The Peptidoglycan Pattern of Staphylococcus carnosus TM300—Detailed Analysis and Variations Due to Genetic and Metabolic Influences
Abstract
:1. Introduction
2. Results
2.1. Peptidoglycan Analysis
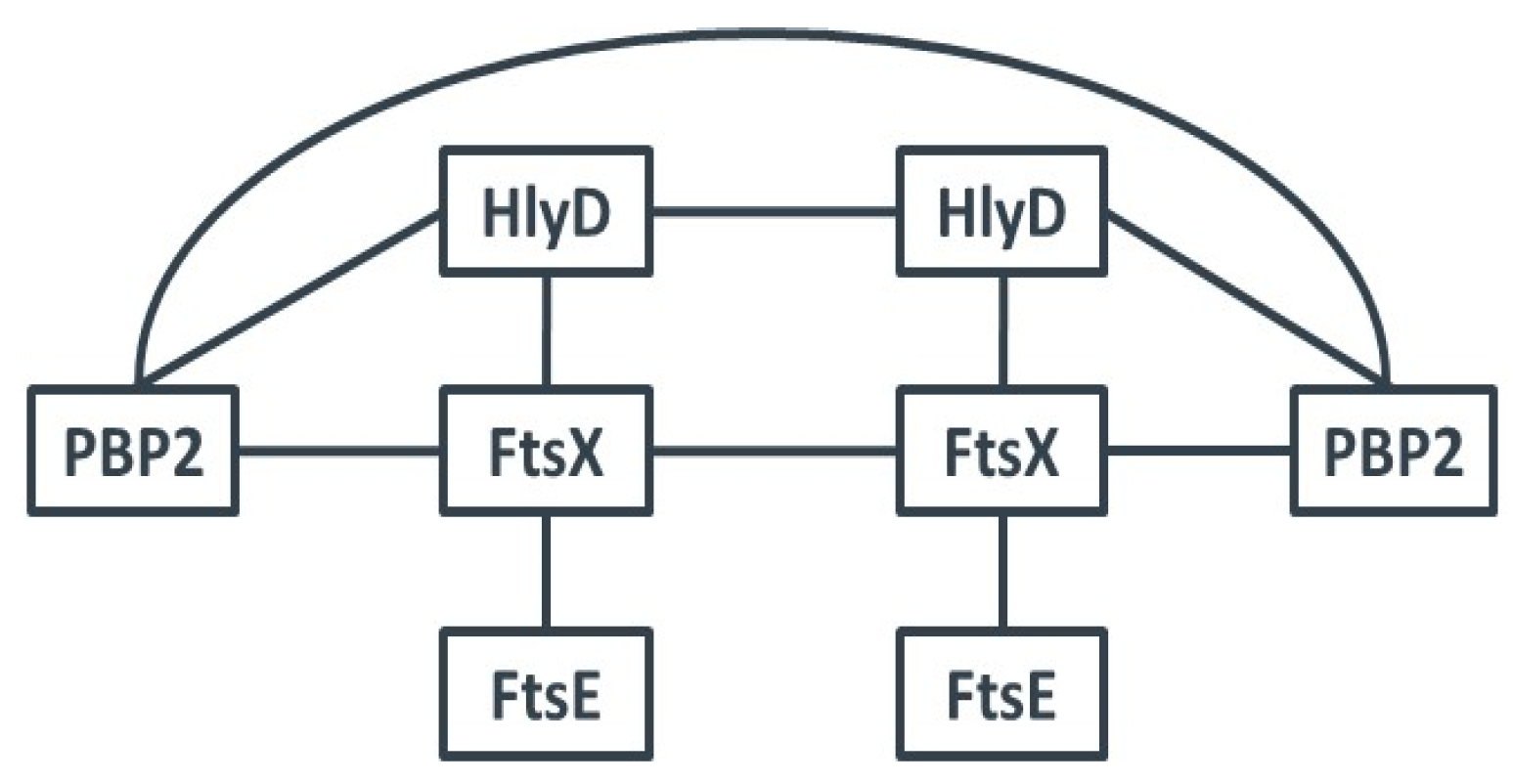
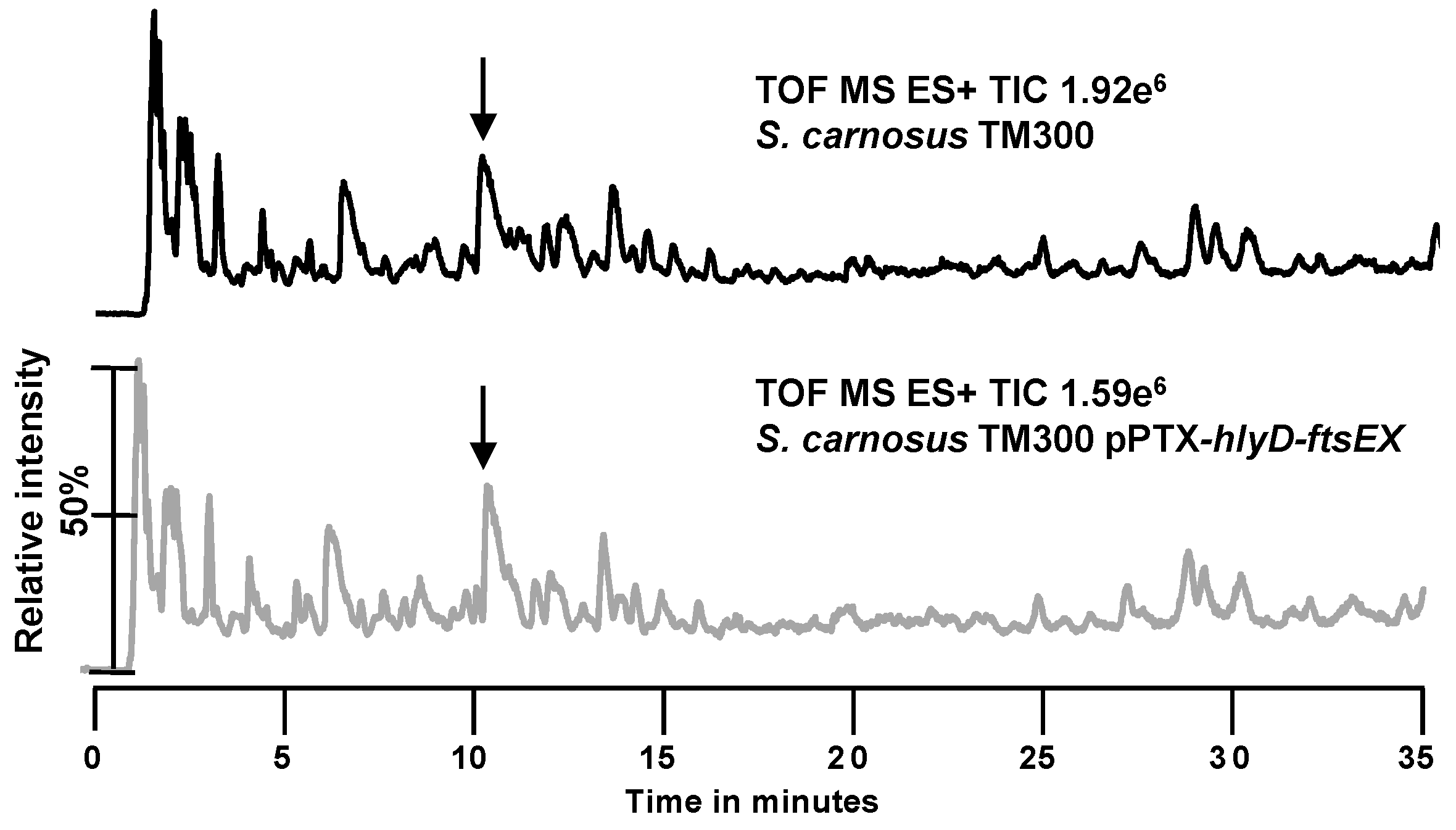
2.2. Search for New Proteins Involved in PGN Biosynthesis
2.3. Precursor Analysis
2.4. Investigation of a Putative l,d-Carboxypeptidase
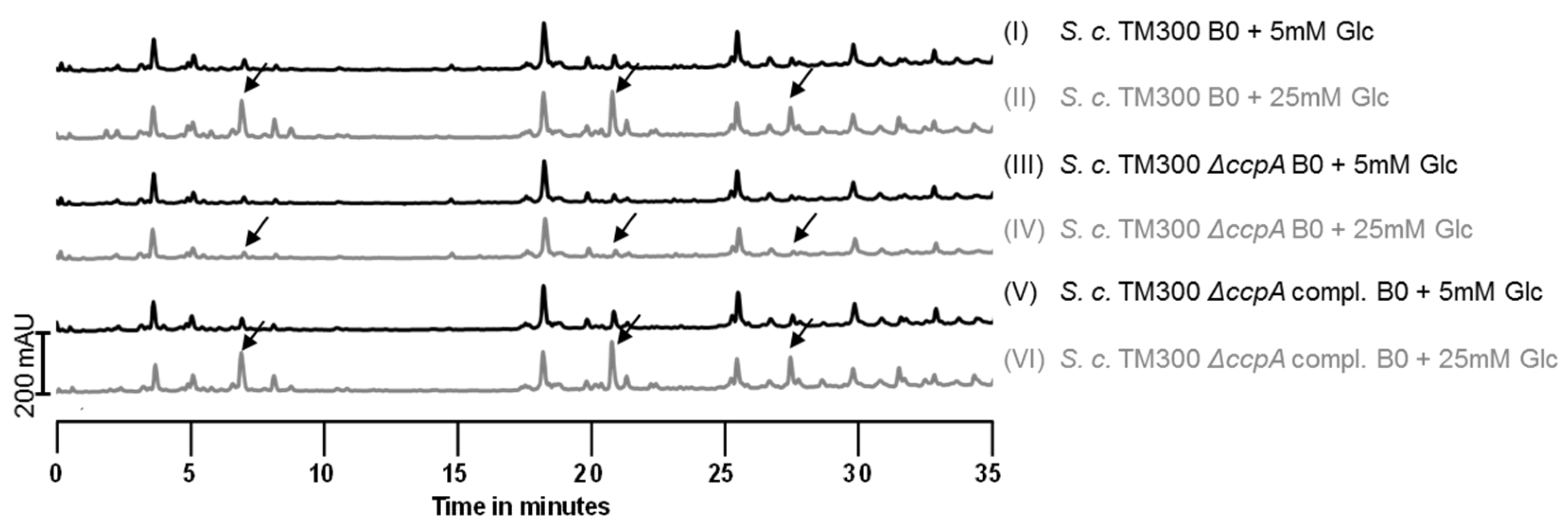
2.5. Influence of Sugars on PGN Composition
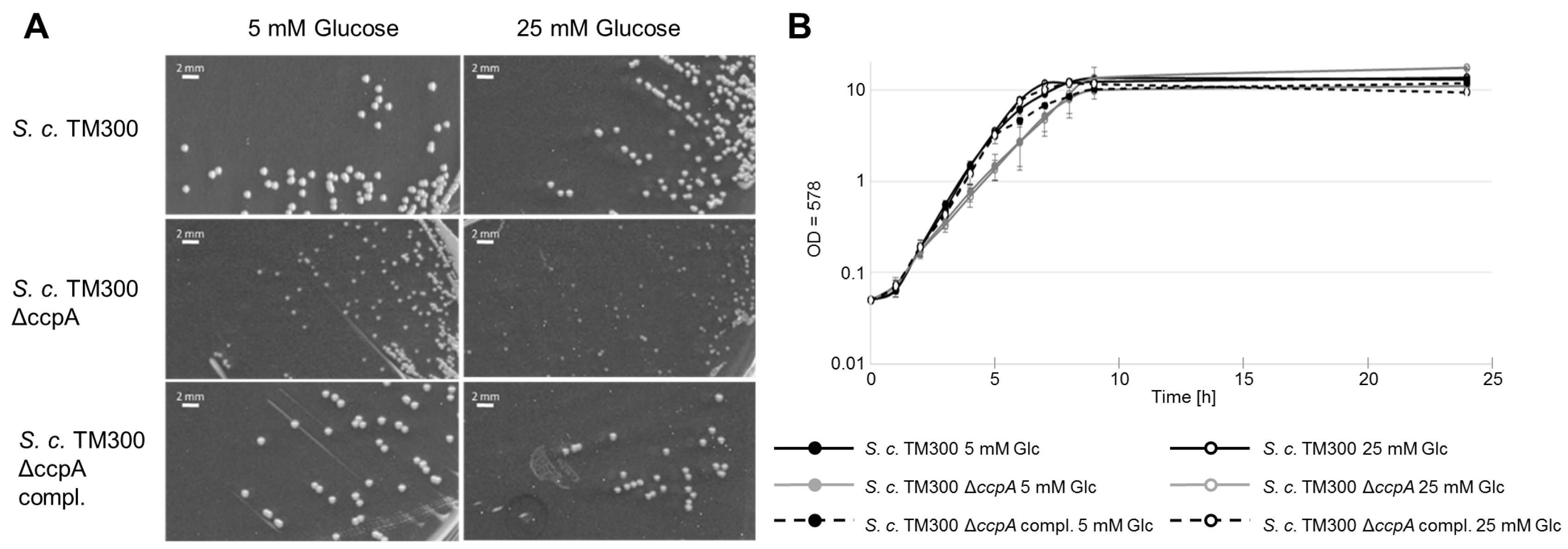
2.6. Deletion of ccpA Causes a Decrease in Colony Size and Growth Rate
3. Discussion
3.1. The Peptidoglycan Composition of S. carnosus TM300
3.2. The hlyD-ftsE-ftsX Operon
3.3. Investigation of a Putative l,d-Carboxypeptidase (SCA_0214)
3.4. Influence of Sugars on the Muropeptide Composition
4. Materials and Methods
4.1. Media
4.2. Reagents
4.3. Plasmid Construction for Deletion Mutants and Overexpression
4.4. Gene Deletion
4.5. Genomic Library
4.6. Bacterial-Two-Hybrid Assays
4.7. Peptidoglycan Analysis
4.8. Overproduction of SCA_0214
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BTH | Bacterial-Two-Hybrid |
| PGN | peptidoglycan |
