the most frequent tetracycline resistance determinant in C. difficile isolates []. Interestingly, most RT078 isolates carry the transposon Tn916 harboring tet(M) []. Nevertheless, other tet genes [tet(W), tet(44)] have been identified in C. difficile [,]. The tet(W) gene has been found together with tet(M) in C. difficile isolates recovered from animals and humans []. The tet(44) gene has been associated to the transposon Tn6164 in human and environmental isolates [,].
4.2. Enterococcus faecium
Enterococci are commensal bacteria of the gastrointestinal tract of mammals and other animals that can also be detected in the environment [,]. In adult people, enterococci account for about 1% of the intestinal microbiota, being E. faecium and Enterococcus faecalis the most prevalent enterococci in the human gastrointestinal tract []. These species are opportunistic human pathogens that are implicated in life-threatening hospital acquired infections such as bacteremia and infective endocarditis [,].
Enterococci are intrinsically resistant to a number of first-line antimicrobial agents. They have resistance against cephalosporins and cotrimoxazole, as well as low-level resistance to β-lactams and aminoglycosides []. Moreover, clinical and animal Enterococcus isolates with resistance to other antimicrobials such as macrolides, tetracyclines, streptogramins and glycopeptides have been described [,]. Antimicrobials such as linezolid, daptomycin and tigecycline can be used in the treatment of enterococcal infections [,]. Nevertheless, resistance against these former antimicrobials has been reported, mainly in the clinical setting [,].
Nowadays, the vancomycin-resistant enterococci (VRE) pose a major therapeutic challenge due to their difficult treatment []. VRE were found in the hospital setting in the 80s [,], while the first description of animal reservoirs of vancomycin-resistant E. faecium was published in 1993 []. Later on, VRE have been described in diverse animals and environmental sources []. As vancomycin has not been used in veterinary medicine, it was hypothesized that the use of another glycopeptide, the avoparcin, as additive in farm animals feed, has influenced the emergence of VRE in animals since the 70s []. This correlation has been criticized and several studies have shown that vancomycin-resistant E. faecium persisted in animals for an extended time after the banning of avoparcin in the 90s [,]. However, this persistence has been related to co-selection with other antimicrobials (such as tetracycline or the macrolide tylosine) and metals (such as copper sulphate) [,]. It has been suggested that VRE strains are predominately host-specific, being hospital isolates genetically different from animal strains []. However, some research, based on multilocus sequence typing (MLST) showed that certain isolates from diverse clonal complexes (CCs) are present in animals, healthy humans and patients [,,]. Moreover, the same variants of Tn1546 carrying the glycopeptide-resistant gene vanA have been detected in enterococci from human and animal origin, underlying that E. faecium from animals can act as a donor of AMR-genes for other pathogenic enterococci [,].
Similar to glycopeptides, the use of other antimicrobials as grown promotors may have influenced the emerging of enterococci resistant strains in food animals. In 1999, the streptogramin combination quinupristin/dalfopristin (RP59500, Synercid) was approved for clinical use. The mixture of type B streptogramin (quinupristin) and type A streptogramin (dalfopristin) in a 30:70 ratio showed good activity against multiresistant E. faecium strains []. Nevertheless, before its approval for clinical use, in 1997, quinupristin/dalfopristin-resistant E. faecium isolates were found in both human patients and chicken samples []. The use of virginiamycin, a streptogramin licensed for growth promotion in animals (including chickens and other poultry), was associated to the development of resistance to streptogramin combinations by enterococci []. In this sense, diverse streptogramin A resistance genes [such as vat(D), vat(E), vga(D) and vat(H)] have been detected in E. faecium from animals in Europe, USA and/or Asia [].
Regarding other antimicrobials, human and animal E. faecium isolates frequently harbored genes conferring resistance against aminoglycosides [aph(3′)IIIa], tetracycline [tet(M)], and macrolides [erm(B)] []. Recently, a novel gene, optrA, which encodes for an ABC transporter that confers resistance to oxazolidinones and phenicols has been found in E. faecalis and E. faecium of human and animal origin []. This gene has been found chromosomally and plasmid located in Enterococci [], but also in Staphylococcus sciuri [,]. The optrA-carrying plasmids in enterococci have also other AMR-genes against antibiotics such as MLSB [erm(A)-like)] and/or phenicol (fexA) []. Retrospective analysis of genome sequences has revealed that optrA has a wide dissemination in Gram-positive bacteria and it has also been found in diverse Streptococcus species (including S. suis, S. agalactiae and S. pyogenes) [].
Some genes (cfr and dfrK) first discovered in animal-related staphylococci have recently been found in enterococci (see Section 4.3.2 and Section 4.3.3). The trimethoprim resistance gene dfrK, encoding a trimethoprim resistant dihydrofolate reductase, was recently detected in E. faecium [] (see Section 4.3.3). The multi-resistance gene cfr encodes a RNA methyltransferase that modifies the 23S rRNA gene conferring resistance to ribosome-targeting antimicrobials []. It has been detected in E. faecalis from animals and humans, E. thailandicus from pigs and farm environment, E. casseliflavus and E. gallinarum from pigs [,]. A non-functional cfr, due to a deletion in the regulatory region upstream, has been detected together with optrA in a clinical E. faecium isolate []. As for C. difficile, a functional cfr(B) has recently been found in E. faecium clinical isolates [] (see Section 4.3.2).
4.3. Staphylococcus aureus and Related Species
S. aureus is part of the normal and transit human microbiota, and it is usually present in the nasopharyngeal mucosa, but also in the skin and other corporal areas [,,]. The general carrier rate in humans is estimated between 20 and 30 percent for persistent colonization []. S. aureus is an opportunistic pathogen, which produces a wide spectrum of diseases, ranging from minor’s skin to deep infections, as well as potentially-fatal diseases such as diverse invasive infections and toxin-mediated diseases [,]. As in humans, S. aureus is a commensal bacterium for animals but it is also able to cause diverse infections including abscesses, chondronecrosis, dermatitis, mastitis, pyaemia, osteomyelitis, pneumonia, septicemia and skin and wound infections [].
Staphylococci of animal origin harbor a wide variety of AMR-genes [,,,,,,,]. Recent reviews by Wendlandt et al. [,] have underlined that at least 44 AMR-genes in staphylococci have been detected in both human and animal isolates (Table 3). Interestingly, some shared genes were firstly described in isolates from animal origin [such as tet(L), erm(T), dfrK, fexA, cfr] or possibly originated from Staphylococcus species related to animals (as the methicillin resistance gene mecA). Next sub-sections are focused on those genes originated from animal related staphylococci.

Table 3.
Examples of AMR-genes identified in Staphylococci from animal and/or human origin.
4.3.1. mec Genes in Staphylococci: Origin and Reservoirs
One of the most important acquired resistance in staphylococci is methicillin resistance. This resistance is mainly due to the acquisition of the mecA gene, encoding a β-lactam low affinity penicillin binding protein (PBP) called PBP2a. This gene is carried on a MGE termed staphylococcal cassette chromosome mec (SCCmec). This MGE has been more extensively studied for S. aureus, since this is the most important Staphylococcus species for the human health. To date, eleven major SCCmec types carrying mecA have been described in methicillin resistant S. aureus (MRSA), which have been assigned on the characterization of its two essential components the mec and ccr complexes [,,]. The mec-gene complex corresponded to the mecA operon variants, which can include functional and/or truncated regulatory genes. The ccr complex includes the recombinase(s) involve in the excision and integration of the element into the chromosome. The remaining regions of the SCCmec on which subtypes are based are called junkyard or joining regions, and they can contain MGEs, as well as heavy metal [arsenate (ars operon), cadmium (cadD-cadX), cadmium and zinc (ccrC), copper (copB), mercury (mer operon)] and other AMR (amynoglicosides (aadD, aad9/spc, aacA-aphD), bleomycin (ble), erythromycin [erm(A)], tetracycline [tet(K)]) resistance genes. Many non-typable SCCmec cassettes exist, and non-S. aureus staphylococci carry similar SCCmec and novel mec-ccr combinations as those found in S. aureus [,,,,]. This suggests horizontal transfer and recombination events within Staphylococci species. In fact, the mecA is widely distributed among methicillin resistant coagulase negative Staphylococci (MRCoNS) [,,,,,].
Closed related mecA allotypes have been described in members of the S. sciuri group [,,]. The members of the S. sciuri group are mainly considered commensal animal-associated bacteria with a broad range of hosts, although they have also been found in the environment and occasionally causing disease in humans and other hosts [,,]. The mecA variants found in members of the S. sciuri species group are chromosomal located without being part of a SCCmec, and are usually phenotypically susceptible to β-lactams or show a heterogeneous resistance []. In this sense, a member of the S. sciuri group, the commensal animal-related S. fleurettii, has been suggested as the highly probable origin of the mecA gene []. The mecA-containing regions of S. fleurettii strains recovered from animals or its products are similar to the mec region of the S. aureus SCCmec type II. Moreover, the analysis of the corresponding gene loci region (with mecA gene homologues) of S. sciuri and S. vitulinus (which evolved from a common ancestor with that of S. fleurettii), probed that the mecA gene of S. fleurettii descended from its ancestor and was not recently acquired []. These findings suggested that the SCCmec in S. aureus was generated by acquiring this intrinsic mecA region of S. fleurettii.
Recently, two mecA gene homologs (mecB and mecC) have been discovered [,,]. The gene mecB (formerly named mecAm) was found in 2009 in a Macrococcus caseolyticus isolated from a chicken []. This gene is located in a typical mec operon (blaZb-mecB-mecR1b-mecIb) with similar regulatory genes but also accompanied of a blaZ homologue encoding for a putative β-lactamase []. The mecB operon has been found on a transposon (Tn6045) either plasmid or chromosomal located [], as well as in SCCmec elements [,]. Nowadays, mecB has not been detected in staphylococci. However, there is a potential risk of transmission since staphylococci and macrococci are closely related members of the same family (Staphylococcaceae). Moreover, these bacteria share a common niche, since M. caseolyticus is a commensal bacterium colonizing animal skin as staphylococci. Additionally, this mec variant is located in different MGEs, including a plasmid, which is a more mobile vehicle within bacteria than the SCCmec [].
The gene mecC (formerly named mecALGA251) was discovered in 2011 in S. aureus by two different working groups [,]. It was described from isolates originating from mastitis in cows and from humans in the United Kingdom, Ireland and Denmark [,]. Even if this gene was first described in 2011, a retrospective analysis in Denmark has shown that mecC-positive MRSA strains have been circulating before []. The mecC gene has a wide distribution and positive strains have been found in humans from both infection cases and carriage state, as well as in animals including livestock (dairy cattle, beef cattle, sheep, farmed rabbits), companion (cats, dogs, guinea pigs), wildlife (birds, mammals) and zoo (mara) animals [,]. Generally, there is a low occurrence of mecC-positive human isolates (ex. less than 1% in clinical MRSA from Belgium, []). This gene has mainly been associated with S. aureus lineages related to infections and colonization in animals []. The gene mecC in S. aureus is located on the SCCmec XI, which as few other SCCmec types also carried heavy metal resistance genes []. mecC homologues have been found in other staphylococci (S. xylosus, S. sciuri, and S. stepanovicii), associated to cassettes similar to SCCmec XI or to composite SCCmec together with mecA [,,]. New mecC allotypes have been identified: mecC1 in S. xylosus related to bovine mastitis [] and mecC2 in Staphylococcus saprophyticus from common shrew [].
