Abstract
Complete genome comparisons, transcriptomic and metabolomic studies were performed on two laboratory-selected, well-characterized vancomycin-intermediate Staphylococcus aureus (VISA) derived from the same parent MRSA that have changes in cell wall composition and decreased autolysis. A variety of mutations were found in the VISA, with more in strain 13136p−m+V20 (vancomycin MIC = 16 µg/mL) than strain 13136p−m+V5 (MIC = 8 µg/mL). Most of the mutations have not previously been associated with the VISA phenotype; some were associated with cell wall metabolism and many with stress responses, notably relating to DNA damage. The genomes and transcriptomes of the two VISA support the importance of gene expression regulation to the VISA phenotype. Similarities in overall transcriptomic and metabolomic data indicated that the VISA physiologic state includes elements of the stringent response, such as downregulation of protein and nucleotide synthesis, the pentose phosphate pathway and nutrient transport systems. Gene expression for secreted virulence determinants was generally downregulated, but was more variable for surface-associated virulence determinants, although capsule formation was clearly inhibited. The importance of activated stress response elements could be seen across all three analyses, as in the accumulation of osmoprotectant metabolites such as proline and glutamate. Concentrations of potential cell wall precursor amino acids and glucosamine were increased in the VISA strains. Polyamines were decreased in the VISA, which may facilitate the accrual of mutations. Overall, the studies confirm the wide variability in mutations and gene expression patterns that can lead to the VISA phenotype.
1. Introduction
The first report of a clinical isolate of Staphylococcus aureus showing decreased susceptibility to vancomycin, minimum inhibitory concentration (MIC) 8 μg/mL, appeared in 1997 [1]. Since that time there have been many reports of laboratory-derived and clinically-isolated vancomycin-intermediate S. aureus (VISA), and these have been reviewed [2,3,4]. Despite considerable effort, the mechanism(s) underlying decreased vancomycin susceptibility is not entirely clear. Although VISA typically show a number of phenotypic traits in common, such as increased cell wall thickness, decreased autolysis and alterations in peptidoglycan structure, a wide variety of mutations and transcriptomes have been found in VISA [2,3,4]. Exposure of S. aureus to cell wall-active antibiotics induces the expression of a set of genes that comprises a cell wall stress stimulon [5]. Altered stress response gene expression without antimicrobial exposure, including members of the cell wall stress stimulon, is characteristic of VISA [6].
In 2000, Pfeltz et al. [7] reported on the characteristics of a number of VISA strains generated by in vitro step selection from vancomycin-susceptible S. aureus (VSSA) in the presence of increasing concentrations of vancomycin. Among these VSSA was heterogeneously methicillin-resistant S. aureus (MRSA) strain 13136p−m+ (penicillinase-negative, mecA-positive). This strain was originally isolated in 1960 as MRSA 13136, one of the handful of MRSA among several thousand clinical S. aureus isolates surveyed [8]. Strain 13136p−m+ is a penicillinase-negative derivative selected from 13136 [9]. Like the well-studied penicillinase-negative, homogeneous MRSA strain Colindale 9204 (COL), the 13136 lineage is lysogenized by prophage L54a inserted near the 3' end of the glycerol ester hydrolase gene (geh). The result is a truncated, inactive gene product (lipase) [10]. L54a, also known as ϕCOL, was only present in COL in a comparison of six staphylococcal genome sequences [11]. Although COL and 13136 were isolated in the same time period (1960–1964) by the Cross-infection Reference Laboratory, Colindale, London, a comparison of genome sequences between the two strains is not available. COL was isolated from a hospital in Colindale, whereas the separate survey identifying 13136 drew isolates from hospitals primarily in southeastern England, but also elsewhere within and outside of that country [8,12].
MRSA 13136p−m+ was the parent strain from which VISA 13136p−m+V5 was derived, and subsequently VISA 13136p−m+V20 was then derived from 13136p−m+V5. The MICs of the VISA strains were 8 and 16 μg/mL for 13136p−m+V5 and 13136p−m+V20, respectively [7]. Strain 13136p−m+V5 was isolated after two selection cycles in one week and strain 13136p−m+V20 after a total of five cycles of selection across five weeks. Each selection cycle consisted of growth in liquid medium that was inoculated onto solid medium containing the same concentration of vancomycin. The 64 µg/mL minimum bactericidal concentration (MBC) of strain 13136p−m+V20 was significantly higher than that of 13136p−m+V5 (16 µg/mL) or the parent strain (8 µg/mL). These strains have been extensively studied for their cell wall-related properties [7,13]. The VISA strains grew slower than the VSSA parent [7]. Muropeptide analysis showed an increase in muropeptides with a single L-alanine residue in the interpeptide bridge in the two VISA strains compared to the VSSA parent strain. Cell wall teichoic acid content was significantly decreased in the VISA strains, in strain 13136p−m+V20 in particular, as judged by cell wall phosphorus content. The VISA strains showed decreased whole cell autolysis, decreased lysostaphin susceptibility and decreased isolated crude cell wall autolytic activity. Differences were noted in the amount and profile of peptidoglycan hydrolase activities in the VISA strains [13]. atl, the gene encoding two major S. aureus autolysins [14], showed significantly reduced expression in 13136p−m+V20 compared to 13136p−m+.
We have used these strains to further probe the mechanism(s) of decreased vancomycin susceptibility by a combination of genomic, transcriptomic and metabolomic approaches. Metabolomics typically involves the comparative analysis of small, cellular organic compounds of a molecular weight less than 1000 daltons [15]. A metabolome is a direct reflection of the physiological status of a cell and thus is another approach to understanding cell function. VISA strains are believed to be activated for cell wall synthesis [16] and to display impaired acetate catabolism [17]. A recent metabolomic analysis of two series of VISA isolates indicated alterations to a wide range of central metabolic pathway intermediates, but did not elucidate the underlying mechanism of decreased vancomycin susceptibility [18].
2. Results
2.1. Mutations in Strains 13136p−m+V5 and 13136p−m+V20 Compared to 13136p−m+
The mutations present in the two VISA strains are summarized in Table 1. A total of nine single-nucleotide polymorphisms (SNPs) across eight genes that resulted in protein amino acid changes were detected in strain 13136p−m+V5 and 16 SNPs across nine genes in 13136p−m+V20, relative to VSSA parent strain 13136p−m+. Although the MIC of 13136p−m+V20 was only double that of 13136p−m+V5, strain 13136p−m+V20 was subjected to three more selection cycles than 13136p−m+V5, and this was reflected in the higher number of SNPs. Six of the nine 13136p−m+V5 SNPs were also present in 13136p−m+V20, but three were not.

Table 1.
Mutations resulting in protein amino acid changes present in VISA strains 13136p−m+V5 and 13136p−m+V20 (yellow cells) following in-vitro passage-selection from VSSA parent 13136p−m+.
| SNP | Amino acid change | Locus | Gene Name | Known or Predicted Gene Product | Mutation Present In: | Function of Predicted Gene Product Based on Published Studies and Similarities to Other Proteins, and Known Stress-Response Associations [references] | ||
|---|---|---|---|---|---|---|---|---|
| 13136p−m+V5 | 13136p−m+V20 | |||||||
| 223 | G→G | K→E | SACOL1005 | pepF | Oligoendopeptidase F | + | - | Cytoplasmic endopeptidase releasing amino acids from internalized peptides; involved in protein turnover [19] |
| 143 | C→T | A→V | SACOL1231 | stp1 | Eukaryotic-like serine / threonine phosphatase | + | - | Influences the regulation of virulence, cell wall structure, autolysis, and susceptibility to some cell wall-active antibiotics [20] |
| 554 | G→A | S→F | SACOL1600 | comGB | ComGB, competence protein | + | - | A membrane protein associated with the SOS response that transports exogenous DNA into the Gram-positive cell [21] |
| 126 | A→C | E→D | SACOL0593 | fusA | Translation elongation factor G | - | + | Protein synthesis roles in tRNA translocation during elongation and post-termination ribosome dissociation; upregulated by acid adaptation [22,23] |
| 121 | G→T | G→C | SACOL0593 | + | + | |||
| 6596 | G→A | A→V | SACOL2150 | fmtB | FmtB protein | + | + | β-lactam resistance-related surface protein with cell wall anchoring and spanning domains, putatively involved in cell wall biosynthesis, cell adhesion, biofilms [6] |
| 171 | C→A | M→I | SACOL2217 | infA | Translation initiation factor IF-1 | + | + | RNA-binding protein that binds the 30S ribosomal subunit, required for correct translation initiation, and downregulated by cell wall-active antibiotics [5,24] |
| 311 | A→T | Y→F | SACOL1319 | glpF | Glycerol uptake facilitator protein | + | + | Housekeeping gene product that transports glycerol or small uncharged molecules into the cell; down-regulated by oxidative stress [25] |
| 353 | G→C | G→P | SACOL0339 | ssb1 | Prophage L54a single-stranded DNA-binding protein | + | + | One of a family of proteins that bind with high affinity to ssDNA intermediates during DNA replication, recombination and repair [26,27,28] |
| 352 | G→C | G→P | SACOL0339 | + | + | |||
| 350 | A→C | N→P | SACOL0339 | - | + | |||
| 349 | A→C | N→P | SACOL0339 | - | + | |||
| 345 | A→C | Q→P | SACOL0339 | - | + | |||
| 344 | A→C | Q→P | SACOL0339 | - | + | |||
| 329 | A→C | K→T | SACOL0339 | - | + | |||
| 670 | C→T | L→F | SACOL0810 | tarO | glycosyl transferase, group 4 family protein (TarO) | - | + | Teichoic acid synthesis enzyme; inactivation increases autolysis rates, reduces β-lactam resistance, and disrupts sepatation and cell separation [29] |
| 11 | G→A | A→V | SACOL1495 | dinG | Damage inducible gene G (DinG) | - | + | 3'→5' ssDNA and ssRNA exonuclease; proposed functions: R-loop resolution, other unspecified recombination repair systems, anitviral defense [30] |
| 191 | C→T | C→Y | SACOL1690 | apt | Adenine phosphoribosyl transferase | - | + | Enzyme in an adenine recycling pathway upregulated by acid adaptation, the stringent response, and in some organisms polyamine metabolism [23,31] |
| 574 | A→C | F→V | SACOL2451 | none | Amino acid ABC transporter binding protein | - | + | Homolog of OpuBC, the glycine betaine/choline binding lipoprotein of an osmoprotectant uptake system [32] |
A total of 12 genes were associated with 19 protein amino acid changes in one or both VISA (Table 1). Five of these genes encode products that interact with nucleic acids in protein synthesis or are associated with DNA damage/SOS responses: comGB, fusA, infA, ssb1 and dinG [21,22,23,24,26,27,28,30]. Three more are housekeeping genes encoding products involved in central intermediary metabolism: pepF encodes an endopeptidase that hydrolyzes internalized peptides to provide nutrients or endogenous peptides for protein turnover [19]; glpF encodes a glycerol transporter (carbon and energy acquisition) [25]; and apt encodes an enzyme in a purine salvage pathway catalyzing the production of AMP from adenine and phosphoribosyl pyrophosphate [23,31]. apt is associated with the stringent response in other Gram-positive bacteria [29,33,34,35]. The fmtB and tarO gene products are cell envelope proteins involved in cell wall biosynthesis, β-lactam resistance and virulence [6,29]. fmtB is located downstream of glmM and is also known as mrp and sasB [36]. FmtB has a C-terminal LPXTG cell wall anchoring motif [37]. The indirect effect of FmtB on methicillin resistance can be relieved by increasing the production of the cell wall precursor glucosamine-l-phosphate [38]. tarO, also known as tagO, llm and farO, catalyzes the initial step of cell wall teichoic acid synthesis. Locus SACOL2451 encodes a putative membrane protein, a homolog of OpuBC, the permease component of an ABC-type osmoprotectant transport system [32].