| UPLC/MS | Ultra Performance Liquid Chromatography coupled to mass spectrometry |
| MurNAc | N-acetylmuramic acid |
| GlcNAc | N-acetylglucosamine |
| CP | carboxypeptidase |
References
- Schleifer, K.H.; Fischer, U. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 1982, 32, 153–156. [Google Scholar] [CrossRef]
- Niinivaara, F.P.; Pohja, M.S. Über die Reifung der Rohwurst. I. Mitt: Die Veränderung der Bakterienflora während der Reifung. Z. Lebensm. Unters. Forsch. 1956, 104, 413–422. (In German) [Google Scholar] [CrossRef]
- Rosenstein, R.; Götz, F. Genomic differences between the food-grade Staphylococcus carnosus and pathogenic staphylococcal species. Int. J. Med. Microbiol. 2010, 300, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Nerz, C.; Biswas, L.; Resch, A.; Raddatz, G.; Schuster, S.C.; Götz, F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 2009, 75, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [PubMed]
- Bera, A.; Biswas, R.; Herbert, S.; Götz, F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 2006, 74, 4598–4604. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, N. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 2015, 8, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.; van Dam, V.; Sijbrandi, R.; Vernet, T.; Zapun, A.; Bouhss, A.; Diepeveen-de Bruin, M.; Nguyen-Disteche, M.; de Kruijff, B.; Breukink, E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011, 30, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.; Sijbrandi, R.; Lutters, M.; Verheul, J.; Martin, N.; den Blaauwen, T.; de Kruijff, B.; Breukink, E. Specificity of the transport of Lipid II by FtsW in Escherichia coli. J. Biol. Chem. 2014. [Google Scholar] [CrossRef] [PubMed]
- Sham, L.-T.; Butler, E.K.; Lebar, M.D.; Kahne, D.; Bernhardt, T.G.; Ruiz, N. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 2014, 345, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Butler, E.K.; Tan, W.B.; Joseph, H.; Ruiz, N. Charge requirements of Lipid II flippase activity in Escherichia coli. J. Bacteriol. 2014, 196, 4111–4119. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, T.A.; Sobral, R.G.; Ludovice, A.M.; Almeida, J.M.; Bui, N.K.; Vollmer, W.; de Lencastre, H.; Tomasz, A. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 2012, 8, e1002508. [Google Scholar] [CrossRef] [PubMed]
- Münch, D.; Roemer, T.; Lee, S.H.; Engeser, M.; Sahl, H.G.; Schneider, T. Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog. 2012, 8, e1002509. [Google Scholar] [CrossRef] [PubMed]
- Nakel, M.; Ghuysen, J.M.; Kandler, O. Wall peptidoglycan in Aerococcus viridans strains 201 Evans and ATCC 11563 and in Gaffkya homari strain ATCC 10400. Biochemistry 1971, 10, 2170–2175. [Google Scholar] [PubMed]
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar] [PubMed]
- Pinho, M.G.; Errington, J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 2005, 55, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Filipe, S.R.; de Lencastre, H.; Tomasz, A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.F.F.; Henriques, A.O.; Pinho, M.G.; de Lencastre, H.; Tomasz, A. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 2007, 189, 3525–3531. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 2000, 182, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Leski, T.A.; Tomasz, A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 2005, 187, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Memmi, G.; Filipe, S.R.; Pinho, M.G.; Fu, Z.; Cheung, A. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 2008, 52, 3955–3966. [Google Scholar] [CrossRef] [PubMed]
- KEGG: Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/ (accessed on 6 August 2016).