4.3.2. The Multi-Resistance Gene cfr
Linezolid, the first member of the oxazolidinone class of antibiotics, is considered to be a last-resort antimicrobial for the treatment of infections caused by VRE, MRSA and penicillin-resistant pneumococci []. Several mechanisms conferring linezolid resistance have been described in staphylococci, including point mutations in genes encoding 23S rRNA and mutations in ribosomal proteins L3, L4 and L22 [,]. In 2000, the gene cfr was identified in a bovine S. sciuri recovered in 1997 []. This gene encodes an RNA methyltransferase that modifies the 23S rRNA gene conferring combined resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins and streptogramin A antimicrobials (known as PhLOPSAA phenotype). It also confers decreased susceptibility to the 16-membered macrolides (josamycin and spiramycin) []. Afterwards, this gene has been identified in methicillin-susceptible S. aureus (MSSA), MRSA, various coagulase negative staphylococci (CoNS) and in the coagulase-variable S. hyicus [].
Although linezolid resistance mediated by cfr is not frequent in the clinical environment [], reports of clinical outbreaks by cfr-containing S. aureus strains have been reported [,]. Recently, a clinical case due to a cfr-positive livestock-associated (LA-) MRSA CC398 has been described []. This gene has been found in important MRSA pandemic lineages such as the sequence type (ST)22/SCCmec IV, the Panton-Valentine leukocidin (PVL) positive ST8/SCCmec IV/USA300 or the ST125-MRSA-IVc [,,]. It has also been detected in clinical strains of S. capitis [] and methicillin-resistant S. epidermidis (MRSE) [].
In staphylococci from animals, the cfr gene has been identified in isolates from different sources including pigs (in S. aureus, S. arlettae, S. cohnii, S. equorum, S. haemolyticus, S. hyicus, S. saprophyticus, S. sciuri, S. simulans and S. warneri), bovine (in S. aureus, S. lentus, S. sciuri and S. simulans), poultry (in S. arlettae, S. cohnii, S. equorum, S. rostri, S. sciuri and S. simulans), and companion animals (in S. pseudintermedius) [,,,]. The cfr gene has been described as chromosomally and plasmid located, linked to specific insertions sequences (IS) []. Moreover, the genetic elements of staphylococci with cfr usually carried additional resistance genes, including β-lactam (blaZ), aminoglycosides [aacA-aphD, aadD, ant(4′)-Ia], aminocylitols (spc), bleomycin (ble), fosfomycin (fosB), MLSB [erm(A), erm(B), erm(C), erm(33)], lincosamides-pleuromutilins-streptogramin A [lsa(B)], phenicol (fexA), tetracycline [tet(L)], oxazolidinones-phenicols (optrA) and/or trimethoprim (dfrK) resistance genes (Table 4). The immediate genetic environment within the diverse cfr-carrying elements is similar, therefore it has been suggested that a limited number of acquisition events explain the diversity seen among the plasmidic or chromosomal structures [].

Table 4.
Examples of the genetic environments of cfr genes.
Recent studies have described the cfr gene or a variant in other Gram-positive (including Bacillus spp., C. difficile, Enterococcus spp., Macrococcus caseolyticus, Jeotgalicoccus pinnipedialis and S. suis) and Gram-negative (Proteus vulgaris and E. coli) species (Table 4). With the exception of Clostridium and Enterococcus isolates, the cfr-positive isolates of the other non-Staphylococcus genera were obtained exclusively from livestock and related farm environments [,,,].
In Bacillus spp., the gene cfr has been identified in three related plasmids from pig isolates from China, one (PBS-01) of these plasmids being previously reported in S. cohnii []. The cfr-carrying plasmid PSS-03 has been detected in both M. caseolyticus and S. cohnii [], while the plasmid pJP1 or variants type pJP1-like have been found in M. caseolyticus, J. pinnipedialis, and S. lentus isolates []. Interestingly, some of these cfr-plasmids found in Gram-positive bacteria carried additional resistance genes against aminoglycosides (aadD, aadY), bleomycin (ble), MLSB [erm(B), erm(C)] and/or phenicol (fexB). In fact, a new multiresistance plasmid pWo28-3 related to pJP1, harbouring aacA-aphD, aadD, ble, optrA, cfr and fexA, has been identified in S. sciuri []. The cfr has also been found associated to a novel IS (ISEnfa5) truncating a novel lincosamide resistance gene lnu(E) in the plasmid pStrcfr of S. suis [].
Recently, the cfr variant gene, designated cfr(B), was identified in C. difficile and E. faecium clinical isolates [,,]. The nomenclature and functionally of this gene has been extensively discussed []. The Cfr proteins detected in other Gram-positive (Bacillus, Macrococcus, Jeotgalicoccus, Staphylococcus and Streptococcus) and in Gram negative (Proteus and Escherichia) bacteria were indistinguishable or similar (99% identity) from the original S. sciuri Cfr protein []. However, the cfr(B) shared only 75% amino acid identity with the original Cfr protein [,,]. Schwarz et al. [] suggested that the protein encoded by cfr(B) was structurally distantly related to the original Cfr. However, further research proved that the cfr(B) product does function as a Cfr protein [], and therefore may be considered as a variant.
Regarding Enterococcus, cfr-carrying plasmids have been described in E. faecalis from animals and humans, as well as in pig-related isolates of E. thailandicus, E. casseliflavus and E. gallinarum [,]. The cfr gene has also been detected chromosomally located in E. casseliflavus from pigs []. As mentioned in Section 4.2, a non-functional cfr gene has been detected together with optrA in clinical E. faecium isolates []. However, recently a cfr(B) gene sharing 99.9% sequence identity with the corresponding gene in C. difficile has been identified in E. faecium clinical isolates [,]. Deshpande et al. [] described two copies of cfr(B) chromosomally located and embedded in a Tn6218 similar to the cfr-carrying transposon described in C. difficile. While the cfr(B) described by Bender et al. [] was found in Tn6218-like elements possible linked to plasmids.
It has been shown that the gene cfr is functionally active in Gram-negative hosts []. Furthermore, it has been identified chromosomally located (inserted into the chromosomal fimD gene) in P. vulgaris from pigs, and in diverse plasmids in E. coli from pigs [,,,,,] or food of animal origin []. The IS26 appears to play an important role in the transfer of this multiresistance gene in Gram-negative bacteria, since it appears in the cfr-carrying plasmids detected in E. coli [,,,,,,]. One of these cfr-carrying plasmids from E. coli of swine origin carried also the extended-spectrum-β-lactamase (ESBL) gene blaCTX-M-14b [].
4.3.3. Other Genes in Animal-Associated S. aureus
S. aureus from animals can be related to various CCs [,,]. However, animals are considered the main reservoir of the specific S. aureus lineage CC398, which has been the subject of numerous studies during recent years [,,,,,,,,]. Studies based on the phylogenetic analysis of genome-wide single nucleotide polymorphisms (SNPs) supported the existence of two subpopulations in CC398: an ancestral human-adapted clade and an animal-associated clade [,]. Nevertheless, LA-MRSA CC398 isolates of the animal-associated clade are able to infect humans [,]. Analysis of genes present in CC398 has revealed numerous genes in common with clinical S. aureus and/or other staphylococci, but also some novel or rare resistance genes have been found in animal-related isolates from this lineage [,]. Some of these rare or novel genes are often on plasmids encoding for other AMR-genes such as the multi-resistance gene cfr (Table 5).

Table 5.
Examples of resistance genes co-located on plasmids from Staphylococci.
One interesting gene, found in animal-associated S. aureus, is the phenicol exporter gene fexA. It was first described in a bovine S. lentus isolate, but later it has been found in diverse plasmids of LA-MRSA CC398 from pigs, cattle and horses, as well as on plasmids from the also animal-related S. aureus CC9 [,]. This gene has been related to the non-conjugative transposon Tn558 that has been detected (partially or complete) in plasmids or in the chromosome of various CoNS of animal origin []. The fexA is co-located in plasmids with additional resistance genes (Table 5). Interestingly, the fexA gene has been described in cfr-carrying plasmids of clinical important MRSA pandemic lineages [,].
The tetracycline gene tet(L) was found initially in the 80s in diverse plasmids from Bacillus. In staphylococci, it was first described in the early 90s on a plasmid from a porcine S. hyicus. In addition, subsequently, it was detected on structurally diverse plasmids of staphylococci of animal origin [,]. This gene has been found co-located on plasmids from staphylococci with other additional antimicrobial and heavy metals resistance genes (Table 5). Regarding LA-MRSA CC398, the gene tet(M) is the most frequent tetracycline-resistant gene found in these isolates, although it is accompanied frequently by tet(K) and tet(L) []. The gene tet(L) has also been described in clinical S. aureus in the 90s [], and recently in a cfr-carrying plasmid with additional antimicrobial genes [dfrK and aadD] in a clinical S. aureus ST125 strain [].
The trimethoprim resistance gene dfrK was discovered co-located on a plasmid with tet(L) in LA-MRSA CC398 [,]. Similar to other genes in staphylococci, the dfrK has been found on diverse plasmids co-located with other antimicrobial and heavy metals resistance genes (Table 5). The dfrK gene is widely disseminated in LA-MRSA CC398 and it has been found in isolates from pigs, cattle and poultry []; but it has also been found chromosomally located on the transposon Tn559 of MSSA CC398 []. Additionally, the Tn559 carrying dfrK has been found in E. faecium [].
The erm(T) gene has been previously identified in other Gram-positive bacteria (such as Streptococcus and Lactobacillus) []. In staphylococci, this gene has been originally found in animal-associated CC398, but further research suggested that it is particularly associated to the MRSA and MSSA CC398 from the ancestral human clade [,,]. Nevertheless, a recent study comparing erm(T)-carrying plasmids from S. aureus ST398 from pig and humans, showed that these plasmids are quite similar and all carried additional tetracycline [tet(L)] and heavy metal (cadD-cadX, copA, mco) resistance genes []. The plasmid (pUR1902) recovered from a pig isolate carried also aadD as a plasmid (pUR2941) from a human isolate, while the remaining plasmid (pUR2940) from a human isolate additionally carried erm(C) and dfrK [].
A recent review by Wendlandt et al. [] underlined the presence of diverse multidrug resistance (MDR) genes co-located together or with other resistance genes on plasmids in staphylococci from animal origin. These genes included MLSB [erm(A), erm(B), erm(C), erm(T), erm(33)], lincosamides-pleuromutilins-streptogramin A [lsa(E), vga(A), vga(C)] and PhLOPSAA (cfr) resistance genes []. The main resistance mechanisms conferred by these MDR genes included target modification by methylation (erm and cfr genes) and active efflux via ABC transporters [vga and lsa(E) genes] []. Their location on plasmids, co-located with other AMR-genes, may potentiate their co-selection and persistence in animal staphylococci, and furthermore represent an important pool of resistance against critically and highly important antimicrobial agents [].