The final mutated gene, stp1, encodes a eukaryotic-like serine-threonine phosphatase and is located in tandem with its cognate serine-threonine kinase gene, stk1 [39]. These two enzymes comprise a two-component regulatory system that plays important roles in bacterial signal transduction [20], in addition to roles in cell wall structure and autolysis [39,40]. Mutations in stp1 have previously been associated with laboratory-selected glycopeptide resistance, along with cell wall alterations characteristic of the VISA phenotype [40,41,42,43]. Passalacqua et al. [42] reported reduced autolytic activity in addition to decreased susceptibility to vancomycin, daptomycin and linezolid due to an stp1 mutation. Additionally, Cameron et al. [43] demonstrated that the deletion of this gene alone is sufficient to slightly reduce vancomycin susceptibility. A eukaryotic-like serine-threonine phosphatase-kinase pair regulates an osmotic stress sensory system in Mycobacterium tuberculosis that leads to an adaptive response involving modifications to cell wall structure and virulence factor production [44]. Eleven of the 12 genes listed in Table 1, all except pepF, can be linked to stress response either by expression change or gene product function.
2.2. Comparison of Transcriptional Profiles between 13136p−m+, 13136p−m+V5 and 13136p−m+V20
Gene expression differences in VISA 13136p−m+V5 and 13136p−m+V20 relative to VSSA progenitor strain 13136p−m+ are listed in Supplementary Tables S1 and S2. These changes are presented graphically by gene functional groupings in Figure 1. Supplementary Table S3 contains the data used to generate Figure 1. For both VISA, approximately twice as many gene expression differences versus VSSA 13136p−m+ were downregulated as were upregulated. Unfortunately, but not unexpectedly, the gene functional group with the largest number of changes was hypothetical functions, and along with the unclassified and unknown function categories accounted for 91 of the 338 genes (27%) with expression changes in VISA strains. Among genes with assigned functions, seven functional groups each had at least 5% of the genes each with expression changes in VISA vs. VSSA 13136p−m+: cell envelope (48 genes, or 14%), transport and binding proteins (39 genes, or 12%), purines, pyrimidines, nucleosides and nucleotides (28 genes, or 8%), protein synthesis (27 genes, or 8%), regulatory functions (24 genes, or 7%), and biosynthesis of cofactors, prosthetic groups and carriers (18 genes) and central intermediary metabolism (16 genes) with 5% each. Together these eight functional groups accounted for 86% of the genes represented in Figure 1. Downregulation was the clear trend for all of these functional groups, except biosynthesis of cofactors, prosthetic groups and carriers, where the numbers of down- and up-regulation were comparable.
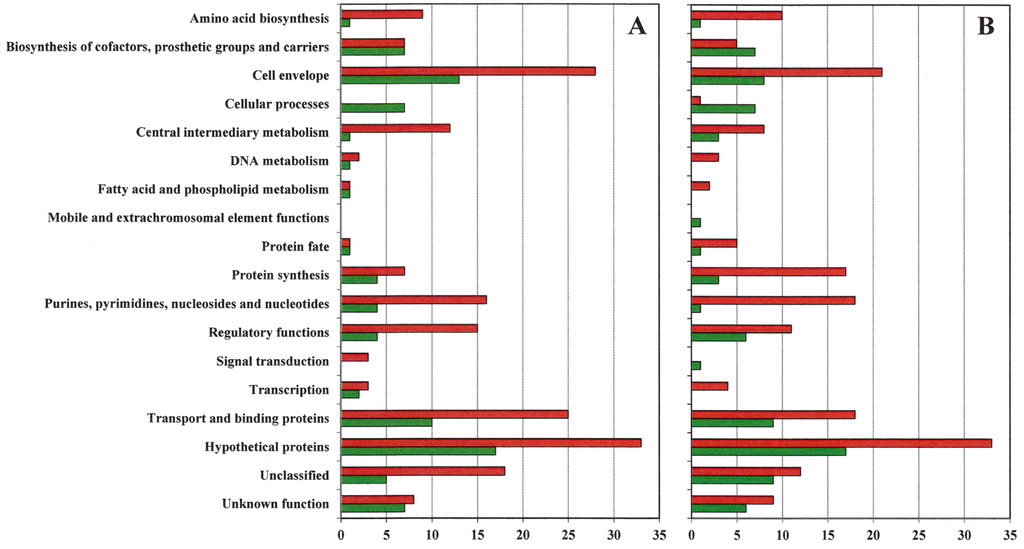
Figure 1.
Gene expression patterns. The number of genes by functional group upregulated (green bars) and downregulated (red bars) at least two-fold in VISA 13136p−m+V5 (A) and 13136p−m−V20 (B) relative to gene expression in vancomycin-susceptible S. aureus (VSSA) parent 13136p−m+.
Supplementary Table S3 presents the gene expression change concordance between VISA13136p−m+V5 and 13136p−m+V20. Approximately the same number of genes were upregulated in each VISA strain, 83 in VISA13136p−m+V5 and 80 13136p−m+V20. Of these, 53%–55% of those upregulated in one VISA were also upregulated in the other VISA strain. Approximately the same number of genes were also downregulated in each VISA strain, 187 in VISA13136p−m+V5 and 177 in 13136p−m+V20. Expression concordance was greater for underexpression patterns, as 70%–73% of genes downregulated in one VISA were also downregulated in the other VISA strain. As shown in Figure 1, VISA 13136p−m+V5 had noticeably more expression changes than 13136p−m+V20 among the cell envelope (41 vs. 29) and transport and binding protein (11 vs. 20) genes and fewer than 13136p−m+V20 among protein fate (two vs. six) and protein synthesis (11 vs. 20) genes, with downregulated genes outnumbering upregulated genes at least 2:1 for each VISA and these functional groups, except the 1:1 ratio for the two 13136p−m+V5 protein fate expression changes. Total numbers of regulatory function gene expression changes were comparable between the two VISA, at 19 for VISA13136p−m+V5 and 17 for 13136p−m+V20, but the ratio of the number of upregulated to downregulated genes varied between the strains at 4:15 and 6:11, respectively.
Although overall gene expression trends were similar between the two VISA versus their progenitor VSSA, specific patterns within these data were not consistent between the two comparisons. For example, 13 of the 16 ribosomal proteins listed in Supplementary Table S1 were downregulated for 13136p−m+V20, with three unchanged, compared to 13136p−m+. In contrast, four were downregulated and 12 unchanged for the 13136p−m+V5 to 13136p−m+ comparison. However, for the expression of 18 pur and pyr genes associated with central intermediary biosynthetic pathways, 12 were downregulated and six unchanged in 13136p−m+V5, and 15 were downregulated and three unchanged in 13136p−m+V20. For either ribosomal protein or pur and pyr genes, when expression was unchanged in one VISA, it was always downregulated in the other VISA versus 13136p−m+. Genes encoding secreted proteases were generally downregulated in both VISA (sec3, sspA, sspB2 and sspC), as were cap genes encoding capsular polysaccharide biosynthesis, but the pattern of expression changes was inconsistent between the two VISA. The expression of genes encoding cell surface protein, regulatory, biofilm-related and cell wall metabolism gene products was more variable, with more frequent upregulation and greater ranges of expression fold-change values. A large number of genes previously identified as associated with the VISA strain, including members of the cell wall stress stimulon, showed expression changes in one or both VISA versus 13136p−m+. These include vraR and prsA, which are overexpressed in both VISA and are part of the cell wall stress stimulon [5], as well as penicillin binding proteins. McAleese et al. [6] previously reported overexpression of the cell wall stress stimulon in VISA.
Three genes, which were altered in expression, as determined by microarray analysis, were validated using qRT-PCR. The expression ratios of cmk, agrA and ddh were shown to be in good agreement with microarray results (data not shown).
2.3. Genes with at Least One Eight-Fold Expression Change among VISA and VSSA
Table 2 is the subset of genes from Supplementary Table S1 with large expression differences among the three strains, as defined by at least one expression change eight-fold or greater in magnitude (≥8-fold) between either VISA and the progenitor VSSA. Supplementary Figure S1 is a graphical representation of the data in Table 2. In Table 2 genes are sorted by attributes related to the VISA phenotype instead of generic functional groupings. Additionally, the Table 2 V20-V5 column indicates fold-change expression differences between VISA 13136p−m+V20 and 13136p−m+V5. A total of 60 changes ≥8-fold were present between the three strains across 52 different genes. The hypothetical/ unclassified/unknown function gene functional group accounted for 21 (40%) of the 52 genes with ≥8-fold changes, followed by 11 cell envelope genes (21%), four genes each (8%) in regulatory functions and biosynthesis of cofactors, prosthetic groups and carriers and three genes each (6%) for purines, pyrimidines, nucleosides, nucleotides and transport and binding proteins; together, these groups accounted for 89% of the ≥8-fold change gene subset. This pattern differed from that in Figure 1 and Supplementary Table S2 in that some functional groups had no genes among the ≥8-fold change gene subset, including the amino acid biosynthesis, transcription and, most notably, the protein synthesis groups. The general central intermediary metabolism functional group had two genes in the ≥8-fold change gene subset, just below 4%, but comparable to the total in Supplementary Table S1.

Table 2.