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Tzagoloff, H.; Novick, R. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 1977, 129, 343–350. [Google Scholar] [PubMed]
- Monteiro, J.M.; Fernandes, P.B.; Vaz, F.; Pereira, A.R.; Tavares, A.C.; Ferreira, M.T.; Pereira, P.M.; Veiga, H.; Kuru, E.; VanNieuwenhze, M.S.; et al. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Aarsman, M.E.; Piette, A.; Fraipont, C.; Vinkenvleugel, T.M.; Nguyen-Disteche, M.; den Blaauwen, T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005, 55, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Söderström, B.; Daley, D.O. The bacterial divisome: More than a ring? Curr. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.L.; Peterson, N.D.; Kustusch, R.J.; Wissel, M.C.; Graham, B.; Phillips, G.J.; Weiss, D.S. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 2004, 186, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, R.; Bruce, K.E.; Davidson, A.L.; Rued, B.E.; Stauffacher, C.V.; Winkler, M.E. Biochemical characterization of essential cell division proteins FtsX and FtsE that mediate peptidoglycan hydrolysis by PcsB in Streptococcus pneumoniae. Microbiol. Open 2016. [Google Scholar] [CrossRef] [PubMed]
- Sham, L.-T.; Jensen, K.R.; Bruce, K.E.; Winkler, M.E. Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae D39. mBio 2013. [Google Scholar] [CrossRef] [PubMed]
- Sham, L.T.; Barendt, S.M.; Kopecky, K.E.; Winkler, M.E. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. USA 2011, 108, E1061–E1069. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Peters, N.T.; Parzych, K.R.; Uehara, T.; Markovski, M.; Bernhardt, T.G. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. USA 2011, 108, E1052–E1060. [Google Scholar] [CrossRef] [PubMed]
- Meisner, J.; Montero Llopis, P.; Sham, L.T.; Garner, E.; Bernhardt, T.G.; Rudner, D.Z. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 2013, 89, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Glauner, B.; Höltje, J.-V.; Schwarz, U. The composition of the murein of Escherichia coli. J. Biol. Chem. 1988, 263, 10088–10095. [Google Scholar] [PubMed]
- Kühner, D.; Stahl, M.; Demircioglu, D.D.; Bertsche, U. From cells to muropeptide structures in 24 h: Peptidoglycan mapping by UPLC-MS. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, B.L.; Chang, Y.S.; Gage, D.; Tomasz, A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 1992, 267, 11248–11254. [Google Scholar] [PubMed]