5. AMR-Genes in Gram-Negative Bacteria from Animals
The Gram-negative members of the ESC(K)APE group are A. baumannii, P. aeruginosa, and the members of Enterobacteriaceae. A large number of antimicrobial resistance genes have been described in Gram-negative bacteria [,]. Since this area is extremely large and complex, in this section, we summarize part of the current knowledge about AMR in A. baumannii, P. aeruginosa and Enterobacteriaceae from animals, with special attention to the emergence of carbapenemase-producing Gram-negative bacteria in animals (Table 6) and the emergence of the colistin resistance.

Table 6.
Examples of carbapenemase genes found in Gram-negative bacteria from animals.
5.1. Acinetobacter baumannii
A. baumannii is an important nosocomial pathogen that usually affects immunocompromised patients suffering from various underlying diseases []. Nosocomial infection with this bacterium has been associated with increased morbidity, mortality and health care costs []. It is responsible of hospital outbreaks, and it has a remarkable ability to survive for prolonged periods throughout hospital environments []. It is well established that the members of the genus Acinetobacter are ubiquitous microorganisms. However, A. baumannii as a highly prevalent microorganism in nature, is a misconception because the difficulties encountered in its identification [,]. In fact, A. baumannii is phenotypically and genotypically closely related to other Acinetobacter species (A. pittii, A. nosocomialis and A. calcoaceticus), making the species identification challenging [].
A. baumannii has been related with community-acquired infections (single events or case series) [], and has been isolated from various environmental locations: soils contaminated with petroleum hydrocarbons, vegetables, inanimate surfaces in contact with humans, manured agricultural soil, pig slurry and aquaculture environments []. Multi-susceptible Acinetobacter isolates were commonly found in the 70s–80s in soil and in the hospital setting, clinical A. baumannii being easily treated with common antibiotics during this period [,]. However, this bacterium has an extraordinary ability to upregulate or acquire resistance determinants, and nowadays infections with multidrug or even pandrug resistant isolates are increasing [].
During the last decade, A. baumannii strains have also been isolated from animals, mainly causing outbreaks in veterinary clinics or hospitals []. A. baumannii strains have been isolated from diverse animals including ducks, pigeons, chickens, donkeys, rabbits, pets (cats and dogs), mules, livestock (cattle, caws, goats, pigs), horses, lice and arthropods []. In a study based on PFGE, it was seen that isolates from livestock were different than A. baumannii human strains []. Yet, isolates recovered from pig fecal samples harbored blaOXA-51, which has already been reported in human clinical isolates []. In the same study new blaOXA-51-like genes (blaOXA-148, blaOXA-149 and blaOXA-150), not previously detected in human isolates, were described in bacteria from cattle []. However, it is important to underline that the blaOXA-23 and blaOXA-51-like genes may be naturally occurring in A. baumannii []. On the other hand, the studies in companion animals have reported A. baumannii isolates genetically similar to the nosocomial European (also called International) clones I, II and III, suggesting its spread from humans to animals directly or via the environment [,,,].
Regarding the distribution of AMR-genes in A. baumannii in animals, some studies have reported the emergence of carbapenemase producing A. baumannii in livestock and companion animals [,] (Table 6). The blaOXA-23 is a wide distributed carbapenemase gene in A. baumannii isolates, which has been described recently in livestock and/or pets [,,,,]. The blaOXA-23 gene has also been found in A. lwoffi from poultry []. In A. baumannii this gene has been found in STs not previously reported in humans, as well as in A. baumannii strains belonging to ST2, which has been previously related to hospitals outbreaks []. The blaOXA-58 has been found together with blaOXA-23 in A. baumannii from fowl []. Similarly, a blaNDM-1 positive A. baumannii isolate has been recovered from a pig suffering from pneumonia and sepsis []. This blaNDM-1 was harbored by a plasmid also carrying other AMR-genes [aphA6, ble and msr(E)-mph(E)] []. The blaNDM-1 has been found in other Acinetobacter species [A. junii (from a pig farm) and A. calcoaceticus (from around a cow farm)] recovered from environmental samples and farm animals [,]. A new carbapenemase (blaOXA-497) has been found in A. baumannii from dairy cattle []. A class 1 integron similar to an integron of a human isolate has been also identified in an equine A. baumannii isolate [].
Currently, there are some reports underlying the emergence of A. baumannii strains resistant to both carbapenems and polymyxins (colistin and polymyxin B) in the clinical setting []. Resistance rates against polymyxins varied from 0.7% to 6.5% depending on the country []. Most polymyxin resistant strains from Europe have been recovered in Greece and Italy []. Interestingly, recent reports have also underlined the presence of colistin and polymyxin B resistant A. baumannii and other Acinetobacter spp. isolates in meat (chicken, turkey, beef and pork) [,]. Resistance to polymyxins in A. baumannii is mediated by mutations in the genes pmrA and/or pmrB []. Nevertheless, a new colistin resistance mechanism mediated by plasmids (the mcr-genes, see Section 5.3.4) has emerged in Enterobacteriaceae []. Although no clinical or environmental A. baumannii isolates carrying mcr-genes have yet been described, a recent study has proved that A. baumannii transformed with mcr-1 carrying plasmids had increased colistin resistance []. This finding highlights the threat of a possible dissemination of these mcr-genes to multidrug resistant A. baumannii [].
5.2. Pseudomonas aeruginosa
P. aeruginosa is an opportunistic pathogen often found in water and soil that is pathogenic to plants, humans, farm animals and companion animals. In humans, it is a cause of community and nosocomial infections, especially in patients immunocompromised and/or with cystic fibrosis. In animals, it caused pyoderma, otitis and urinary tract infections in companion animals, mastitis in dairy cows, endometritis in horses and hemorrhagic pneumoniae in fur-bearing animals [,,,,]. Due to the presence of several drug efflux systems and porins, as well as its cell wall with low permeability, P. aeruginosa is intrinsically resistant to a wide range of antimicrobials including benzylpenicillins, aminobenzylpenicillins, carboxypenicillins, first and second generation cephalosporins, chloramphenicol and tetracycline [,]. Moreover, this bacterium is able to form biofilms and to acquire diverse resistance mechanisms.
The studies about AMR in animals are scarce, and mainly have focused in companion animals. A study has shown that P. aeruginosa isolates recovered from diverse veterinary samples during 1994–2003 have high resistance rates against β-lactams (70–100%) and sulphonamides (80–90%), while resistance against quinolones an aminoglycosides are more variable, with resistances ranging from 5 to 98% depending of the antibiotic []. Similar results were found in a study with canine isolates recovered between 2003 and 2006, although an increase in quinolone resistance was seen []. Resistance rates to the former antimicrobials are also similar in current studies among companion animals, although higher resistance rates against the aminoglycoside gentamicin [] and the quinolone enrofloxacin [] have been found. In P. aeruginosa from companion animals, the resistance to quinolones has been related to point mutations in gyrA, gyrB, parC and/or parE genes [,], and the resistance to aminoglycoside has been associated to diverse resistance genes (such as aacA4 and aadA6) [,]. Resistance rates are generally lower in P. aeruginosa from livestock comparing to strains from companion animals [,].
Regarding its potential as zoonotic pathogen, a recent study by Haenni et al. [] confirmed that the P. aeruginosa population in veterinary samples from France has a non-clonal epidemic structure. There was a poor association between an animal species and a specific clone, even though certain clones, possibly correlating with higher pathogenicity, seem to be more prevalent than others []. Clones associated to human outbreaks were detected, but not the most frequent epidemic clones associated to MDR in humans (also called “high risk clones”) [].
The presence of ESBLs (via phenotypic methods) has been confirmed in P. aeruginosa isolates from animals [,]. Moreover, some reports have underlined the emergence of P. aeruginosa isolates with carbapenemases in animals (Table 6). The gene blaIMP-4 has been found in P. aeruginosa from dog [], and the gene blaVIM-2 has been found in P. aeruginosa from cattle and fowl [].
Similar to A. baumannii, recent reports have underlined the emergence of polymyxin-resistant P. aeruginosa []. Resistance rates are low (0.5% to 1.1%) in most countries, although, the situation is worrying in China where 22.2% of extensively drug-resistant bacteraemic P. aeruginosa isolates are resistant to polymyxin B []. Diverse mechanisms of polymyxin resistance have been described in P. aeruginosa, even though no clinical or environmental strains carrying mcr-genes have been reported []. Nevertheless, the recent study by Liu et al. [] showed that P. aeruginosa isolates transformed with plasmids carrying mcr-1 only had moderate changes in colistin susceptibility.
5.3. Enterobacteriaceae
As some Enterobacteriaceae (Ex. Salmonella enterica, Yersinia enterocolitica) are typically food-borne pathogens, this section will mainly focus on some transmissible AMRs-genes that have emerged in Enterobacteriaceae from animals (Table 7). The recent discover of the cfr gene in Enterobacteriaceae has been discussed at Section 4.3.2.

Table 7.
Examples of important plasmid-associated resistance mechanisms in Enterobacteriaceae from animals.
5.3.1. Emergence of Streptothricin-Resistant E. coli in the 1980s
Similarly to Gram-positive bacteria, the use of antibiotics as grown promotors in the animal husbandry may potentiate the emergence of resistant Enterobacteriaceae isolates in food animals. A good example is the emergence of streptothricin-resistant E. coli in the 1980s. The aminoglycosidic growth promoter nourseothricin (streptothricin) was used in farm animals in Germany during the 1980s, while no equivalent antimicrobials were used in humans over this period []. Resistance emerged in E. coli from pigs the second year after the introduction of this antibiotic. It was mediated by a plasmid containing a transposon coding for a streptothricin acetyltransferase []. Subsequently, this resistance was found in E. coli isolated from pig farmers, in the community and in other Enterobacteriaceae from humans (Salmonella and Shigella) []. Nowadays, streptothricin-resistance is extensively extended and it is also characteristic of Gram-positive bacteria such as enterococci and staphylococci [].
5.3.2. ESBL/AmpC-Carrying Enterobacteriaceae in Animals
Bacteria carrying ESBLs are a worldwide clinical problem. ESBLs are mainly plasmid-encoded enzymes providing an extended resistance to β-lactam antibiotics, due to their ability to inactivate cephalosporins [,]. They can be produced by a variety of different bacteria including Enterobacteriaceae or no-fermenting bacteria (such as P. aeruginosa), E. coli and K. pneumoniae being the most frequently found ESBL-producing bacteria. ESBL-producing bacteria are known as nosocomial pathogens and since the late 1990s they have been increasingly found as a causal agent of infections in the community []. Until the 90s, the vast majority of ESBLs identified in human clinical isolates were SHV (sulfhydryl-variable) or TEM (named Temoneira for the first patient from whom the pathogen was isolated) types []. However, later, CTX-M (cefotaximase) β-lactamases have emerged and currently they are the most prevalent ESBLs in human Enterobacteriaceae [].