Expression patterns and assigned functional roles for genes with at least one ≥ 8-fold expression increase or decrease (yellow cells) between VSSA 13136p-m+ and VISA or VISA 13136p−m+V5 and 13136p−m+V20.
| Locus ID | Gene | Protein | 13136p−m+V5 vs. VSSA | 13136p−m+V20 vs. VSSA | VISA V20 vs. VISA V5 | VISA-Related Categories | Stress Response Association | Gene and Protein Functional Role Comments [references] |
|---|---|---|---|---|---|---|---|---|
| SAV2009 | sec3 | enterotoxin typeC3 | −24.4 | −21.5 | 2.9 | Virulence Factor-Associated | Secreted exotoxin, same as gene sea in MRSA strain COL [11] | |
| SACOL0907 | seb | staphylococcal enterotoxin B | −17.2 | −18.3 | −1.1 | Nutritional | Secreted cytotoxin upregulated during the stringent response [45] | |
| SACOL1871 | epiG | epidermin immunity protein F | −11.2 | 11.2 | Nutritional | Virulence factor upregulated during the stringent response [45] | ||
| SAV2472 | NA | short chain dehydrogenase | −8.5 | −5.2 | 3.3 | Capsular biosynthesis enzyme from the SDR protein super-family [46] | ||
| SAV0372 | NA | predicted PepSY family membrane peptidase propeptide | −4.1 | −10.5 | −6.4 | Unknown function but likely to have a protease inhibitory function based on homology to peptidase propeptides | ||
| SAV1046 | sspC | cysteine protease | −3.4 | −8.6 | −5.2 | Secreted virulence factor Staphostatin B that inhibits sspB2-encoded serine protease Staphopain B activity [47] | ||
| SACOL2026 | agrA | accessory gene regulator protein A | −2.8 | −9 | −6.2 | Response regulator of the agr operon which generally upregulates secreted proteins and downregulates cell surface proteins [48] | ||
| SACOL0096 | sarS | staphylococcal accessory regulator S | 8.9 | 8.9 | Nutritional | SarA global regulator family that generally upregulates virulence factor genes; sarS is downregulated by agr and upregulated by Rot and during the stringent response [45,48,49] | ||
| SAV0111 | spa | Immunoglobulin G binding protein A precursor | 11.5 | 11.5 | General | Cell surface adhesion protein upregulated by heat shock and downregulated by cell wall-active antibiotics [5,47,50] | ||
| SAV1764 | rot | repressor of toxins Rot | 12.6 | 12.6 | Virulence Factor-Associated | Homolog of the SarA transcriptional regulator that often exerts effects opposite of agr [48] | ||
| SAV0320 | geh | glycerol ester hydrolase | 2.7 | 12 | 9.3 | rot-regulated lipase translated as an inactive, truncated form due bacteriophage L54a integration [10,49] | ||
| SAV2637 | aur | zinc metalloproteinase aureolysin | 8.2 | −8.2 | Secreted virulence factor [51] | |||
| SAV2667 | icaD | intercellular adhesion protein D | 15.9 | −15.9 | Oxidative | Cell surface virulence factor downregulated by oxidative stress [25] | ||
| SACOL2689 | icaA | intercellular adhesion protein A | 25.2 | −25.2 | Oxidative | Cell surface virulence factor downregulated by oxidative stress [25] | ||
| SAS0236 | scdA | cell wall metabolism protein ScdA | −2 | 9.5 | 11.5 | Cell Wall Metabolism | Oxidative | ScdA is involved in peptidoglycan cross-linking and cell division, and upregulated by oxidative stress [25] |
| SACOL0034 | mecR1 | methicillin-resistance MecR1 regulatory protein | 38.5 | −38.5 | Cell Wall-Active Antibiotics | Integral membrane metalloprotease acting as a beta-lactam sensing signal transducer [52] | ||
| SAV0041 | mecA | penicillin binding protein 2A | 48.5 | −48.5 | Nutritional & Cell Wall-Active Antibiotics | Alternative PBP imparting mec-mediated resistance to beta-lactam antibiotics | ||
| SACOL2147 | NA | transcriptional antiterminator, BglG family/DNA-binding protein | −12 | −4.4 | 7.6 | Central Intermediary Metabolism | Oxidative | Regulatory functions; downregulated by oxidative stress [25] |
| SACOL1573 | NA | integrase/recombinase, core domain family | −10.6 | −12.1 | −1.5 | DNA Damage | Pseudogene not located within a prophage or pathogenicity island | |
| SAV2328 | NA | dehydrogenase | −9.9 | −4.4 | 5.5 | Unknown function; from the SDR protein super-family [53] | ||
| SAV2182 | asp23 | alkaline shock protein 23 | −9.8 | −19.7 | −9.9 | Nutritional & General | σB-regulated general stress response gene upregulated in VISA, by alkaline or heat shock, and during the stringent response [45,50,54] | |
| SACOL1114 | NA | Mn2+/Fe2+ transporter, NRAMP family | −9.5 | −9.4 | 0.1 | Oxidative & Nutritional | Gene upregulated by oxidative stress - Mn2+ and Fe2+ are important for oxidative stress resistance, and Mn may have a role in virulence related competition with hosts for limited nutrient [25,55] | |
| SAV1074 | purD | phosphoribosylamine-glycine ligase | −9.5 | −2.3 | 7.2 | Nutritional | Purine ribonucleotide biosynthesis gene downregulated during the stringent response [45] | |
| SAV1072 | purN | phosphoribosylglycinamide formyltransferase | −9.5 | −2.5 | 7 | Purine ribonucleotide biosynthesis gene [56] | ||
| SAV2185 | NA | glycine betaine transporter opuD homolog | −9.2 | −14.5 | −5.3 | Osmotic & General | The opuCABCD operon is upregulated by osmotic stress and part of the general stress response [57] | |
| SAS0678 | NA | glutamine amidotransferase class-I protein | −8.6 | −5.4 | 3.2 | Subunit of anthranilate synthase, an enzyme from the glutamate-consuming folate biosynthetic pathway | ||
| SACOL0872 | NA | OsmC/Ohr family protein | −8.5 | −5.7 | 2.8 | Osmotic | Membrane protein of unknown function induced by osmotic stress | |
| SAV1071 | purM | phosphoribosylaminoimidazole synthetase | −8.4 | −2.2 | 6.2 | Nutritional | Purine ribonucleotide biosynthesis gene downregulated during the stringent response [45] | |
| SACOL0630 | NA | amino acid permease | −6 | −9.5 | −3.5 | Transmembrane amino acid transporter protein | ||
| SACOL2428 | bioD | dethiobiotin synthase | 8.7 | 8.7 | Biotin biosynthesis enzyme; competition for biotin may play an important role in phagosome escape [58] | |||
| SACOL0032 | maoC | (R)-specific enoyl-CoA hydratase | 23.7 | −23.7 | Amino acid degradation enzyme in aerobic phenylalanine/phenylacetate catabolism [59] | |||
| SACOL0866 | NA | hypothetical protein | −12.9 | −3.8 | 9.1 | Hypothetical Proteins | ||
| SAR0592 | NA | hypothetical protein | −12.3 | −15.8 | −3.5 | |||
| SAV0823 | NA | hypothetical protein | −11.3 | −3.1 | 8.2 | |||
| SAR2275 | NA | hypothetical protein | −9.8 | −20.4 | −10.6 | |||
| SACOL2547 | NA | hypothetical protein | −9.5 | 4.2 | 13.7 | |||
| SACOL2720 | NA | hypothetical protein | −9.4 | −7 | 2.4 | |||
| SAS2396a | NA | hypothetical protein | −8.3 | −6.4 | 1.9 | |||
| SAV2565 | NA | hypothetical protein | −8.2 | −2.3 | 5.9 | |||
| SAS2047 | NA | hypothetical protein | −8.1 | −3.9 | 4.2 | |||
| SACOL2174 | NA | hypothetical protein | −7.4 | −20.2 | −12.8 | |||
| SACOL1679 | NA | hypothetical protein | −7.3 | −10.3 | −3 | |||
| SACOL2175 | NA | hypothetical protein | −6.8 | −17.3 | −10.5 | |||
| SACOL1680 | NA | hypothetical protein | −6.3 | −8.4 | −2.1 | |||
| SAV2474 | NA | hypothetical protein | −5.5 | −10.8 | −5.3 | |||
| SACOL0912 | NA | hypothetical protein | −5.4 | −8.3 | −2.9 | |||
| SACOL1574 | NA | hypothetical protein | −5.4 | −8.6 | −3.2 | |||
| SACOL0908 | NA | hypothetical protein | −3.8 | −8.8 | −5 | |||
| SAS0281 | NA | hypothetical protein | 10 | 3.6 | −6.4 | |||
| SACOL0625 | NA | hypothetical protein | 11.2 | 2.5 | −8.7 | |||
| SACOL0067 | NA | hypothetical protein | 12.6 | 5 | −7.6 | |||
| SAV2556 | NA | hypothetical protein | 12.8 | 3.6 | −9.2 |
Five genes in the ≥8-fold change subset that encode regulatory proteins (agrA, sarS, rot, mecR1 and locus SACOL2147) all had very different expression patterns between the two VISA versus parent VSSA3136p−m+. The only two genes with ≥8-fold expression changes between both VISA and the parent VSSA and between the two VISA themselves were locus SAR2275, which encodes a hypothetical protein, and asp23, whose gene product is the σB-regulated alkaline shock protein 23. This gene is downregulated by cell wall-active antibiotics and often upregulated during the general stress response and many specific stress responses, but its function is not known [5].
Genes with ≥8-fold expression changes were grouped into four VISA-related categories in Table 2: virulence factor-associated (14 genes, 12 up- and 12 down-regulated changes), cell wall metabolism (three genes, four up- and two down-regulated changes), central intermediary metabolism (14 genes, three up- and 18 down-regulated changes) and hypothetical proteins (21 genes, seven up- and 25 down-regulated changes). Of the 31 genes that did not encode hypothetical functions, 17 have one or more known stress response associations that are listed in the Table 2 Stress Response Association column. Eight genes were associated with nutritional stress (stringent response), five with oxidative stress, three with the general stress response, two each with osmotic and cell wall-active antibiotic stresses, and one with DNA damage response (recombination repair function and/or SOS response).
Ten of the ≥8-fold expression changes were upregulations (29%), and 24 were downregulations (71%) for the 13136p−m+V5 versus VSSA 13136p−m+ comparison. These figures were six and 20 (23% and 77%) and 10 and 13 (43% and 57%) for the 13136p−m+V20 versus VSSA 13136p−m+ and 13136p−m+V20 versus 13136p−m+V5 comparisons, respectively.
2.4. Metabolomics
Ninety-eight total metabolites were identified in VSSA 13136p−m+ and its two VISA derivatives, 13136p−m+V5 and 13136p−m+V20. Twenty-four were in the amines and polyamines metabolite class, 25 were amino acids, 26 were polar organic acids and the remaining 23 were sugars. The concentrations of 43 metabolites were increased and 42 were decreased in 13136p−m+V5 cells compared to the parent strain 13136p−m+ (12 were unchanged). In 13136p−m+V20 cells, the concentrations of 47 metabolites were increased and 48 were decreased compared to their concentrations in the parent strain 13136p−m+ (only two were unchanged). Thirteen metabolites were increased in both VISA and twelve others were decreased in both VISA compared to the parent VSSA. The relative concentrations of the identified metabolites in the three strains are shown in Table 3, which also contains calculated fold-changes for both VISA versus parent VSSA relative concentrations, as well as fold-changes for 13136p−m+V20 compared to its immediate passage-selection predecessor, 13136p−m+V5. In 13136p−m+V20 cells, the concentrations of 46 metabolites were increased, and 48 were decreased compared to their concentrations in predecessor 13136p−m+V5 13136p−m+ (four relative concentrations were unchanged).