- Murakami, K.; Fujimura, T.; Doi, M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol. Lett. 1994, 117, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Lovering, A.L.; de Castro, L.H.; Lim, D.; Strynadka, N.C. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 2007, 315, 1402–1405. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, E.; Graham, B.; Phillips, G.J.; ten Hagen-Jongman, C.M.; Oudega, B.; Luirink, J. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 1999, 31, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Shivers, R.P.; Dineen, S.S.; Sonenshein, A.L. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: Establishing a potential hierarchy in carbon flow. Mol. Microbiol. 2006, 62, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Sonenshein, A.L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007, 5, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.M.; Thoms, B. Role of sugar uptake and metabolic intermediates on catabolite repression in Bacillus subtilis. J. Bacteriol. 1977, 129, 217–224. [Google Scholar] [PubMed]
- De Jonge, B.L.; Chang, Y.S.; Gage, D.; Tomasz, A. Peptidoglycan composition in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 1992, 267, 11255–11259. [Google Scholar] [PubMed]
- Takacs, C.N.; Hocking, J.; Cabeen, M.T.; Bui, N.K.; Poggio, S.; Vollmer, W.; Jacobs-Wagner, C. Growth medium-dependent glycine incorporation into the peptidoglycan of Caulobacter crescentus. PLoS ONE 2013, 8, e57579. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cegelski, L. Nutrient-dependent structural changes in S. aureus peptidoglycan revealed by solid-state NMR spectroscopy. Biochemistry 2012, 51, 8143–8153. [Google Scholar] [CrossRef] [PubMed]
- Schülein, R.; Gentschev, I.; Möllenkopf, H.J.; Goebel, W. A topological model for the haemolysin translocator protein HlyD. Mol. Gen. Genet. 1992, 234, 155–163. [Google Scholar] [PubMed]
- Steele, A.L.; Young, S. A descriptive study of Myers-Briggs personality types of professional music educators and music therapists with comparisons to undergraduate majors. J. Music Ther. 2011, 48, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Hoyland, C.N.; Aldridge, C.; Cleverley, R.M.; Duchene, M.C.; Minasov, G.; Onopriyenko, O.; Sidiq, K.; Stogios, P.J.; Anderson, W.F.; Daniel, R.A.; et al. Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition. Structure 2014, 22, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Ottenwalder, B.; Götz, F. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 1996, 137, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Popella, P.; Krauss, S.; Ebner, P.; Nega, M.; Deibert, J.; Götz, F. VraH is a third component of the Staphylococcus aureus VraDEH system involved in gallidermin and daptomycin resistance and pathogenicity. Antimicrob. Agents Chemother. 2016. [Google Scholar] [CrossRef] [PubMed]