The occurrence of ESBL-producing bacteria has been broadly recognized in veterinary medicine since the 2000s [,]. ESBL-producing bacteria have been found as disease agents and/or colonizers in livestock, companion animals, zoo animals and wild animals [,,,,]. E. coli and Klebsiella spp. are cause of mastitis in dairy cattle, but most often livestock animals are asymptomatic carriers of ESBLs producers [,]. The first ESBL-producing E. coli was isolated from a dog with urinary tract infection and it carried SHV-12 []. Later on diverse CTX-M, TEM and SHV types have been observed in E. coli, Salmonella spp. and K. pneumoniae from livestock and companion animals [,,,,]. The β-lactamases CTX-M-1, CTX-M-14, CTX-M-15, SHV-12 and TEM-52 are the most frequent types in Enterobacteriaceae from animals [,,,,,]. This distribution is similar in humans, where CTX-M types are the major β-lactamases in E. coli and Klebsiella spp. [,]. Host-range plasmids of different incompatibility groups (such as IncN, IncI, incF and IncK) have been related to these blaCTX-M genes [].
Identical phylogenic lineages (such as ST131) have been found in E. coli isolates from humans and animals []. The risk of zoonotic transfer from livestock to people with close contact to these animals is still largely unknown, but some studies have implicated a transfer of ESBL-producing E. coli or ESBLs genes from poultry or pigs to farm workers [] Besides this direct zoonotic transfer, other routes as foods of animal origin may be a risk factor for human colonization or infection. In a recent study 70.6% of tested farms were ESBL-positive []. In one case the same isolate was detected in human and cattle samples, indicating a zoonotic transfer. In few other cases, pig and human isolates shared the same ESBLs genes, although the isolates belonged to different lineages suggesting horizontal gene transfer.
In contrast to the situation in Europe, ESBL genes have not been commonly reported in animal isolates in North America []. In North America, the plasmid-encoded AmpC B-lactamases genes are more frequently found [], but these genes have also been described in Europe []. The broad-spectrum cephalosporinases (AmpC) were the first β-lactamases described in E. coli [], and nowadays they have emerged worldwide []. As for ESBLs, E. coli carrying AmpC (blaCMY-2) has been identified in animals during the 2000s []. These β-lactamases have been detected in E. coli and Salmonella spp. from livestock and companion animals, being CMY-2 the most frequent one [,]. Other AmpC β-lactamases encoding by blaACC type genes have also been detected in Salmonella spp. from livestock [,].
5.3.3. Carbapenemase-Producing Enterobacteriaceae in Animals
The carbapenemases are β-lactamases able of degrading carbapenems []. The epidemiologically most important carbapenemases are class B metallo-β-lactamases (MBLs) such as VIM (Verona integron-encoded MBL), IMP (imipenemase) and NDM (New Delhi MBL), class A, such as, KPC (K. pneumoniae carbapenemase), and class D including OXA (Carbapenem-hydrolysing oxacillinase) carbapenemases []. Resistance due to carbapenemases is mainly linked to the nosocomial setting, since carbapenems are not approved for use in veterinary medicine. However, they may be use for companion animals under certain conditions []. Although they are not used in livestock, carbapenem-resistant bacteria have been isolated from farm animals in recent years []. In animals, the first carbapenemase gene, blaVIM-1, was found in a porcine E. coli and it was linked to a multiresistance class 1 integron []. Later on, this and other carbapenemases have been found in livestock, wildlife and companion animals (Table 6), notably blaVIM-1 in S. enterica serovar Infantis and E. coli from livestock; blaNDM-1 in S. enterica sevorar Corvallis and E. coli from wild animals and/or livestock; blaOXA-48 in E. coli and K. pneumoniae from companion animals [,]. Carbapenemases have been detected in other Gram-negative bacteria including A. baumannii (see Section 5.1) and P. aeruginosa (see Section 5.2). However, the limited reports suggested that carbapenem-resistant bacteria are still at a very low prevalence in livestock [].
5.3.4. mcr-Genes Mediating Colistin Resistance
Colistin, also named polymyxin E, is currently used as a last-line drug against MDR Gram-negative bacteria [,]. However, resistance against colistin has even emerged in humans without contact to this antibiotic []. Moreover, since the 60s colistin has been used in pig production for therapeutic (in monotherapy), prophylactic and grown promotions purposes []. In addition, to its use in pigs, polymyxins (especially polymyxin B), are used in some countries for the treatment of coliform and Pseudomonas mastitis in cows []. Polymyxins are used in companion animals (dogs and cats) for topical indications such as otitis and ophthalmic diseases [].
Colistin resistance is frequently due to chromosomal mutations and it has been detected in P. aeruginosa, A. baumannii and Enterobacteriaceae [,]. Most mechanisms conferring resistance against colistin are related to modifications of its primary target, the lipid A moiety of lipopolysaccharide (LPS) [,]. Colistin resistance mechanisms are different across bacterial genera, but most implied lipid A modifications with 4-amino-4-deoxy-l-arabinose (l-ara4N) and/or phosphoethanolamine (PEtN) [,].
The chromosomal-related colistin resistance mechanisms have no possibly of horizontal transfer, but plasmid-related genes (mcr-1, mcr-2) coding for PEtN transferases have recently emerged. The first mcr-gene, corresponded to mcr-1. It was initially described in an IncI2 plasmid from an E. coli from a pig, but in the same study it was soon after found in E. coli and K. pneumoniae from patients []. This gene was discovered in November 2015, but recent research has identified this gene in E. coli collections from the 80s []. The amino-acid sequence of MCR-1 showed that it was closely related (63%) to the PEtN transferase EptA found in Paenibacillus sophorae and Enhydrobacter aerosaccus []. Interestingly, the polymyxin is biosynthesized in Paenibacillus spp. []. Structural analysis of the MCR-1 complete protein showed that it was closely related to the PEtN transferases LptA of Neisseria meningitidis and EptC of Campylobacter jejuni, which are also known to be intrinsically resistant to polymyxin [,]. Further research showed that the prevalence of the mcr-1 gene was 20% in animal strains and 1% in human strains in China, and nowadays it has been also detected in animal and human isolates recovered from other countries covering Europe, Africa and America [,,].
The mcr-1 gene has been identified in Enterobacteriaceae from humans, food, farm animals, wildlife, and environment samples []. Although it has been mainly detected (80%) in E. coli, it has also been described in Klebsiella, Salmonella, Shigella and Enterobacter [,]. In addition, it has recently been detected in Cronobacter sakazakii []. Interestingly, the mcr-1 gene has been related to diverse plasmid incompatibility groups including IncFIA, IncFIB, IncFIC, IncFI, IncFII, IncHI2, IncI1, IncI2, IncN, IncP, IncQ1, IncX1, IncX4, IncY and pVT553 [,,]. This gene has also been related to a class 1 integron within an IncFII plasmid []. Moreover, mcr-1 has been identified in ESBL and carbapenamase producing Enterobacteriaceae from animals and humans (Table 8).

Table 8.
Associated β-lactam resitances in colistin-resistant mcr-1 carrying isolates.
A variant (with only a SNP of difference) of mcr-1, mcr-1.2 has been detected on a transferable IncX4 plasmid in a K. pneumoniae recovered from a patient []. This mcr-1.2 strain also carried β-lactamases genes (blaTEM-1, blaSHV-11, and blaKPC-3) []. Very recently, a novel plasmid mediated colistin resistance gene, mcr-2 has been discovered []. This mcr-2 has been found in an IncX4 plasmid of E. coli from porcine and bovine origin in Belgium []. This gene corresponded to a new mcr-gene, since MCR-1 and MCR-2 shared 80.7% of similarity []. It has been seen that the mcr-2 gene had higher prevalence than mcr-1 in E. coli isolates from porcine origin [].
Food animals seem the main source of human contamination by the mcr-genes []. However, the mcr-1 gene has also been detected in E. coli carried or infecting humans without animal contact []. These findings underlined that this gene, which probably has emerged in the animal sector, is already widespread in the environment and it is transmissible via various routes to humans [,].
5.3.5. Other AMR-Genes in Enterobacteriaceae from Animals
Increasing levels of quinolone resistance among Enterobacteriaceae and other bacteria (Campylobacter spp.) have been a particular case of concern since the 90s []. The extended use of quinolones to treat poultry infections has increased the quinolone resistance among E. coli strains in poultry industry []. Although quinolone resistance is generally not highly frequent in animal farming, high and/or moderate levels of quinolone resistance have been reported in poultry [].
The mechanisms of quinolone resistance in Enterobacteriaceae from animals are similar to those described in isolates from humans []. Mutations at gyrA (DNA gyrase) and/or parC (topoisomerase IV) genes were responsible of quinolone resistance in E. coli and Salmonella spp. isolates from animals [,]. Moreover, reports about plasmid-mediated quinolone resistance (PMQR) genes [qnr, acc(6)-Ib-cr, qepA, oqxAB] in bacteria from animal origin have been published since 2000s [,].
The Qnr proteins are encoded by several variants of qnr genes (qnrA, qnrB, qnrC, qnrD, qnrS), being the qnrS1 gene variant the most frequently reported and worldwide diffused in animal and human samples [] (Table 9). Some qnr genes have been described in Enterobacteriaceae (mainly Salmonella spp. and E. coli) of animal origin [,,]. Their wide distribution suggests an origin prior to the inclusion of quinolones in medicine []. It has been suggested that these genes have originated in bacteria from the natural environment []. In fact, qnr genes have been found in aquatic and waterborne organisms such as Shewanella, Aeromonas and Citrobacter species, and in the Vibrionaceae family [,].

Table 9.
Examples of distribution of certain PMQR-genes.
As the qnr genes, PMQR efflux pump genes (qepA and oqxAB) have also a wide distribution, and they have been found in diverse Enterobacteriaceae from animals and humans [,] (Table 9). These genes are related to transmissible plasmids, although oqxAB is also commonly found in the chromosome of K. pneumoniae []. Other plasmid-mediated efflux pumps affecting quinolones have been described sporadically [].
The cr variant of the aminoglycoside aac(6′)-Ib resistance gene was discover in 2006 []. The aac(6′)-Ib gene encodes a aminoglycoside acetyltransferase able to confer resistance against kanamycin, tobramycin and amikacin. The two mutations present at aac(6′)-Ib-cr confer low level ciprofloxacine resistance, with a slightly cost to the levels of aminoglycoside resistance []. Hence, it acts additively together with Qnr proteins to generate quinolone resistance []. This gene is widely distributed in Gram-negative bacteria (Table 9).
Regarding other aminoglycoside resistance genes, the gene aac(3)-IV, which confers cross-resistance between gentamicin and apramycin, need to be underlined []. It was originally isolated in 1981 from an E. coli recovered from farm animals in France. Although apramycin has only veterinary use, this gene has been detected in Enterobacteriaceae from human patients and wastewater from a residential area []. It has been suggested that apramycin consumption at farm level has increased the occurrence of aac(3)-IV positive E. coli in pigs [].