Supplementary Figure S2 is a graphical presentation by metabolite class of the fold-change of all 98 detected metabolites’ relative concentrations for 13136p−m+V5 in comparison to the parent VSSA, 13136p−m+V20 in comparison to the parent VSSA, and 13136p−m+V20 in comparison to 13136p−m+V5. In general, a decrease in sugars was evident in strain 13136p−m+V20 versus its VISA predecessor 13136p−m+V5. Relative concentrations of 10 sugars increased and 12 decreased (one unchanged) in 13136p−m+V5 versus the parent VSSA, but the tallies were only five increased and 18 decreased for the 13136p−m+V20 versus parent VSSA and five increased and 16 decreased in 13136p−m+V20 in comparison to 13136p−m+V5. Most of the increased relative concentrations of sugars in 13136p−m+V20 in comparison to 13136p−m+V5 were still below those in VSSA 13136p−m+. In contrast, the relative concentrations of 15 amino acids increased and eight decreased in 13136p−m+V5 in comparison to 13136p−m+, 11 increased and 13 decreased in 13136p−m+V20 in comparison to 13136p−m+, and 12 increased and 11 decreased in 13136p−m+V20 in comparison to 13136p−m+V5.

Table 3.
The 98 metabolites identified by metabolomic analyses, relative concentrations, and fold-change differences between strains. Empty cells: no changes (one-fold changes); ND: Not Detected, with values of 0.01 used for < and > fold-change estimates. Metabolites within each class sorted alphabetically.
| Metabolite Class | Metabolite | Metabolite relative concentration per 10 mg dry weight (mean ± SD) | Metabolite Relative Concentration Fold-Change | ||||
|---|---|---|---|---|---|---|---|
| VSSA 13136p−m+ | 13136p−m+V5 | 13136p−m+V20 | VSSA → V5 | VSSA → V20 | V5 → V20 | ||
| Amines & Polyamines | 2-Amino-4,6-dihydroxypyrimidine | 1.4 ± 0.2 | 1.9 ± 0.4 | 1.3 ± 0.3 | 1.4 | −1.1 | −1.4 |
| 4,5-Dimethyl-2,6-hydroxypyrimidine | 0.3 ± 0.1 | 0.3 ± 0.0 | 0.2 ± 0.0 | −1.5 | −1.5 | ||
| 5-Methylthioadenosine | 2.7 ± 0.2 | 2.4 ± 0.6 | 4.6 ± 1.0 | −1.3 | 1.7 | 1.9 | |
| Adenine | 181.5 ± 21.5 | 138.5 ± 27.2 | 74.4 ± 5.5 | −1.3 | −2.4 | −1.9 | |
| Adenosine | 125.0 ± 21.5 | 300.9 ± 94.4 | 353.6 ± 10.3 | 2.4 | 2.8 | 1.2 | |
| Adenosine-5-monophosphate | ND | ND | 5.7 ± 1.2 | >100 | >100 | ||
| Cytosine | 5.5 ± 0.6 | 5.7 ± 1.7 | 3.3 ± 0.3 | −1.7 | −1.7 | ||
| Dihydroorotic acid | ND | 3.8 ± 0.7 | 171.8 ± 27.9 | >100 | >100 | 45.2 | |
| Ethanolamine | 2.7 ± 0.7 | 4.0 ± 0.8 | 1.4 ± 0.1 | 1.5 | −2 | −2.9 | |
| Glucosamine | ND | 6.9 ± 1.5 | 129.4 ± 30.5 | >100 | >100 | 18.8 | |
| Guanine | 16.2 ± 1.2 | 2.7 ± 0.8 | 1.7 ± 0.3 | −5 | −10 | −1.6 | |
| Guanosine | 10.9 ± 2.3 | ND | 42.7 ± 8.6 | <−100 | 3.9 | >100 | |
| Hydroxylamine | 1.0 ± 0.1 | 4.3 ± 0.8 | 1.9 ± 0.2 | 4.3 | 1.9 | −2.3 | |
| Hypoxanthine | ND | ND | 0.1 ± 0.0 | >10 | >10 | ||
| Inosine | ND | ND | 3.3 ± 0.7 | >100 | >100 | ||
| Nicotinamide | 19.0 ± 1.7 | 13.0 ± 1.6 | 25.9 ± 1.1 | −1.4 | 1.4 | 2 | |
| Nicotinic acid | 4.0 ± 0.9 | 0.5 ± 0.1 | 9.5 ± 1.5 | −8 | 2.4 | 19 | |
| Orotic acid | 1.7 ± 0.1 | 1.5 ± 0.1 | 38.7 ± 1.3 | −1.1 | 23 | 25.8 | |
| Putrescine | 63.3 ± 3.7 | 63.5 ± 6.5 | 11.8 ± 1.8 | −5.4 | −5.4 | ||
| Spermidine | 42.1 ± 5.7 | 4.5 ± 0.5 | 3.7 ± 0.6 | −9.4 | −11.4 | −1.2 | |
| Thymine | 15.0 ± 2.4 | 4.4 ± 1.1 | 3.4 ± 0.7 | −3.4 | −4.4 | −1.3 | |
| Uracil | 23.7 ± 2.1 | 4.4 ± 0.7 | 6.5 ± 0.8 | −5.4 | −3.6 | 1.5 | |
| Urea | 5.6 ± 0.5 | 11.4 ± 2.2 | 3.7 ± 0.6 | 2 | −1.5 | −3.1 | |
| Uridine | 9.3 ± 1.0 | 57.6 ± 7.4 | 13.2 ± 0.9 | 6.2 | 1.4 | −4.4 | |
| Amino Acids | 4-hydroxyproline | 5.5 ± 0.6 | 6.3 ± 1.7 | 3.9 ± 1.1 | 1.1 | −1.4 | −1.6 |
| Alanine | 498.0 ± 47.1 | 525.3 ± 67.1 | 493.2 ± 41.0 | 1.1 | −1.1 | ||
| Asparagine | 18.9 ± 3.0 | 36.8 ± 4.5 | 14.7 ± 2.5 | 1.1 | −1.3 | −2.5 | |
| Aspartic acid | 975.4 ± 127.0 | 2203.8 ± 446.5 | 2303.0 ± 397.1 | 2.3 | 2.4 | ||
| Cystathionine | 4.5 ± 0.8 | ND | ND | <−100 | <−100 | ||
| Glutamic acid | 20.5 ± 4.0 | 284.7 ± 76.8 | 308.5 ± 36.4 | 14 | 15 | 1.1 | |
| Glycine | 33.8 ± 10.3 | 45.7 ± 6.2 | 109.5 ± 9.8 | 1.4 | 3.2 | 2.4 | |
| Homocysteine | 1.5 ± 0.3 | 3.7 ± 0.8 | 5.7 ± 1.0 | 2.5 | 3.8 | 1.5 | |
| Homoserine | 1.0 ± 0.2 | ND | 0.6 ± 0.1 | <−100 | −1.7 | >100 | |
| Isoleucine | 26.5 ± 3.5 | 36.0 ± 7.1 | 88.9 ± 9.9 | 1.4 | 3.4 | 2.4 | |
| Leucine | 118.1 ± 24.2 | 330.4 ± 40.9 | 451.9 ± 68.2 | 2.8 | 3.8 | 1.4 | |
| Lysine | 354.5 ± 39.9 | 276.8 ± 20.6 | 121.9 ± 27.5 | −1.3 | −2.9 | −2.3 | |