- Henkin, T.M.; Grundy, F.J.; Nicholson, W.L.; Chambliss, G.H. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 1991, 5, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.M.; Lascelles, J. The effect of growth conditions on oxidative and dehydrogenase activity in Staphylococcus aureus. J. Gen. Microbiol. 1962, 29, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Strasters, K.C.; Winkler, K.C. Carbohydrate Metabolism of Staphylococcus aureus. J. Gen. Microbiol. 1963, 33, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Seidl, K.; Stucki, M.; Ruegg, M.; Goerke, C.; Wolz, C.; Harris, L.; Berger-Bächi, B.; Bischoff, M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006, 50, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Brückner, R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 151, 1–8. [Google Scholar] [CrossRef]
- Krismer, B.; Nega, M.; Thumm, G.; Götz, F.; Peschel, A. Highly efficient Staphylococcus carnosus mutant selection system based on suicidal bacteriocin activation. Appl. Environ. Microbiol. 2012, 78, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.; Chastanet, A.; Debarbouille, M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70, 6887–6891. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Lofblom, J.; Kronqvist, N.; Uhlen, M.; Stahl, S.; Wernerus, H. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 2007, 102, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Dagert, M.; Ehrlich, S.D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 1979, 6, 23–28. [Google Scholar] [CrossRef]
- Leibig, M.; Krismer, B.; Kolb, M.; Friede, A.; Götz, F.; Bertram, R. Marker removal in Staphylococci via Cre recombinase and different lox sites. Appl. Environ. Microbiol. 2008, 74, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef] [PubMed]
- Kohlrausch, U.; Höltje, J.-V. One-step purification procedure for UDP-N-acetylmuramyl-peptide murein precursors from Bacillus cereus. FEMS Microbiol. Lett. 1991, 62, 253–257. [Google Scholar] [CrossRef] [PubMed]
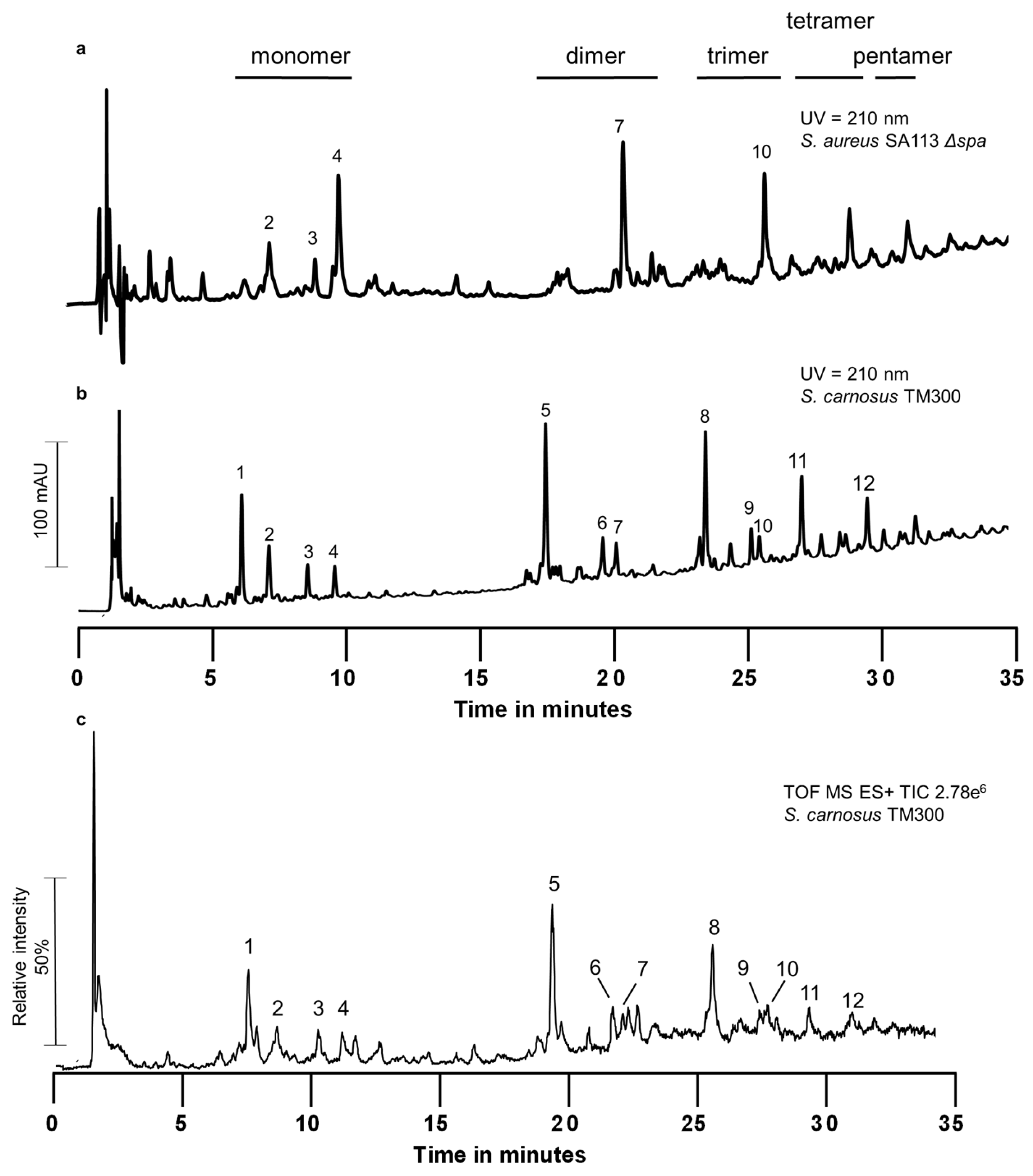

| Peak | Retention Time (min) | M + H+ (Da) | Proposed Molecular Formula | Lengths of the Stem Peptides | Inter-Peptide Bridges |
|---|---|---|---|---|---|
| Gly | |||||
| 1 | 7.5 | 1111.5192 | C43H75N12O22+ | Tri | 5 |
| 2 | 8.6 | 1036.4937 | C40H70N13O19+ | Tetra w/o GlcNAc | 6 |
| 1093.5164 | C42H73N14O20+ | 7 | |||
| 1150.5417 | C44H76N15O21+ | 8 | |||
| 1207.5699 | C46H79N16O22+ | 9 | |||
| 1239.5699 | C48H83N14O24+ | Tetra | 6 | ||
| 1296.5939 | C50H86N15O25+ | 7 | |||
| 1353.6135 | C52H89N16O26+ | 8 | |||
| 1410.6370 | C54H92N17O27+ | 9 | |||
| 3 | 10.2 | 979.4691 | C38H67N12O18+ | Tetra w/o GlcNAc | 5 |