Several other aminoglycoside resistance genes [such as aac(3)-II, aac(3)-III, aadA1, aadA2, aadA5 and ant(2″)-I] have been described in E. coli from animal origin []. Other aminoglycoside resistance genes [aac(3)-I, ant(2″)-Ia and aac(6)-Ib] are more related to human E. coli isolates [].
Enterobacteriaceae from animals carry other resistance genes. For example, a moderate incidence of chloramphenicol resistance is registered in E. coli from food animals in Europe, mainly mediated by genes such as catA1, floR and cmlA1 []. Other chloramphenicol resistance genes (such as catB) are more frequent in human E. coli isolates [].
E. coli isolates from animals carry also diverse sulfonamide (sul1, sul2, sul3), tetracycline [tet(A), tet(B)], and trimethoprim (dfrA1, dfrA12, dfrA17) resistance genes [,]. Interestingly, the gene sul3 was first detected in an E. coli isolate from a pig, and later it was found in both healthy and diseased humans [].
6. Conclusions
The continuous antibiotic selective pressure in human and animal health may contribute to the mobilization of acquired resistance genes. This is illustrated by several facts: (i) some studies have suggested that the mecA gene may have originated in animal related staphylococci; (ii) it has recently been observed that two mutiresistance genes (cfr and optrA) associated to MGEs such as plasmids along with other AMR-genes, have a wide dissemination in Gram-positive bacteria from animals and humans; (iii) typical nosocomial resistances linked to ESBLs and carbapenemases are emerging in Gram-negative bacteria from animals; and (iv) the mcr-genes, which may probably have emerged in the animal sector are currently spreading among human and animal E. coli isolates. These examples underline the fact that bacteria from animals represent an important pool of resistance genes for human pathogens.
Resistance to antibiotics is escalating, and at the same time the current pipeline of new antimicrobials is running dry, creating an ever increasing gap. Since a complete ban on the use of antimicrobials in farm animals would have serious repercussions for animal health, welfare and productivity, we need to use these agents more prudently in both human and animal medicine []. Rapid diagnosis tools are needed to determine therapy strategies more quickly and accurately, as well as the further examination of alternatives to antimicrobials for farm animals (such as phage therapy or vaccines) []. We are facing the possibility of a future without effective antibiotics for some infections and a scenario where infections that hitherto were considered harmless, are now a serious health problem and a major cause of morbidity, mortality, together with major financial and social repercussions.
Acknowledgments
We thank members of our laboratories and our microbiologist colleagues for their support to the Belgian National Reference Centre—Staphylococcus aureus.
Author Contributions
Maria Angeles Argudín conceived, designed and wrote the manuscript, Ariane Deplano, Alaeddine Meghraou, Magali Dodémont, Amelie Heinrichs, Olivier Denis, Claire Nonhoff and Sandrine Roisin revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| l-ara4N | 4-amino-4-deoxy-l-arabinose |
| ABC | ATP-binding cassette |
| AMR | antimicrobial resistance |
| CC | clonal complex |
| CoNS | coagulase negative staphylococci |
| CTX-M | cefotaximase |
| ECDC | European Centre for Disease Prevention and Control |
| EFSA | European Food Safety Authority |
| erm | erythromycin ribosomal methylase |
| ESBLs | extended spectrum beta-lactamases |
| ESC(K)APE | E. faecium, S. aureus, Clostridium difficile, A. baumannii, P. aeruginosa, and Enterobacteriaceae |
| ESKAPE | Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. |
| HIV | human immunodeficiency virus |
| IMP | imipenemase |
| IS | insertion sequence |
| KPC | K. pneumoniae carbapenemase |
| QRDR | quinolone-resistance determining region |
| LA-MRSA | livestock-associated methicillin resistant Staphylococcus aureus |
| LPS | lipopolysaccharide |
| MBL | metallo-β-lactamase |
| MFS | Major Facilitator Superfamily |
| MGE | mobile genetic element |
| MLSB | macrolide-lincosamide-streptogramin B |
| MLST | multilocus sequence typing |
| MRCoNS | methicillin resistant coagulase negative staphylococci |
| MDR | multidrug resistance |
| MRSA | methicillin resistant Staphylococcus aureus |
| MRSE | methicillin-resistant Staphylococcus epidermidis |
| MSSA | methicillin susceptible S. aureus |
| NDM | New Delhi metallo-β-lactamases |
| OIE | World Organization for Animal Health |
| OXA | carbapenem-hydrolysing oxacillinase |
| PBP | penicillin binding protein |
| PCR | polymerase chain reaction |
| PEtN | phosphoethanolamine |
| PFGE | pulsed-field gel electrophoresis |
| PMQR | plasmid-mediated quinolone resistance |
| PVL | Panton-Valentine leucocidin |
| REA | restriction enzyme analysis |
| RND | resistance-nodulation-cell division family |
| RT | ribotype |
| SCCmec | staphylococcal cassette chromosome mec |
| SHV | sulfhydryl-variable β-Lactamase |
| SNP | single nucleotide polymorphism |
| ST | sequence type |
| TEM | Temoneira β-Lactamase |
| VIM | Verona integron-encoded metallo-β-lactamase |
| VRE | vancomycin-resistant enterococci |
| WHO | World Health Organization |
References
- Butaye, P.; Argudín, M.A.; Threlfall, J. Introduction to antimicrobial-resistant foodborne pathogens. In Antimicrobial Resistance and Food Safety: Methods and Techniques, 1st ed.; Chen, C.Y., Yan, X., Jackson, C.R., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 1–18. ISBN 978-0-12-801214-7. [Google Scholar]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance; HM Government and the Wellcome Trust: London, UK, 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 27 March 2017).
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, F.G.; Corcione, S.; Pagani, N.; Di Perri, G. From ESKAPE to ESCAPE, from KPC to CCC. Clin. Infect. Dis. 2015, 60, 1289–1290. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- One Health Initiative will unite human and veterinary medicine. Available online: http://www.onehealthinitiative.com/index.php (accessed on 5 January 2017).
- Da Costa, P.M.; Loureiro, L.; Matos, A.J. Transfer of multidrug-resistant bacteria between intermingled ecological niches: The interface between humans, animals and the environment. Int. J. Environ. Res. Public Health 2013, 10, 278–294. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Fedorka-Cray, P.J. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002, 34, S93–S106. [Google Scholar] [CrossRef] [PubMed]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. mBio 2016, 7, e02227. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 5th ed.; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-151222-0. [Google Scholar]
- World Organisation for Animal Health. Available online: http://www.oie.int/en/ (accessed on 27 March 2017).
- Anonymous. OIE List of Antimicrobial Agents of Veterinary Importance. World Organisation for Animal Health (OIE), 2015. Available online: http://www.oie.int/en/our-scientific-expertise/veterinary-products/antimicrobials/ (accessed on 27 March 2017).
- FDA. Summary Report on Antimicrobials Sold or Distributed for Use in Food Producing Animals. 2015. Available online: http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM534243.pdf (accessed on 27 March 2017).
- FAO. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. 2016. Available online: http://www.fao.org/3/a-i6209e.pdf (accessed on 27 March 2017).
- Yazdankhah, S.; Rudi, K.; Bernhoft, A. Zinc and copper in animal feed—Development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 2014, 25. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Butaye, P. Dissemination of metal resistance genes among animal methicillin-resistant coagulase-negative staphylococci. Res. Vet. Sci. 2016, 105, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Lauzat, B.; Kraushaar, B.; Alba, P.; Agerso, Y.; Cavaco, L.; Butaye, P.; Porrero, M.C.; Battisti, A.; Tenhagen, B.A.; et al. Heavy metal and disinfectant resistance genes among livestock-associated methicillin-resistant Staphylococcus aureus isolates. Vet. Microbiol. 2016, 191, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Nunez-Garcia, J.; Kearns, A.M.; Doumith, M.; Butaye, P.R.; Argudín, M.A.; Lahuerta-Marin, A.; Pichon, B.; AbuOun, M.; Rogers, J.; et al. Livestock-associated methicillin resistant Staphylococcus aureus (LA-MRSA) clonal complex (CC) 398 isolated from UK animals belong to European lineages. Front. Microbiol. 2016, 7, 1741. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.G.; McEwen, S.A.; Li, X.Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Agga, G.E.; Arthur, T.M.; Durso, L.M.; Harhay, D.M.; Schmidt, J.W. Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS ONE 2015, 10, e0132586. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yuan, K.; Chen, X.; Yang, Y.; Zhang, T.; Wang, Y.; Luan, T.; Zou, S.; Li, X. Metagenomic analysis revealing antibiotic resistance genes (ARGs) and their genetic compartments in the Tibetan environment. Environ. Sci. Technol. 2016, 50, 6670–6679. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K. Antibiotic resistance gene discovery in food-producing animals. Curr. Opin. Microbiol. 2014, 19, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G. The structure and diversity of human, animal and environmental resistomes. Microbiome 2016, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.; Walsh, F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol. Ecol. 2016, 92, 168. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Harhay, G.P.; Bono, J.L.; Smith, T.P. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Method 2011, 84, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Qu, A.; Brulc, J.M.; Wilson, M.K.; Law, B.F.; Theoret, J.R.; Joens, L.A.; Konkel, M.E.; Angly, F.; Dinsdale, E.A.; Edwards, R.A. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 2008, 3, e2945. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.W.; Yanke, J.L.; Reuter, T.; Topp, E.; Read, R.R.; Selinger, B.L.; McAllister, T.A. Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics. BMC Microbiol. 2011, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Chambers, L.; Yang, Y.; Littier, H.; Ray, P.; Zhang, T.; Pruden, A.; Strickland, M.; Knowlton, K. Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin. PLoS ONE 2015, 10, e0133764. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse antibiotic resistance genes in dairy cow manure. mBio 2014, 5, e01017. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Miller, D.N.; Wienhold, B.J. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and nonagricultural metagenomes. PLoS ONE 2012, 7, e48325. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczak, K.A.; Scott, K.P.; Kelly, D.; Aminov, R.I. Tetracycline resistome of the organic pig gut. Appl. Environ. Microbiol. 2009, 75, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Brown Kav, A.; Sasson, G.; Jami, E.; Doron-Faigenboim, A.; Benhar, I.; Mizrahi, I. Insights into the bovine rumen plasmidome. Proc. Natl. Acad. Sci. USA 2012, 109, 5452–5457. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Binh, C.T.; Jechalke, S.; Kopmann, C.; Zimmerling, U.; Krögerrecklenfort, E.; Ledger, T.; González, B.; Top, E.; Smalla, K. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: Diversification driven by class 1 integron gene cassettes. Front. Microbiol. 