| Methionine | 10.4 ± 2.1 | 11.7 ± 2.8 | 8.5 ± 1.2 | 1.1 | −1.2 | −1.4 | |
| N-Acetylglutamic acid | ND | ND | 25.6 ± 1.5 | >100 | >100 | ||
| O-Acetyl-serine | 5.4 ± 1.3 | 1.0 ± 0.2 | ND | −5.4 | <−100 | <−100 | |
| Ornithine | 30.8 ± 4.8 | 16.6 ± 3.1 | 1.2 ± 0.2 | −2 | −26 | −13.8 | |
| Phenylalanine | 165.9 ± 29.1 | 237.0 ± 30.3 | 116.5 ± 9.3 | 1.4 | −1.4 | −2 | |
| Proline | 10.9 ± 1.5 | 154.6 ± 30.0 | 239.8 ± 8.0 | 14 | 22 | 1.6 | |
| Pyroglutamic acid | 1017.3 ± 159.3 | 1017.6 ± 136.4 | 1659.0 ± 164.2 | 1.6 | 1.6 | ||
| Serine | 11.5 ± 1.3 | 7.2 ± 1.4 | 9.5 ± 2.9 | −1.6 | −1.2 | 1.3 | |
| Threonine | 52.8 ± 10.6 | 20.0 ± 8.5 | 7.8 ± 1.2 | −2.6 | −6.8 | −2.6 | |
| Tryptophan | 29.7 ± 4.8 | 8.5 ± 1.3 | 11.9 ± 3.0 | −3.5 | −2.5 | 1.4 | |
| Tyrosine | 33.6 ± 7.4 | 40.8 ± 5.7 | 26.3 ± 2.2 | 1.2 | −1.3 | −1.6 | |
| Valine | 63.1 ± 14.3 | 152.9 ± 25.8 | 170.6 ± 35.3 | 2.4 | 2.7 | 1.1 | |
| β-Alanine | 57.4 ± 7.6 | 272.3 ± 42.6 | 95.1 ± 12.9 | 4.7 | 1.7 | −2.9 | |
| Polar Organic Acids | 2-Hydroxyglutaric acid | 6.1 ± 0.4 | 2.5 ± 0.3 | 2.8 ± 0.2 | −2.4 | −2.2 | 1.1 |
| 2-Phosphoglycerate | 14.6 ± 2.0 | 7.5 ± 1.9 | 3.0 ± 0.6 | −2 | −5 | −2.5 | |
| 3-Hydroxybenzoic acid | 0.3 ± 0.0 | 0.4 ± 0.1 | 0.5 ± 0.0 | 1.3 | 1.7 | 1.3 | |
| 3-Phosphoglycerate | 242.6 ± 45.1 | 62.7 ± 10.9 | 40.1 ± 7.5 | −3.9 | −6 | −1.6 | |
| Aminomalonic acid | 0.6 ± 0.1 | 0.21 ± 0.1 | 0.7 ± 0.1 | 2.9 | 1.2 | 3.3 | |
| Benzoic acid | 1.6 ± 0.1 | 1.9 ± 0.2 | 1.6 ± 0.4 | 1.2 | −1.2 | ||
| cis-Aconitic acid | ND | 2.1 ± 0.2 | ND | >100 | <−100 | ||
| Citramalic acid | 14.8 ± 2.7 | 19.1 ± 4.1 | 27.3 ± 5.4 | 1.3 | 1.8 | 1.4 | |
| Citric acid | 15.6 ± 2.0 | 243.7 ± 22.6 | 35.1 ± 8.4 | 16 | 2.3 | −6.9 | |
| Fumaric acid | 19.8 ± 4.5 | 1.5 ± 0.4 | 28.6 ± 4.0 | −12.5 | 1.4 | 19.1 | |
| Glucaric acid | 1.2 ± 0.3 | 2.7 ± 0.4 | 3.0 ± 0.1 | 2.3 | 2.6 | 1.1 | |
| Gluconic acid | 0.6 ± 0.1 | 11.8 ± 2.2 | 5.8 ± 2.1 | 20 | 10 | −2 | |
| Glyceric acid | 7.0 ± 0.9 | 6.9 ± 1.3 | 3.7 ± 0.2 | −2 | −1.9 | ||
| Glycolic acid | 5.2 ± 0.6 | 5.1 ± 0.9 | 20.2 ± 1.8 | 3.9 | 4 | ||
| Lactic acid | 358.9 ± 52.4 | 320.6 ± 36.2 | 682.9 ± 60.6 | −1.1 | 1.9 | 2.1 | |
| Malic acid | 3.4 ± 0.6 | ND | 12.9 ± 2.3 | <−100 | 3.7 | >100 | |
| Monomethylphosphate | 104.8 ± 24.0 | 161.3 ± 2.0 | 258.1 ± 17.9 | 1.5 | 2.5 | 1.6 | |
| Oxyphosphinyloxyacetate | 3.8 ± 0.1 | 3.4 ± 0.3 | 6.6 ± 1.0 | −1.1 | 1.7 | 1.9 | |
| Pantothenate | ND | ND | 2.2 ± 0.0 | >100 | >100 | ||
| Phenylpyruvic acid | ND | ND | 3.4 ± 0.4 | >100 | >100 | ||
| Pyruvic acid | 58.1 ± 10.5 | 38.9 ± 15.8 | 31.4 ± 4.8 | −1.4 | −2 | −1.2 | |
| Succinic acid | 36.1 ± 8.7 | 31.6 ± 8.2 | 42.9 ± 9.6 | −1.1 | 1.2 | 1.4 | |
| α-Glycerophosphate | 824.3 ± 131.3 | 1487.9 ± 121.8 | 920.7 ± 76.9 | 1.8 | 1.1 | −1.6 | |
| α-Ketoglutaric acid | 30.7 ± 6.9 | 11.1 ± 2.1 | 18.2 ± 2.7 | −2.8 | −1.7 | 1.6 | |
| β-Lactate | 3.6 ± 0.8 | 2.0 ± 0.2 | 8.5 ± 1.7 | −1.7 | 2.3 | 4.3 | |
| β-Phenyllactic acid | 3.0 ± 0.6 | 1.7 ± 0.2 | 7.3 ± 0.7 | −1.7 | 2.4 | 4.3 | |
| Sugars | Arabitol | 6.3 ± 1.3 | 12.0 ± 2.3 | 4.8 ± 0.7 | 1.9 | −1.3 | −2.5 |
| Fructose | 40.5 ± 7.5 | 4.0 ± 0.8 | 1.8 ± 0.2 | −10 | −23 | −2.2 | |
| Fructose-6-P | 6.7 ± 2.0 | 5.2 ± 1.0 | 3.0 ± 0.7 | −1.3 | −2.5 | −1.7 | |
| Galactitol | ND | 2.1 ± 0.2 | 2.0 ± 0.3 | >100 | >100 | −1.1 | |
| Galactopyranose | 2.2 ± 0.5 | 3.4 ± 0.7 | 2.0 ± 0.1 | 1.5 | −1.1 | −1.7 | |
| Galactose | 5.1 ± 1.3 | 7.9 ± 0.8 | 1.5 ± 0.3 | 1.5 | −3.3 | −5.3 | |
| Glucose-1-P | 20.3 ± 5.2 | 13.8 ± 3.4 | 2.3 ± 0.6 | −1.4 | −10 | −6 | |
| Glucose-6-P | 13.6 ± 0.7 | 1.6 ± 0.3 | 0.5 ± 0.1 | −8.5 | −27.2 | −3.2 | |
| Glycerol | 569.8 ± 71.4 | 668.7 ± 76.0 | 611.0 ± 103.9 | 1.2 | 1.1 | −1.1 | |
| Glycerol-2-P | 28.2 ± 7.4 | 12.1 ± 3.0 | 7.6 ± 1.5 | −2.5 | −3.3 | −1.6 | |
| Inositol | 3.1 ± 1.0 | 12.8 ± 1.1 | 0.1 ± 0.0 | 4.1 | −31 | −128 | |
| Inositol, -chiro- | 0.8 ± 0.1 | 1.5 ± 0.3 | 1.9 ± 0.1 | 1.8 | 2.2 | 1.3 | |
| Mannitol | 178.1 ± 15.6 | 29.5 ± 7.2 | 32.3 ± 3.1 | −6 | −5.5 | 1.1 | |
| Mannitol-P | 36.6 ± 3.2 | 9.0 ± 1.3 | 14.3 ± 2.8 | −4.1 | −2.6 | 1.6 | |
| Mannose | 7.2 ± 1.0 | 12.8 ± 2.4 | 1.2 ± 0.2 | 1.8 | −6 | −10.7 | |
| Mannose-6-P | 13.2 ± 2.9 | ND | ND | <−100 | <−100 | ||
| Ribitol | 122.9 ± 22.0 | 24.2 ± 4.4 | 76.2 ± 15.4 | −5 | −1.6 | 3.1 | |
| Ribose | 47.9 ± 9.4 | 42.0 ± 7.1 | 25.0 ± 3.3 | −1.1 | −2 | −1.7 | |
| Ribose-5-P | 4.3 ± 0.5 | 30.6 ± 1.8 | 12.5 ± 2.0 | 7.2 | 2.9 | −2.4 | |