| 1182.5502 | C46H80N13O23+ | Tetra | 5 | ||
| 4 | 11.1 | 1253.5856 | C49H85N14O24+ | Penta | 5 |
| 5 | 19.2 | 2275.0595 | C89H152N25O44+ | Tri-Tetra | 10 |
| 6 | 21.5 | 2346.0815 | C92H157N26O45+ | Tetra2 | 10 |
| 7 | 21.9 | 2417.1143 | C95H162N27O46+ | Penta-Tetra | 10 |
| 8 | 25.4 | 3438.5927 | C135H229N38O66+ | Tri-Tetra2 | 15 |
| 9 | 27.2 | 3509.6465 | C138H234N39O67+ | Tetra3 | 15 |
| 10 | 27.5 | 3580.6553 | C141H239N40O68+ | Penta-Tetra2 | 15 |
| 11 | 29.1 | 4602.0920 | C181H306N51O88+ | Tri-Tetra3 | 20 |
| 12 | 31.6 | 5765.6142 | C227H383N64O110+ | Tri-Tetra4 | 25 |
| pUT18C | ||||||
|---|---|---|---|---|---|---|
| pKT25 | Units/mg | pbp2 | ftsX | ftsE | hlyD | zip |
| pbp2 | 3769 ± 626 | 4373 ± 105 | 136 ± 13 | 3488 ± 658 | 164 ± 28 | |
| ftsX | 3714 ± 833 | 2502 ± 491 | 2008 ± 289 | 2842 ± 287 | 165 ± 25 | |
| ftsE | 109 ± 21 | 4484 ± 279 | 354 ± 138 | 132 ± 27 | 201 ± 103 | |
| hlyD | 4020 ± 431 | 2280 ± 456 | 205 ± 29 | 3515 ± 113 | 156 ± 36 | |
| zip | 113 ± 20 | 155 ± 4 | 126 ± 7 | 157 ± 59 | 6608 ± 546 | |
| Strain | 5 mM Glucose (mm) | 25 mM Glucose (mm) |
|---|---|---|
| S. carnosus TM300 | 0.91 ± 0.07 | 0.83 ± 0.09 |
| S. carnosus TM300 ΔccpA | 0.39 ± 0.07 | 0.33 ± 0.06 |
| S. carnosus TM300 ΔccpA compl. | 0.92 ± 0.04 | 0.89 ± 0.07 |
| Strain | 5 mM Glucose (td/h) | 25 mM Glucose (td/h) |
|---|---|---|
| S. carnosus TM300 | 1.82 ± 0.16 | 2.10 ± 0.35 |
| S. carnosus TM300 ΔccpA | 0.71 ± 0.31 | 0.67 ± 0.19 |
| S. carnosus TM300 ΔccpA compl. | 1.90 ± 0.40 | 2.00 ± 0.38 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deibert, J.; Kühner, D.; Stahl, M.; Koeksoy, E.; Bertsche, U. The Peptidoglycan Pattern of Staphylococcus carnosus TM300—Detailed Analysis and Variations Due to Genetic and Metabolic Influences. Antibiotics 2016, 5, 33. https://doi.org/10.3390/antibiotics5040033
Deibert J, Kühner D, Stahl M, Koeksoy E, Bertsche U. The Peptidoglycan Pattern of Staphylococcus carnosus TM300—Detailed Analysis and Variations Due to Genetic and Metabolic Influences. Antibiotics. 2016; 5(4):33. https://doi.org/10.3390/antibiotics5040033
Chicago/Turabian StyleDeibert, Julia, Daniel Kühner, Mark Stahl, Elif Koeksoy, and Ute Bertsche. 2016. "The Peptidoglycan Pattern of Staphylococcus carnosus TM300—Detailed Analysis and Variations Due to Genetic and Metabolic Influences" Antibiotics 5, no. 4: 33. https://doi.org/10.3390/antibiotics5040033
APA StyleDeibert, J., Kühner, D., Stahl, M., Koeksoy, E., & Bertsche, U. (2016). The Peptidoglycan Pattern of Staphylococcus carnosus TM300—Detailed Analysis and Variations Due to Genetic and Metabolic Influences. Antibiotics, 5(4), 33. https://doi.org/10.3390/antibiotics5040033