2012, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Looft, T.; Bayles, D.O.; Humphrey, S.; Levine, U.Y.; Alt, D.; Stanton, T.B. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2011, 2, e00260. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Morroni, G.; Vincenzi, C.; Manso, E.; Mingoia, M.; Giovanetti, E.; Varaldo, P.E. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 2016, 71, 1118–1119. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, L.M.; Ashcraft, D.S.; Kahn, H.P.; Pankey, G.; Jones, R.N.; Farrell, D.J.; Mendes, R.E. Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2015, 59, 6256–6261. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Shen, Y.; Schwarz, S.; Cai, J.; Lv, Y.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J.; et al. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 2016, 71, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Li, D.; Wang, Y.; He, T.; Feßler, A.T.; Schwarz, S.; Wu, C. Presence of the optrA gene in methicillin-resistant Staphylococcus sciuri of porcine origin. Antimicrob. Agents Chemother. 2016, 60, 7200–7205. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Dai, L.; Wu, C.; Shen, J. First report of multiresistance gene cfr in Enterococcus species casseliflavus and gallinarum of swine origin. Vet. Microbiol. 2014, 170, 352–357. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Kadlec, K.; Schwarz, S.; Torres, C. First detection of the staphylococcal trimethoprim resistance gene dfrK and the dfrK-carrying transposon Tn559 in enterococci. Microb. Drug Resist. 2012, 18, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Schwarz, S.; Aarts, H.J. Erratum: Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2012, 3, 384. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, Y.; Schwarz, S. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2013, 68, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Spigaglia, P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther. Adv. Infect. Dis. 2016, 3, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Van Hoek, A.H.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed]
- Wendlandt, S.; Feßler, A.T.; Monecke, S.; Ehricht, R.; Schwarz, S.; Kadlec, K. The diversity of antimicrobial resistance genes among staphylococci of animal origin. Int. J. Med. Microbiol. 2013, 303, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.A.; Dawson, S.; French, G.L.; Goossens, H.; Hawkey, P.M.; Kuijper, E.J.; Nathwani, D.; Taylor, D.J.; Teale, C.J.; Warren, R.E.; et al. Antimicrobial-resistant pathogens in animals and man: Prescribing, practices and policies. J. Antimicrob. Chemother. 2010, 65, i3–i17. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Galperin, M.Y. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 2013, 15, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Citron, D.M.; Tyrrell, K.L.; Finegold, S.M. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938. Anaerobe 2016, 40, 95–99. [Google Scholar] [CrossRef] [PubMed]
- March, J.W.; Harrison, L.H. Clostridium difficile: A food safety concern? In Antimicrobial Resistance and Food Safety: Methods and Techniques, 1st ed.; Chen, C.Y., Yan, X., Jackson, C.R., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 181–206. [Google Scholar]
- Keessen, E.C.; Hensgens, M.P.; Spigaglia, P.; Barbanti, F.; Sanders, I.M.; Kuijper, E.J.; Lipman, L.J. Antimicrobial susceptibility profiles of human and piglet Clostridium difficile PCR-ribotype 078. Antimicrob. Resist. Infect. Control 2013, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Dahms, C.; Hübner, N.O.; Wilke, F.; Kramer, A. Mini-review: Epidemiology and zoonotic potential of multiresistant bacteria and Clostridium difficile in livestock and food. GMS Hyg. Infect. Control 2014, 9. [Google Scholar] [CrossRef]
- Norén, T.; Johansson, K.; Unemo, M. Clostridium difficile PCR ribotype 046 is common among neonatal pigs and humans in Sweden. Clin. Microbiol. Infect. 2014, 20, O2–O6. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 2008, 282, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Martín, A.; Alcalá, L.; Cercenado, E.; Iglesias, C.; Reigadas, E.; Bouza, E. Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob. Agents Chemother. 2015, 59, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.H.; Vester, B. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob. Agents Chemother. 2015, 59, 5841–5843. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Thanki, A.M.; Jose, B.R.; Oggioni, M.R.; Clokie, M.R. Use of single molecule sequencing for comparative genomics of an environmental and a clinical isolate of Clostridium difficile ribotype 078. BMC Genom. 2016, 17, 1020. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Zurek, L. Antibiotic resistance in Enterococci: A food safety perspective. In Antimicrobial Resistance and Food Safety: Methods and Techniques, 1st ed.; Chen, C.Y., Yan, X., Jackson, C.R., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 155–180. [Google Scholar]
- Garrido, A.M.; Gálvez, A.; Pulido, R.P. Antimicrobial Resistance in Enterococci. J. Infect. Dis. Ther. 2014, 2, 150. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Bates, J.; Jordens, Z.; Selkon, J.B. Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 1993, 342, 490–491. [Google Scholar] [CrossRef]
- Woodford, N.; Palepou, M.F.; Johnson, A.P.; Chadwick, P.R.; Bates, J. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. Lancet 1997, 350, 738. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Schwarz, S.; Cai, J.; Fan, R.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 2016, 71, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, L.; Wu, Z.; Wang, L. Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J. Antimicrob. Chemother. 2016, 72, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [PubMed]
- Kluytmans, J.A.; Wertheim, H.F. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 2005, 33, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; van Leeuven, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Van Belkum, A.; Verkaik, N.J.; de Vogel, C.P.; Boelens, H.A.; Verveer, J.; Nouwen, J.L.; Wertheim, H.F. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009, 199, 1820–1826. [Google Scholar] [CrossRef] [PubMed]
- Mandell, G.L.; Bennett, J.E.; Dolin, R. Staphylococcus aureus (including Staphylococcal Toxic Shock). In Bennett’s-Principles and Practice of Infectious Diseases; Waldvogel, F.A., Ed.; Churchill Livingstone: Philadelphia, PA, USA, 2000. [Google Scholar]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.R.; Penades, J.R. Staphylococci of Animals. In Staphylococcus Molecular Genetics, 1st ed.; Lindsay, J., Ed.; Caister Academic Press: Haverhill, UK, 2008; pp. 255–269. ISBN 978-1-904455-29-5. [Google Scholar]
- Werckenthin, C.; Cardoso, M.; Martel, J.L.; Schwarz, S. Antimicrobial resistance in staphylococci from animals with particular reference to bovine Staphylococcus aureus, porcine Staphylococcus hyicus, and canine Staphylococcus intermedius. Vet. Res. 2001, 32, 341–362. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Fessler, A.T.; Hauschild, T.; Kehrenberg, C.; Kadlec, K. Plasmid-mediated resistance to protein biosynthesis inhibitors in staphylococci. Ann. N. Y. Acad. Sci. 2011, 1241, 82–103. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Fessler, A.T.; Hauschild, T.; Schwarz, S. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012, 18, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Shen, J.; Wendlandt, S.; Fessler, A.T.; Wang, Y.; Kadlec, K.; Wu, C.M. Plasmid-mediated antimicrobial resistance in Staphylococci and other firmicutes. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Wendlandt, S.; Shen, J.; Kadlec, K.; Wang, Y.; Li, B.; Zhang, W.J.; Feßler, A.T.; Wu, C.; Schwarz, S. Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol. 2015, 23, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Tenhagen, B.A.; Fetsch, A.; Sachsenröder, J.; Käsbohrer, A.; Schroeter, A.; Hammerl, J.A.; Hertwig, S.; Helmuth, R.; Bräunig, J.; et al. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 2011, 277, 3052–3060. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Vanderhaeghen, W.; Butaye, P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res. Vet. Sci. 2015, 99, 10–16. [Google Scholar] [CrossRef] [PubMed]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Ito, T.; Tsubakishita, S.; Sasaki, T.; Takeuchi, F.; Morimoto, Y.; Katayama, Y.; Matsuo, M.; Kuwahara-Arai, K.; Hishinuma, T.; et al. Genomic Basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 2013, 45, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, F.; Liu, D.; Xue, H.; Zhao, X. Novel Type XII Staphylococcal Cassette Chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob. Agents Chemother. 2015 59, 7597–7601. [CrossRef] [PubMed]
- Vanderhaeghen, W.; Vandendriessche, S.; Crombé, F.; Dispas, M.; Denis, O.; Hermans, K.; Haesebrouck, F.; Butaye, P. Species and staphylococcal cassette chromosome mec (SCCmec) diversity among methicillin-resistant non-Staphylococcus aureus staphylococci isolated from pigs. Vet. Microbiol. 2012, 158, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Vanderhaeghen, W.; Butaye, P. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from pig farms in Belgium. Vet. J. 2015, 203, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Vanderhaeghen, W.; Vandendriessche, S.; Vandecandelaere, I.; André, F.X.; Denis, O.; Coenye, T.; Butaye, P. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from animals and humans. Vet. Microbiol. 2015, 178, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Butaye, P.; Argudín, M.A.; Smith, T.C. Livestock-associated MRSA and its current evolution. Curr. Clin. Microbiol. Rep. 2016, 3, 19–31. [Google Scholar] [CrossRef]
- Becker, K.; Ballhausen, B.; Köck, R.; Kriegeskorte, A. Methicillin resistance in Staphylococcus isolates: The “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int. J. Med. Microbiol. 2014, 304, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Mendoza, M.C.; González-Hevia, M.A.; Bances, M.; Guerra, B.; Rodicio, M.R. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Environ. Microbiol. 2012, 78, 2930–2935. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Mendoza, M.C.; Martín, M.C.; Rodicio, M.R. Molecular basis of antimicrobial drug resistance in Staphylococcus aureus isolates recovered from young healthy carriers in Spain. Microb. Pathog. 2014, 74, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Seah, C.; Alexander, D.C.; Louie, L.; Simor, A.; Low, D.E.; Longtin, J.; Melano, R.G. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1916–1920. [Google Scholar] [CrossRef] [PubMed]
- Strauss, C.; Hu, Y.; Coates, A.; Perreten, V. A Novel erm(44) gene variant from a human Staphylococcus saprophyticus isolate confers resistance to macrolides and lincosamides but not streptogramins. Antimicrob. Agents Chemother. 2016, 61, e01655. [Google Scholar] [CrossRef] [PubMed]
- Wipf, J.R.; Schwendener, S.; Perreten, V. The novel macrolide-lincosamide-streptogramin B resistance gene erm(44) is associated with a prophage in Staphylococcus xylosus. Antimicrob. Agents Chemother. 2014, 58, 6133–6138. [Google Scholar] [CrossRef] [PubMed]