| Sedoheptulose | 2.3 ± 0.7 | 2.2 ± 0.0 | ND | <−100 | <−100 | ||
| Sedoheptulose-7-P | 1.0 ± 0.0 | ND | ND | <−100 | <−100 | ||
| Sucrose | 14.5 ± 2.5 | 1.4 ± 0.7 | 6.4 ± 0.2 | −10 | −2.3 | 4.6 | |
| Trehalose | 2.5 ± 0.3 | 17.6 ± 0.7 | 3.4 ± 0.8 | 7 | 1.4 | −5.2 | |
Figure 2 is a set of diagrams of central intermediary metabolic pathways with detected metabolites indicated along with increases and decreases in concentration for all three comparisons of relative concentrations (each VISA versus the parent VSSA and VISA 13136p−m+V20 vs. VSSA 3136p−m+V5). For clarity, only metabolite relative concentration changes of five-fold or greater are presented. Forty five of the 98 metabolites identified had at least one relative concentration change of five-fold or greater among the three strain comparisons. By metabolite class, these were: 13 amines and polyamines, eight amino acids, nine polar organic acids and 15 sugars. These 45 metabolites, their relative concentrations per strain, and relative concentration fold-changes for all three strain comparisons are listed in Supplementary Table S4. The Figure 2 diagrams are not intended to be complete pathways, and many metabolites are involved in numerous metabolic processes; and for some, such as polyamines and unphosphorylated sugars, there is uncertainty around their metabolic roles. The pathways presented represent those that seem relevant given the context of the other metabolites identified and the results of the genomic and transcriptomic analyses.
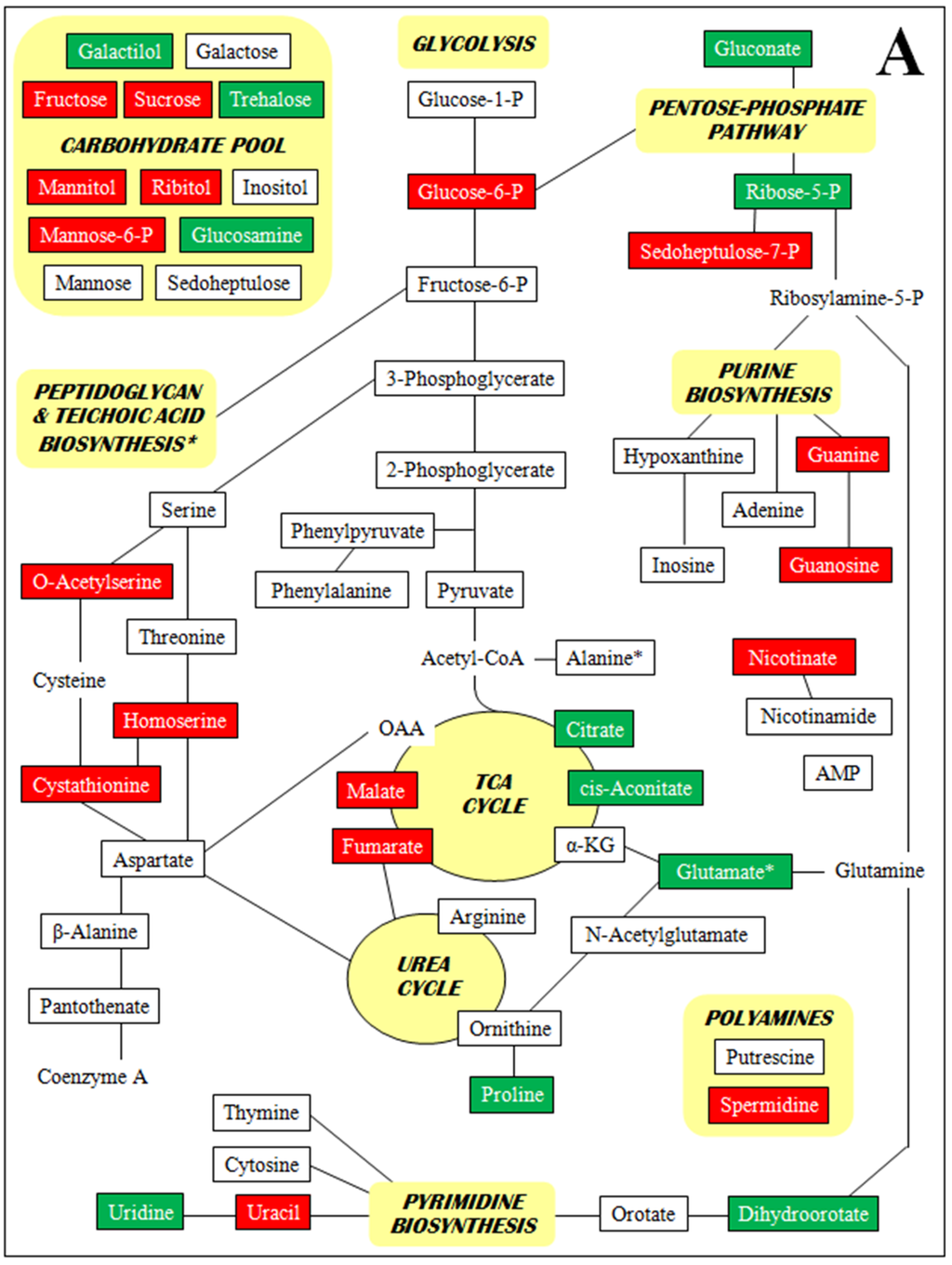
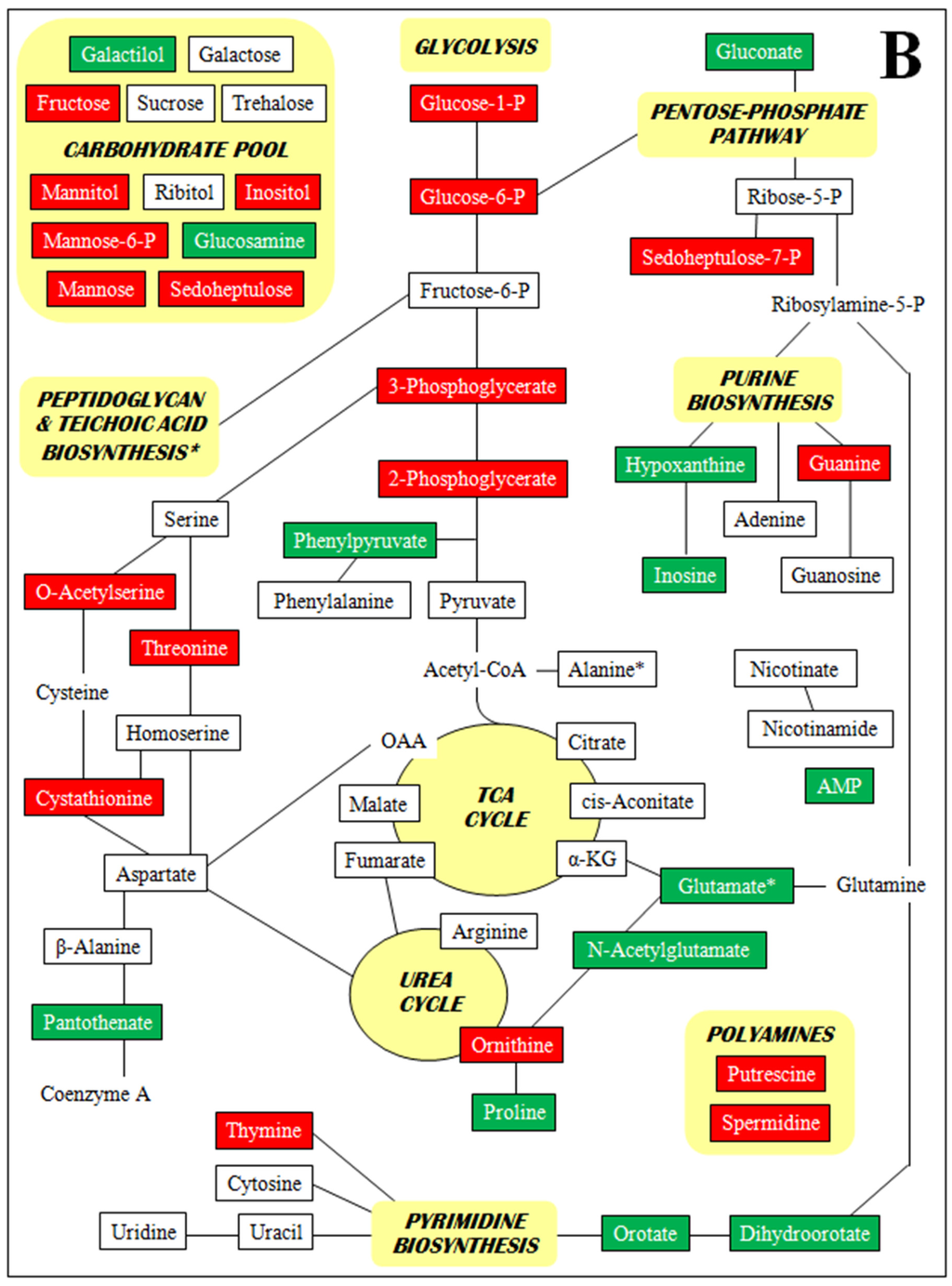
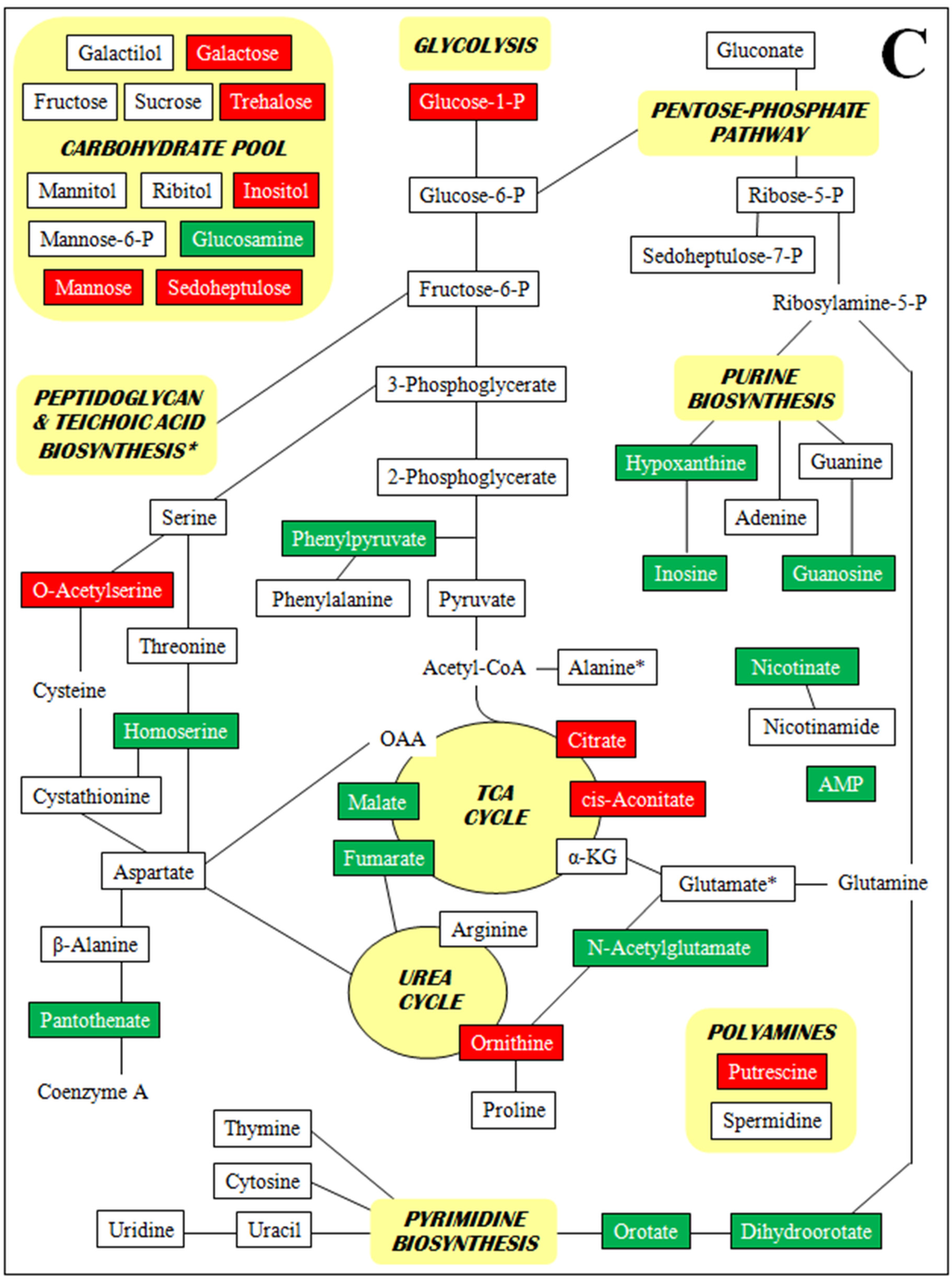
Figure 2.
Metabolites with at least one five-fold relative concentration difference between (A) VISA 13136p−m+V5 relative to VSSA 13136p−m+, (B) VISA 13136p−m+V20 relative to VSSA 13136p−m+ and (C) VISA 13136p−m+V20 relative to VISA 13136p−m+V5, as well as their positions in metabolic pathways. Bordered terms: metabolites identified by metabolomic analyses; green shading: five-fold or greater relative concentration increases; red shading: five-fold or greater relative concentration decreases.
A number of metabolites were only detected in one or two of the three strains employed in this study. Particularly striking were the absence of guanosine from 13136p−m+V5 and the accumulation of AMP, N-acetylglutamic acid, pantothenate and phenylpyruvic acid in 13136p−m+V20. Polyamine relative concentrations were sequentially diminished as greater vancomycin tolerance accrued over the course of VISA selection. Initially, the spermidine relative concentration was reduced 9.4-fold by the selection of 13136p−m+V5 from VSSA 13136p−m+, while putrescine’s was unchanged; then the selection of 13136p−m+V20 from 13136p−m+V5 led to a 5.4-fold reduction in putrescine that was accompanied by a further, small reduction in spermidine. Several sugars were sequentially diminished in a similar fashion, notably glucose-1-phosphate, glucose-6-phosphate, and fructose.
The amino acids consistently present in the highest concentrations were alanine, aspartic acid, glutamic acid, leucine, lysine, proline and pyroglutamic acid. In general, the concentrations of these amino acids were increased in strains 13136p−m+V5 and 13136p−m+V20 compared to the parent strain. Aspartic acid was increased 2.3- and 2.4-fold, respectively, in the two VISA strains, glutamic acid 14- and 15-fold, leucine 2.8- and 3.8-fold and proline 14- and 22-fold.
There were high concentrations at a similar level in all three strains of pyroglutamate, which is the cyclic lactam of glutamic acid [60]. Although pyroglutamic acid is almost ubiquitously present in cells from Archaea to humans, it is a neglected metabolite. Pyroglutamate is formed by a number of different routes, including from the degradation of glutathione, spontaneous cyclization of glutamate phosphate and non-enzymatically from glutamate and glutamine. We are unaware of previous reports of its occurrence in S. aureus.
Glycerol was present at similar high levels in all three strains. Alpha glycerophosphate was present in all three strains. Mannitol relative concentration was significantly decreased in the VISA strains. Acid production from mannitol is a species characteristic of S. aureus. Lactic acid was present at high levels in all three strains.