- Wipf, J.R.; Schwendener, S.; Nielsen, J.B.; Westh, H.; Perreten, V. The new macrolide-lincosamide-streptogramin B resistance gene erm(45) is located within a genomic island in Staphylococcus fleurettii. Antimicrob. Agents Chemother. 2015, 59, 3578–3581. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Vanderhaeghen, W.; Argudín, M.A.; Haesebrouck, F.; Butaye, P. Characterization of methicillin-resistant Staphylococcus sciuri isolates from industrially raised pigs, cattle and broiler chickens. J. Antimicrob. Chemother. 2014, 69, 2928–2934. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Haesebrouck, F.; Butaye, P. Molecular epidemiology of methicillin-resistant Staphylococcus sciuri in healthy chickens. Vet. Microbiol. 2014, 171, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Feßler, A.T.; Hauschild, T.; Schwarz, S.; Butaye, P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014, 171, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-Arai, K.; Sasaki, T.; Hiramatsu, K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 2010, 54, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-Arai, K.; Baba, T.; Hiramatsu, K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob. Agents Chemother. 2010, 54, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, L.; Holden, M.T.; Lindsay, H.; Webb, C.R.; Brown, D.F.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Shore, A.C.; Deasy, E.C.; Slickers, P.; Brennan, G.; O’Connell, B.; Monecke, S.; Ehricht, R.; Coleman, D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Schwendener, S.; Thomann, A.; Gobeli Brawand, S.; Perreten, V. First Staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob. Agents Chemother. 2015, 59, 4577–4583. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Stegger, M.; Heltberg, O.; Christensen, J.; Zeuthen, A.; Knudsen, L.K.; Urth, T.; Sorum, M.; Schouls, L.; Larsen, J.; et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin. Microbiol Infect. 2013, 19, E16–E22. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Gongora, C.; Harrison, E.M.; Moodley, A.; Guardabassi, L.; Holmes, M.A. MRSA carrying mecC in captive mara. J. Antimicrob. Chemother. 2015, 70, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Deplano, A.; Vandendriessche, S.; Nonhoff, C.; Denis, O. Genetic diversity among methicillin-resistant Staphylococcus aureus isolates carrying the mecC gene in Belgium. J. Antimicrob. Chemother. 2010, 69, 1457–1460. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.M.; Paterson, G.K.; Holden, M.T.; Morgan, F.J.; Larsen, A.R.; Petersen, A.; Leroy, S.; De Vliegher, S.; Perreten, V.; Fox, L.K.; et al. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob. Agents Chemother. 2013, 57, 1524–1528. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.M.; Paterson, G.K.; Holden, M.T.; Ba, X.; Rolo, J.; Morgan, F.J.; Pichon, B.; Kearns, A.; Zadoks, R.N.; Peacock, S.J.; et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J. Antimicrob. Chemother. 2014, 69, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Loncaric, I.; Kübber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. Antimicrob. Chemother. 2013, 68, 2222–2225. [Google Scholar] [CrossRef] [PubMed]
- Małyszko, I.; Schwarz, S.; Hauschild, T. Detection of a new mecC allotype, mecC2, in methicillin-resistant Staphylococcus saprophyticus. J. Antimicrob. Chemother. 2014, 69, 2003–2005. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.E.; Deshpande, L.M.; Jones, R.N. Linezolid update: Stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 2014, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.C.; Lazaris, A.; Kinnevey, P.M.; Brennan, O.M.; Brennan, G.I.; O’Connell, B.; Feßler, A.T.; Schwarz, S.; Coleman, D.C. First report of cfr-carrying plasmids in the pandemic Sequence Type 22 methicillin-resistant Staphylococcus aureus Staphylococcal Cassette Chromosome mec Type IV Clone. Antimicrob. Agents Chemother. 2016, 60, 3007–3015. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Werckenthin, C.; Kehrenberg, C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 2000, 44, 2530–2533. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.E.; Hogan, P.A.; Jones, R.N.; Sader, H.S.; Flamm, R.K. Surveillance for linezolid resistance via the Zyvox(R) Annual Appraisal of Potency and Spectrum (ZAAPS) programme (2014): Evolving resistance mechanisms with stable susceptibility rates. J. Antimicrob. Chemother. 2016, 71, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Morales, G.; Picazo, J.J.; Baos, E.; Candel, F.J.; Arribi, A.; Peláez, B.; Andrade, R.; de la Torre, M.A.; Fereres, J.; Sánchez-García, M. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 2010, 50, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Sánchez García, M.; De la Torre, M.A.; Morales, G.; Peláez, B.; Tolón, M.J.; Domingo, S.; Candel, F.J.; Andrade, R.; Arribi, A.; García, N.; et al. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 2010, 303, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Paridaens, H.; Coussement, J.; Argudín, M.A.; Delaere, B.; Huang, T.D.; Glupczynski, Y.; Denis, O. Clinical case of cfr-positive MRSA CC398 in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.C.; Brennan, O.M.; Ehricht, R.; Monecke, S.; Schwarz, S.; Slickers, P.; Coleman, D.C. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 2010, 54, 4978–4984. [Google Scholar] [CrossRef] [PubMed]
- Gopegui, E.R.; Juan, C.; Zamorano, L.; Pérez, J.L.; Oliver, A. Transferable multidrug resistance plasmid carrying cfr associated with tet(L), ant(4′)-Ia, and dfrK genes from a clinical methicillin-resistant Staphylococcus aureus ST125 strain. Antimicrob. Agents Chemother. 2012, 56, 2139–2142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Niu, D.; Cao, X.; Ning, M.; Zhang, Z.; Shen, H.; Zhang, K. Clonal dissemination of linezolid-resistant Staphylococcus capitis with G2603T mutation in domain V of the 23S rRNA and the cfr gene at a tertiary care hospital in China. BMC Infect. Dis. 2015, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.; Powell, J.; Finnegan, C.; O’Gorman, A.; Barrett, S.; Hopkins, K.L.; Pichon, B.; Hill, R.; Power, L.; Woodford, N.; et al. 2015. Incidence, management and outcomes of the first cfr-mediated linezolid-resistant Staphylococcus epidermidis outbreak in a tertiary referral centre in the Republic of Ireland. J. Hosp. Infect. 2015, 90, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.L.; Wei, H.K.; Wang, J.; Lin, D.C.; Liu, X.Q.; Liu, J.H. High prevalence of Cfr-producing Staphylococcus species in retail meat in Guangzhou, China. BMC Microbiol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Monchique, C.; Belas, A.; Marques, C.; Gama, L.T.; Pomba, C. Trends and molecular mechanisms of antimicrobial resistance in clinical staphylococci isolated from companion animals over a 16 year period. J. Antimicrob. Chemother. 2016, 71, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, D.; Song, L.; Liu, Y.; He, T.; Liu, H.; Wu, C.; Schwarz, S.; Shen, J. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 2013, 57, 4061–4063. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Wang, Y. Nomenclature and functionality of the so-called cfr gene from Clostridium difficile. Antimicrob. Agents Chemother. 2015, 59, 2476–2477. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Fleige, C.; Klare, I.; Fiedler, S.; Mischnik, A.; Mutters, N.T.; Dingle, K.E.; Werner, G. Detection of a cfr(B) variant in german Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS ONE 2016, 11, e0167042. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Morroni, G.; Pollini, S.; Tiberi, E.; Mingoia, M.; Varaldo, P.E.; Rossolini, G.M.; Giovanetti, E. Characterization of novel conjugative multiresistance plasmids carrying cfr from linezolid-resistant Staphylococcus epidermidis clinical isolates from Italy. J. Antimicrob. Chemother. 2016, 71, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q.; Wang, J.; Li, W.; Zhao, L.Q.; Lu, Y.; Liu, J.H.; Zeng, Z.L. Distribution of cfr in Staphylococcus spp. and Escherichia coli strains from pig farms in China and characterization of a novel cfr-carrying F43:A-:B- Plasmid. Front. Microbiol. 2017, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, H.; Li, L.; Chen, M.Y.; Fang, L.X.; Yang, Q.E.; Liu, Y.H.; Liao, X.P. Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 738–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Xu, X.R.; Schwarz, S.; Wang, X.M.; Dai, L.; Zheng, H.J.; Liu, S. Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J. Antimicrob. Chemother. 2014, 69, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wang, X.M.; Dai, L.; Hua, X.; Dong, Z.; Schwarz, S.; Liu, S. Novel conjugative plasmid from Escherichia coli of swine origin that coharbors the multiresistance gene cfr and the extended-spectrum-β-lactamase gene blaCTX-M-14b. Antimicrob. Agents Chemother. 2015, 59, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Sun, B.; Wang, Y.; Lei, L.; Schwarz, S.; Wu, C. Characterization of a cfr-Carrying Plasmid from Porcine Escherichia coli That Closely Resembles Plasmid pEA3 from the Plant Pathogen Erwinia amylovora. Antimicrob. Agents Chemother. 2016, 60, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Sun, J.; Ma, J.; Li, L.; Fang, L.X.; Zhang, Q.; Liu, Y.H.; Liao, X.P. Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin. PLoS ONE 2014, 9, e102378. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus Sequence Type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef] [PubMed]
- Feltrin, F.; Alba, P.; Kraushaar, B.; Ianzano, A.; Argudín, M.A.; Di Matteo, P.; Porrero, M.C.; Aarestrup, F.M.; Butaye, P.; Franco, A.; et al. A Livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus Clonal Complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl. Environ. Microbiol. 2015, 82, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Rodicio, M.R.; Guerra, B. The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Lett. Appl. Microbiol. 2010, 50, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Fetsch, A.; Tenhagen, B.A.; Hammerl, J.A.; Hertwig, S.; Kowall, J.; Rodicio, M.R.; Käsbohrer, A.; Helmuth, R.; Schroeter, A.; et al. High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I macrorestriction-pulsed-field gel electrophoresis profiles and spa and SCCmec types. Appl. Environ. Microbiol. 2010, 76, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Crombé, F.; Argudín, M.A.; Vanderhaeghen, W.; Hermans, K.; Haesebrouck, F.; Butaye, P. Transmission Dynamics of Methicillin-Resistant Staphylococcus aureus in Pigs. Front. Microbiol. 2013, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Roelandt, S.; Argudín, M.A.; Haesebrouck, F.; Butaye, P. Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathol. 2013, 42, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Haesebrouck, F.; Butaye, P. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet. Res. 2014, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Peeters, L.E.; Argudín, M.A.; Azadikhah, S.; Butaye, P. Antimicrobial resistance and population structure of Staphylococcus aureus recovered from pigs farms. Vet. Microbiol. 2015, 180, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Abdelbary, M.; Layer, F.; Werner, G.; Witte, W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 2015, 177, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Bismuth, R.; Zilhao, R.; Sakamoto, H.; Guesdon, J.L.; Courvalin, P. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob. Agents Chemother. 1990, 34, 1611–1614. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Schwarz, S. dentification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 2010, 54, 3475–3477. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Kadlec, K.; Feßler, A.T.; Zarazaga, M.; Torres, C.; Schwarz, S. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob. Agents Chemother. 2013, 57, 3275–3282. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Layer, F.; Köck, R.; Werner, G.; Witte, W. Methicillin susceptible Staphylococcus aureus (MSSA) of clonal complex CC398, t571 from infections in humans are still rare in Germany. PLoS ONE 2013, 8, e83165. [Google Scholar] [CrossRef] [PubMed]