3. Discussion
It has become apparent that the stress response network and virulence are intrinsically interwoven with central intermediary metabolism. Figure 3 is a diagram of the relationships between these three aspects of bacterial physiology. Secreted virulence factors, such as proteases and hemolysins, destroy host tissue in order to generate nutrients for growing pathogen populations. The released nutrients are transported into the S. aureus cell to serve as substrates for central intermediary metabolic pathways. Metabolic pathways are modulated to minimize damage from host immune response oxidative and nitrosative assaults [61]. Cell surface associated virulence factors, such as adhesions, promote host tissue colonization and aid in the evasion of host immune responses that are triggered by the activities of the secreted proteins. The production of virulence factors in S. aureus is density-dependent and regulated by a complicated network of global regulatory loci, but also by stress-related adjustments of central metabolic pathways, including the tricarboxylic acid (TCA) cycle. Some TCA cycle enzymes are upregulated in biofilm-associated cells versus planktonic culture [62,63]. Many of these global regulators, such as alternative sigma factor σB and members of the staphylococcal accessory regulator (Sar) family, exert regulatory influence over not only virulence factors, but the S. aureus general stress response and expression of antimicrobial resistances, including vancomycin intermediate susceptibility [64].
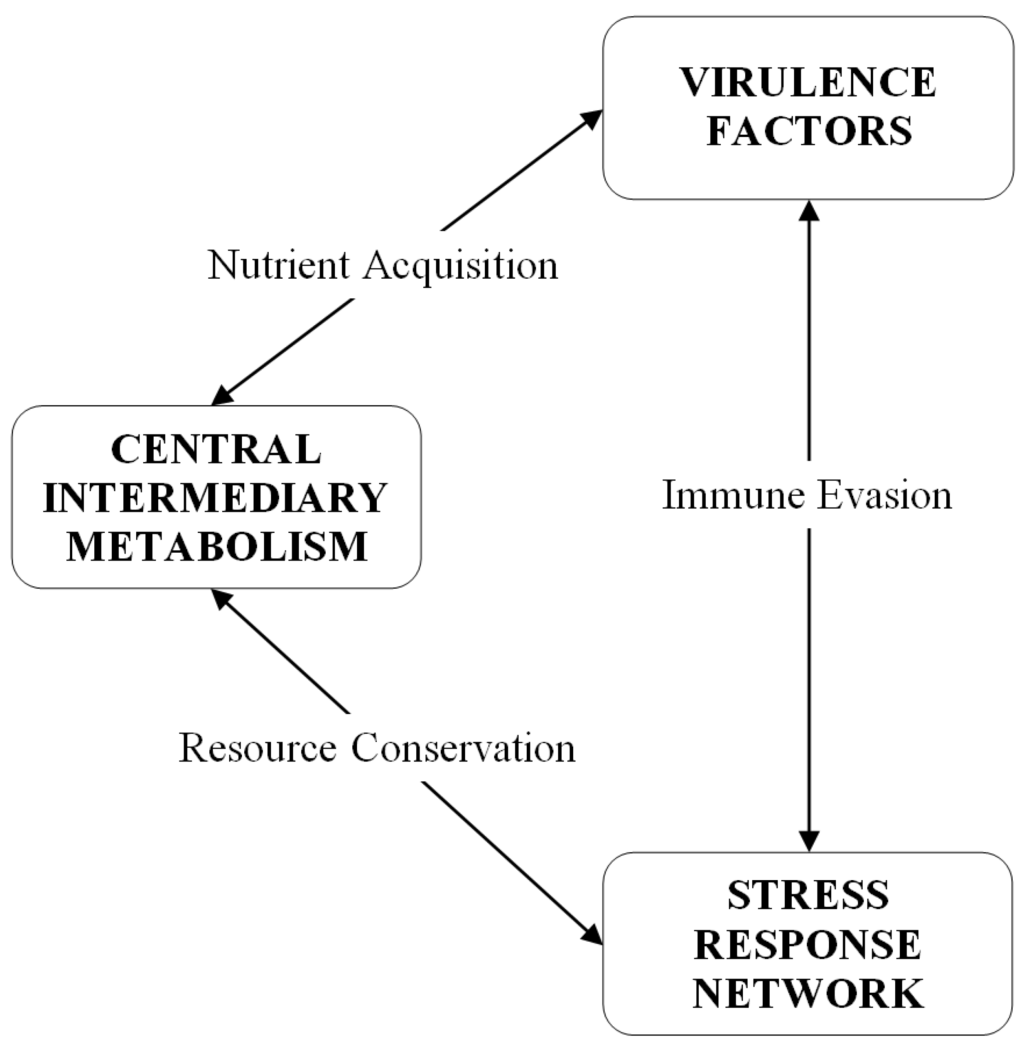
Figure 3.
Conceptual diagram of the relationship between central intermediary metabolism, virulence factors, and the stress response network. Virulence is related to central intermediary metabolism through the production of soluble exoprotein virulence factors, regulated in a cell density-dependent fashion, which destroy host tissue in order to make nutrients available to a growing bacterial population. Cell-associated virulence factors are primarily defenses against host immune responses, working in conjunction with stress responses to immune system oxidative insults and damage sensing and responses to cell wall-active antibiotics. The stringent response is a stress response that coordinates a downregulation of central intermediary metabolism, and protein and nucleic acid synthesis, in order to conserve resources during periods of nutritional scarcity.
The alarmone-mediated nutritional stress response, known as the stringent response, has been linked to activated stress responses and methicillin resistance [45]. The S. aureus starvation response involves expression changes to genes associated with DNA repair and oxidative stress resistance [65]. rpoB mutations associated with conversion from VSSA to VISA also impart characteristics of the stringent response, reduced expression of genes encoding ribosomal proteins and upregulation of genes encoding amino acid biosynthesis enzymes, in addition to a slowed growth rate [66]. RNA polymerase subunit encoding genes rpoB and rpoC were downregulated 3.1- and 2.1-fold in 13136p−m+V20, respectively, but expression was unchanged in 13136p−m+V5. The stringent response is activated in MRSA in response to β-lactam antibiotics and leads to intermediary metabolic pathways optimized for energy production [67,68]. Stringent response induction by mupirocin in a heterogeneous MRSA dramatically increased the production of PBP2A and resulted in homogenous methicillin resistance expression [69]. Additionally, the link between virulence and the stringent response has been described for S. aureus and other bacteria [70]. Despite a dramatic upregulation of the mecA gene in VISA 13136p−m+V5 (48.5-fold vs. parent VSSA or 13136p−m+ V20), the oxacillin MIC of 13136p−m+V5 was comparable to those of the other two strains. Expression of two other genes encoding penicillin-binding proteins changed only slightly, pbp2 down 2.3-fold in 13136p−m+V20 and pbp3 down 2.5-fold in13136p−m+V5, despite the reported importance of PBP2 to laboratory-selected VISA [71].
Multiple studies have identified genes associated with decreased susceptibility to vancomycin. These studies have been summarized by Howden et al. [3,4], who have attempted to make some generalizations. Mutations in the regulatory genes, walKR, vraRS and graRS, and the gene encoding RNA polymerase subunit (rpoB) have been most frequently associated with the VISA phenotype. WalKR is an essential two-component system involved in the control of cell wall metabolism, particularly in controlling the expression of peptidoglycan hydrolases [72]. VraSR controls the cell wall stress stimulon [5,73]. GraSR is a two-component system associated with vancomycin resistance [74] and more generally with sensing antimicrobial peptides [75]. We did not detect mutations in any of these genes in our VISA strains. However, mutations were detected in stp1, fmtB and tarO, which are all involved in cell wall metabolism. Matsuo et al. [76] have identified mutations associated with the transition from hVISA to VISA. These authors have suggested that mutations in genes involved in protein synthesis, such as translation elongation factor G and translation initiation factor IF-1, in our case, may reduce protein synthesis and allow more use of amino acids for peptidoglycan synthesis. Indeed, the levels of several peptidoglycan precursor amino acids were elevated in our VISA strains.
Many of the mutations we encountered in our VISA strains have either not been or have only been rarely associated with the VISA phenotype. Five of the 12 13136p−m+ VISA mutations are in genes encoding products that interact with nucleic acids in protein synthesis or are associated with DNA damage/SOS responses: comGB, fusA, infA, ssb1 and dinG. Often, a single stress will activate components from multiple stress responses. Oxidative stress and heat shock are known to elicit very broad stress responses [25]. However, SOS responses, such as recombination repair and competence mechanisms, are not commonly listed among the stress response alterations associated with the VISA phenotype.
That seven of the nineteen mutant loci identified in this study were in L54a prophage gene ssb1 is intriguing. Single-stranded DNA binding proteins of L54a and similar prophages are thought to function as positive transcriptional regulators of integrase and perhaps other recombination-related phage genes [28,77]. Prophage genomes are known to harbor bacterial virulence determinants that can be regulated by global regulatory elements, such as the agr system [78], and cases have been reported of phage proteins exerting regulatory influence over host gene expression [79,80]. Cell wall biosynthesis disruption in MRSA COL results in downregulation of 33 L54a genes and upregulation of only two (none were ssb1), interpreted as a reinforcement of the lysogenic state, because stress is known to activate the SOS system and thereby the entry of the prophage into the lytic cycle [27]. The fact that mutations were identified in three L54a loci in VISA 13136p−m+V5 and additional mutations accumulated during the selection of increased resistance in VISA 13136p−m+V20, and that there were no mutations in other L54a genes such as those encoding the excisionase, suggests that ssb1 may have a regulatory function that was the target of VISA selection. It seems unlikely that so many mutations in one gene would be necessary to simply disable lytic cycle capabilities. Likewise, mutations in fusA, infA and stp1 may serve to modulate regulatory functions. Competence damage-inducible protein CinA, with functional similarities to both the comGB and dinG gene products, was identified as a member of the cell wall stress stimulon by Utaida et al. [5]. Interestingly, in relation to comGB, L54a itself cannot induce competence as a prophage [81].
Matsuo et al. [66] described the importance of regulation to the VISA resistance mechanism. The greater ratio of up- to down-regulated regulatory function genes for VISA 13136p−m+V20 (6:11) than 13136p−m+V5 (4:15) supports this observation. This is also true for the stp1 SNP reversion when VISA 13136p−m+V20 was selected from VISA 13136p−m+V5. Several regulatory genes showed modest downregulation in 13136p−m+V5: arlSR, glnR, pyrR, rsbU, agrA, agrB, agrC2 (but not agrD), icaR, sarA, sarV, rpoF (encodes stress response RNA polymerase sigma factor σB) and rsbV (encodes an anti-anti-sigma factor). However, saeS, sarR and vraR showed modest upregulation in 13136p−m+V5, and the downregulation of both rpoF and rsbV together was unusual in that this would have opposite effects relative to σB-mediated regulation. In 13136p−m+V20, sarA homologs sarS and rot (repressor of toxins) were highly upregulated at 8.9- and 12-fold versus parent VSSA, respectively. agrA, agrB, agrC2 and agrD genes, as well as saeS, sarA and rsbW were downregulated in 13136p−m+V20, while arlSR, glnR, pyrR, rpoF, rsbV, sarR, sarV and vraR had no expression change versus the progenitor 13136p−m+. saeS is the sensor histidine kinase gene of a two-component regulation system that is upregulated by β-lactams and vancomycin in some other strains [82], and sarA is important for intrinsic antimicrobial resistance [83]. The complexity of the network of regulatory elements that controls virulence factor expression also extends to capsules and biofilms. The general downregulation of capsular polysaccharide biosynthesis genes can be directly interpreted as capsule production inhibition, although typically cap genes have been found to be overexpressed in VISA strains [3,4]. Whether or not biofilm formation was favored or inhibited in the two VISA cannot be discerned from the pattern of expression changes among regulatory loci, although the general downregulation of the protease-encoding genes is favorable to biofilm formation [84].