- Lekkerkerk, W.S.; van Wamel, W.J.; Snijders, S.V.; Willems, R.J.; van Duijkeren, E.; Broens, E.M.; Wagenaar, J.A.; Lindsay, J.A.; Vos, M.C. What is the origin of Livestock-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 398 isolates from humans without livestock contact? An epidemiological and genetic analysis. J. Clin. Microbiol. 2015, 53, 1836–1841. [Google Scholar] [CrossRef] [PubMed]
- Al Bayssari, C.; Dabboussi, F.; Hamze, M.; Rolain, J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J. Antimicrob. Chemother. 2015, 70, 950–951. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Escherichia coli producing VIM-1 carbapenemase isolated on a pig farm. J. Antimicrob. Chemother. 2012, 67, 1793–1795. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Fischer, J.; Helmuth, R. An emerging public health problem: Acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet. Microbiol. 2014, 171, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Freitag, C.; Wendlandt, S.; Eidam, C.; Feßler, A.T.; Lopes, G.V.; Kadlec, K.; Schwarz, S. Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol. 2015, 10, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Enne, V.I.; van Duijkeren, E. 40 years of veterinary papers in JAC—What have we learnt? J. Antimicrob. Chemother. 2016, 71, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Schwarz, S.; Zhang, R.; Lei, L.; Liu, X.; Lin, D.; Shen, J. IMP-45-producing multidrug-resistant Pseudomonas aeruginosa of canine origin. J. Antimicrob. Chemother. 2014, 69, 2579–2581. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.E.; Bugarel, M.; den Bakker, H.C.; Nightingale, K.K.; Granier, S.A.; Scott, H.M.; Loneragan, G.H. Carbapenem-Resistant Bacteria Recovered from Faeces of Dairy Cattle in the High Plains Region of the USA. PLoS ONE 2016, 11, e0147363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Lu, Z.; Schwarz, S.; Zhang, R.M.; Wang, X.M.; Si, W.; Yu, S.; Chen, L.; Liu, S. Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J. Antimicrob. Chemother. 2013, 68, 1681–1682. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Janssen, T.; Wieler, L.H. Multidrug resistant Acinetobacter baumannii in veterinary medicine—Emergence of an underestimated pathogen? Berl. Munch. Tierarztl. Wochenschr. 2014, 127, 435–446. [Google Scholar] [PubMed]
- Eveillard, M.; Kempf, M.; Belmonte, O.; Pailhoriès, H.; Joly-Guillou, M.L. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int. J. Infect. Dis. 2013, 17, e802–e805. [Google Scholar] [CrossRef] [PubMed]
- Al Atrouni, A.; Joly-Guillou, M.L.; Hamze, M.; Kempf, M. Reservoirs of Non-baumannii Acinetobacter Species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Hamouda, A.; Findlay, J.; Al Hassan, L.; Amyes, S.G. Epidemiology of Acinetobacter baumannii of animal origin. Int. J. Antimicrob. Agents 2011, 38, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Cag, Y.; Caskurlu, H.; Fan, Y.; Cao, B.; Vahaboglu, H. Resistance mechanisms. Ann. Transl. Med. 2016, 4, 326. [Google Scholar] [CrossRef] [PubMed]
- Zordan, S.; Prenger-Berninghoff, E.; Weiss, R.; van der Reijden, T.; van den Broeck, P.; Baljer, G.; Dijkshoorn, L. Multidrug-resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg. Infect. Dis. 2011, 17, 1751–1754. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Klotz, P.; Leidner, U.; Stamm, I.; Prenger-Berninghoff, E.; Göttig, S.; Semmler, T.; Scheufen, S. OXA-23 and ISAba1-OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int. J. Antimicrob. Agents 2017, 49, 7–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, D. Detection of NDM-1 carbapenemase-producing Acinetobacter calcoaceticus and Acinetobacter junii in environmental samples from livestock farms. J. Antimicrob. Chemother. 2015, 70, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Abbott, Y.; O’Mahony, R.; Leonard, N.; Quinn, P.J.; van der Reijden, T.; Dijkshoorn, L.; Fanning, S. Characterization of a 2.6kbp variable region within a class 1 integron found in an Acinetobacter baumannii strain isolated from a horse. J. Antimicrob. Chemother. 2005, 55, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, K.; Bolard, A.; Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Vogt, D.; Seiffert, S.N.; Endimiani, A.; Perreten, V. Antibiotic resistance and phylogenetic characterization of Acinetobacter baumannii strains isolated from commercial raw meat in Switzerland. J. Food Prot. 2014, 77, 1976–1981. [Google Scholar] [CrossRef] [PubMed]
- Carvalheira, A.; Casquete, R.; Silva, J.; Teixeira, P. Prevalence and antimicrobial susceptibility of Acinetobacter spp. isolated from meat. Int. J. Food Microbiol. 2017, 243, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Chandler, C.E.; Leung, L.M.; McElheny, C.L.; Mettus, R.T.; Shanks, R.M.; Liu, J.H.; Goodlett, D.R.; Ernst, R.K.; Doi, Y. Structural Modification of Lipopolysaccharide Conferred by mcr-1 in Gram-Negative ESKAPE Pathogens. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Walker, R.D.; Blickenstaff, K.; Bodeis-Jones, S.; Zhao, S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet. Microbiol. 2008, 131, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Beier, R.C.; Foley, S.L.; Davidson, M.K.; White, D.G.; McDermott, P.F.; Bodeis-Jones, S.; Zhao, S.; Andrews, K.; Crippen, T.L.; Sheffield, C.L.; et al. Characterization of antibiotic and disinfectant susceptibility profiles among Pseudomonas aeruginosa veterinary isolates recovered during 1994–2003. J. Appl. Microbiol. 2015, 118, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Hocquet, D.; Ponsin, C.; Cholley, P.; Guyeux, C.; Madec, J.Y.; Bertrand, X. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet. Res. 2015, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Scaccabarozzi, L.; Leoni, L.; Ballarini, A.; Barberio, A.; Locatelli, C.; Casula, A.; Bronzo, V.; Pisoni, G.; Jousson, O.; Morandi, S.; et al. Pseudomonas aeruginosa in Dairy Goats: Genotypic and Phenotypic Comparison of Intramammary and Environmental Isolate. PLoS ONE 2015, 10, e0142973. [Google Scholar] [CrossRef] [PubMed]
- Arais, L.R.; Barbosa, A.V.; Carvalho, C.A.; Cerqueira, A.M. Antimicrobial resistance, integron carriage, and gyrA and gyrB mutations in Pseudomonas aeruginosa isolated from dogs with otitis externa and pyoderma in Brazil. Vet. Dermatol. 2016, 27, 113–e31. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; de Jong, A.; Moyaert, H.; El Garch, F.; Janes, R.; Klein, U.; Morrissey, I.; Thiry, J.; Youala, M. Antimicrobial susceptibility monitoring of dermatological bacterial pathogens isolated from diseased dogs and cats across Europe (ComPath results). J. Appl. Microbiol. 2016, 121, 1254–1267. [Google Scholar] [CrossRef] [PubMed]
- Odumosu, B.T.; Ajetunmobi, O.; Dada-Adegbola, H.; Odutayo, I. Antibiotic susceptibility pattern and analysis of plasmid profiles of Pseudomonas aeruginosa from human, animal and plant sources. Springerplus 2016, 5, 1381. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents, 2016, 48, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updat. 2016, 29, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Tschäpe, H.; Witte, W. Spread of plasmid-mediated nourseothricin resistance due to antibiotic use in animal husbandry. J. Basic Microbiol. 1986, 26, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Hildebrandt, B.; Witte, W. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3267–3269. [Google Scholar] [CrossRef] [PubMed]
- Dahms, C.; Hübner, N.O.; Kossow, A.; Mellmann, A.; Dittmann, K.; Kramer, A. Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE 2015, 10, e0143326. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Praud, K.; Nguyen-Ho-Bao, T.; Argudín, M.A.; Bertrand, S.; Butaye, P.; Cloeckaert, A. Extended-spectrum β-lactamase- and AmpC β-lactamase-producing D-tartrate-positive Salmonella enterica serovar Paratyphi B from broilers and human patients in Belgium, 2008–10. J. Antimicrob. Chemother. 2014, 69, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Pardon, B.; Smet, A.; Butaye, P.; Argudín, M.A.; Valgaeren, B.; Catry, B.; Haesebrouck, F.; Deprez, P. Nosocomial intravascular catheter infections with extended-spectrum Beta-lactamase-producing Escherichia coli in calves after strain introduction from a commercial herd. Transbound. Emerg. Dis. 2017, 64, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.A.; Zarazaga, M.; Ben Sallem, R.; Jouini, A.; Ben Slama, K.; Torres, C. Antibiotic resistance in Escherichia coli in husbandry animals: The African perspective. Lett. Appl. Microbiol. 2017, 64, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Teshager, T.; Domínguez, L.; Moreno, M.A.; Saénz, Y.; Torres, C.; Cardeñosa, S. Isolation of an SHV-12 beta-lactamase-producing Escherichia coli strain from a dog with recurrent urinary tract infections. Antimicrob. Agents Chemother. 2000, 44, 3483–3484. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Emergence of colistin-resistant bacteria in humans without colistin usage: A new worry and cause for vigilance. Int. J. Antimicrob. Agents 2016, 47, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Liu, B.T.; Song, F.J.; Zou, M.; Hao, Z.H.; Shan, H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and Escherichia coli from the same animal. Antimicrob. Agents Chemother. 2016. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 2016, 71, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Blas, J.F.; Ovejero, C.M.; Abadia-Patiño, L.; Gonzalez-Zorn, B. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob. Agents Chemother. 2016, 60, 6356–6358. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, V.; Arena, F.; Tascini, C.; Cannatelli, A.; Henrici De Angelis, L.; Fortunato, S.; Giani, T.; Menichetti, F.; Rossolini, G.M. mcr-1.2, a New mcr Variant Carried on a Transferable Plasmid from a Colistin-Resistant KPC Carbapenemase-Producing Klebsiella pneumoniae Strain of Sequence Type 512. Antimicrob. Agents Chemother. 2016, 60, 5612–5615. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.H.; Lei, C.W.; Ma, S.Z.; Jiang, W.; Liu, B.H.; Wang, Y.X.; Guan, R.; Men, S.; Yuan, Q.W.; Cheng, G.Y.; et al. Various sequence types of Escherichia coli co-harboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 2016. [Google Scholar] [CrossRef]
- Mediavilla, J.R.; Patrawalla, A.; Chen, L.; Chavda, K.D.; Mathema, B.; Vinnard, C.; Dever, L.L.; Kreiswirth, B.N. Colistin- and Carbapenem-Resistant Escherichia coli Harboring mcr-1 and blaNDM-5, Causing a Complicated Urinary Tract Infection in a Patient from the United States. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016, 21. [Google Scholar] [CrossRef]
- Schwarz, S.; Johnson, A.P. Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Szmolka, A.; Nagy, B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Sánchez-Céspedes, J.; Soto, S.; Vila, J. Quinolone resistance in the food chain. Int. J. Antimicrob. Agents 2008, 31, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Veldman, K.; Cavaco, L.M.; Mevius, D.; Battisti, A.; Franco, A.; Botteldoorn, N.; Bruneau, M.; Perrin-Guyomard, A.; Cerny, T.; De Frutos Escobar, C.; et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J. Antimicrob. Chemother. 2011, 66, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Heuer, O.E. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).