Secreted and cell surface-associated virulence factor gene expression patterns were similarly complicated. aur, encoding the zinc metalloproteinase aureolysin, was overexpressed 8.2-fold in 13136p−m+V5. Aureolysin has been shown to cleave complement component C3 and is proposed to mediate immune evasion [85]. Gene sspB encodes a cysteine protease precursor and was upregulated 3.7-fold in 13136p−m+V5, but unchanged in 13136p−m+V20. This gene was reported as downregulated in another VISA strain, JH9 [6]. Genes sspC and sspA, encoding a cysteine protease and V8 serine protease, respectively, were downregulated in both 13136p−m+V5 and 13136p−m+V20. Ingavale et al. [86] suggested that decreased levels of SspA may affect the processing of autolysins, such as Atl, resulting in altered autolytic activity, a characteristic displayed by our VISA strains. Serine protease loss may also affect other proteases, specifically cysteine protease proteolytic maturation [87]. The spa gene, encoding protein A, had no expression change in 13136p−m+V5 but surprisingly was upregulated 11.5-fold in 13136p−m+V20. Downregulation of this gene is the typical observation in VISA strains [3,4].
All 16 SNPs in VISA 13136p−m+V20 were in genes with known associations to stress responses, primarily to cell wall antibiotics and DNA damage. The tarO gene product is a membrane-associated protein that catalyzes the initial step in the biosynthesis of cell wall teichoic acid, a polymer of ribitol phosphate subunits [13]. The functions of teichoic acid are not fully understood, but it has roles in VISA characteristics such as cell wall structure, cell separation, autolysis and β-lactam susceptibility. Teichoic acid D-alanylation, mediated by dlt gene products, promotes biofilm formation and cell adhesion and is associated with the regulation of autolysis and virulence [88]. Both dlt genes were downregulated three-fold, but only in VISA 13136p−m+V5. Only VISA 13136p−m+V5 had the mutation in tarO, and it had the largest decrease in the metabolite ribitol relative concentration (five-fold, versus only 1.6-fold in VISA 13136p−m+V20). VISA 13136p−m+V20 has only 70% of the cell wall phosphorus of the parent VSSA, whereas VISA 13136p−m+V5 has 92% of the parent level [13]. Reductions in ribitol concentration did not correlate with lower phosphorus levels in VISA purified cell walls, which is indicative of lower teichoic acid content.
Fructose levels were 10- and 23-fold lower in 13136p−m+V5 and 13136p−m+V20, respectively, versus the relative concentration in the parent VSSA. Fructose phosphate levels were slightly lower in the two VISA than the parent. Expression of locus SAV0698, encoding a putative fructose utilization (fru) operon repressor, was upregulated 2.3- and 4.3-fold in 13136p−m+V5 and 13136p−m+V20, respectively. Factors contributing to the diminished fructose pool could be the repression of genes encoding enzymes required for uptake and utilization or the diversion of available fructose toward upregulated peptidoglycan synthesis. There are problems with both of these hypotheses, however. The uptake mechanism encoded by the fru operon is a phosphotransferase system, but it was unphosphorylated fructose whose relative concentrations changed dramatically. Furthermore, the thickened cell walls characteristic of the VISA phenotype are generally only produced in the presence of glycopeptides. Analyses in this study were performed on cells grown in antibiotic-free medium. Additionally, in 13136p−m+V5, the glmS gene is downregulated, but not in 13136p−m+V20. GlmS catalyzes the initial step in UDP-N-acetylglucosamine biosynthesis, in which fructose as fructose-6-phosphate is incorporated into the cell wall biosynthetic pathway.
Components of the S. aureus general stress response network were selectively deployed in the two VISA, as three of the eight genes whose expression was highly downregulated (≥8-fold versus parent VSSA) were associated with stress responses. These were asp23, encoding alkaline shock protein 23, locus SAV2185, encoding a putative transporter of osmoprotectant glycine betaine, and SACOL1114, encoding a putative Mn2+/Fe2+ transporter associated with oxidative stress management. The remaining five genes encoded two enterotoxins (sec, seb), a pseudogene with homology to integrase/recombinase proteins and two hypothetical proteins. No genes were upregulated ≥8-fold in both VISA.
There was evidence of stress response activation or modification in metabolomic data. The ica operon was strongly upregulated in VISA 13136p−m+V5, but not VISA 13136p−m+V20. Production of a number of virulence factors, including the intercellular adhesin proteins encoded by the ica operon, are regulated by TCA cycle changes in response to stress. An increase in ribose concentration has been linked to TCA cycle stress and an accompanying pentose phosphate pathway regulation modulation [63]. Ribose-5-phosphate concentration was 7.2-fold greater in VISA 13136p−m+V5 than the parent VSSA, although only up 2.9-fold in VISA 13136p−m+V20, while concentrations of ribose itself were comparable in VISA 13136p−m+V5 and the parent VSSA but two-fold lower in VISA 13136p−m+V20 than the parent strain.
Alcohol dehydrogenase is a fermentation pathway enzyme encoded by the adh1 gene, whose expression was decreased 7.8-fold in VISA 13136p−m+V5 versus the parent VSSA, representing a reduced potential for fermentation, while expression was increased 4.7-fold in VISA 13136p−m+V5 versus the parent VSSA, representing an increased potential for fermentation. The pdhC gene encodes a component of the pyruvate dehydrogenase complex, which converts pyruvate to acetate. Expression of pdhC versus the parent 13136p−m+ was unchanged in 13136p−m+V5 and downregulated 3.1-fold in 13136p−m+V20, indicating that fermentation to acetate was not occurring. A shift away from anaerobic growth is consistent with the fact that acetate was not detected by the metabolomic analysis, pyruvic acid levels declined only slightly in the two VISA in comparison to the VSSA parent, and nitrate extrusion protein gene narK was downregulated in 13136p−m+V5.
Adenosine monophosphate (AMP) was not detected in 13136p−m+ or 13136p−m+V5, but was at detectable levels in 13136p−m+V20. In S. aureus, the PurA protein is a substrate for phosphorylation by Stk1, the cognate serine threonine phosphatase to the Stp1 serine threonine kinase encoded by the stp1 gene [20] containing an SNP in 13136p−m+V5 that reverted during selection of 13136p−m+V20. The purA gene encodes adenylosuccinate synthase, which catalyzes the final step in AMP biosynthesis. However, purA was downregulated 2.4-fold in 13136p−m+V20, but unchanged in 13136p−m+V5, so the mutant Stp1 protein was not responsible for the increased AMP level in 13136p−m+V20. Therefore, the accumulation of AMP and downregulation of a gene encoding its biosynthesis may be due to a reduced demand for ATP in 13136p−m+V20. Energy conservation is a stringent response characteristic, as is the general downregulation of purine and pyrimidine metabolism genes observed in both VISA.
The major proline biosynthetic pathway in S. aureus is through arginine and ornithine via the urea cycle, not from glutamate [89]. Accumulation of osmoprotectants, including trehalose and especially proline, is an important osmotic stress countermeasure employed by S. aureus. Trehalose was present in 13136p−m+V5 and 13136p−m+V20 at relative concentrations seven- and 1.4-fold higher than in the parent strain. This is consistent with an activated stress response network in VISA, but not with the gene expression data for the trehalose-specific phosphotransferase system encoded by treP (downregulated 4.2-fold and upregulated 5.3-fold in 13136p−m+V5 and 13136p−m+V20, respectively). The TreP system is phosphoenolpyruvate (PEP) dependent, however, and PEP was not among the identified metabolites, suggesting that other uptake systems may have been utilized for trehalose and indicating that no significant gluconeogenesis was occurring in any of the three strains.
Proline relative concentrations were extremely elevated versus the parent VSSA in both 13136p−m+V5 (14-fold) and 13136p−m+V20 (22-fold). Citrate, cis-aconitate and glutamate, metabolites prior to the urea cycle branch point in the TCA cycle, all accumulated in 13136p−m+V5, while fumarate and malate concentrations, which are at or after the urea cycle branch point, were reduced. TCA cycle intermediate cis-aconitate was not detected in parent VSSA or 13136p−m+V20, only in 13136p−m+V5. In 13136p−m+V20, the identified TCA cycle intermediates, except cis-aconitate, were all within five-fold of parent strain relative concentrations, but many intermediates in glycolysis were reduced at least five-fold versus the parent. This suggests elevated flux through glycolysis to allow the TCA cycle to maintain such high proline levels. However, S. aureus also imports proline via the ProP low-affinity transport system in response to osmotic stress or a high-affinity PutP transport system in response to nutrient limitation [90]. Gene expression for locus SAV0573, encoding a ProP proline/betaine transporter homolog, was upregulated 3.7-fold in 13136p−m+V5 versus the parent strain, consistent with elevated proline concentrations and an activated stress response network. Data were not available for 13136p−m+V20 proP expression. Overall, the results suggest that 13136p−m+V5 was more reliant on transport into the cell via ProP to accumulate proline than 13136p−m+V20, which, in turn, relied more on proline biosynthesis from arginine via the urea cycle. Osmoprotectant uptake from the environment is a more rapid response to osmotic stress than de novo biosynthesis. However, that becomes irrelevant if proline biosynthesis is an aspect of the stress response network in VISA 13136p−m+V20 that is permanently activated to maintain high proline concentrations. Glutamate accumulation may have been due to reduced demand from nucleotide biosynthesis pathways.
Nelson et al. [17] have suggested that there is decreased flow through the TCA cycle in VISA strains and that this is associated with decreased aconitase activity; however, no expression changes were identified for any of the genes in the cit operon. Of seven pyrimidine ribonucleotide biosynthetic pathway genes with expression changes in this study, two were downregulated and five unchanged in 13136p−m+V5 and six downregulated and one unchanged in 13136p−m+V20. The relative concentrations of pyrimidines, as well as purines, did not correlate well with biosynthetic gene expression patterns. McAleese et al. [6] reported pyr gene downregulation in VISA, and Reiss et al. [45] reported pur gene downregulation in MRSA COL during a mupirocin-induced stringent response. Hypoxanthine and inosine were detected in 13136p−m+V20, but not in 13136p−m+V5 or the parent 13136p−m+. These are purine salvage pathway substrates either taken up from the growth medium, perhaps due to activated elements of the starvation response, or are degradation products from excess adenine and adenosine after downregulated purine biosynthesis [65]. Likewise, the accumulation of pyrimidine biosynthetic intermediates, dihydroorotate and orotate, was consistent with a downregulated pyrimidine biosynthetic pathway. Their accumulation was sequential, as dihydroorotate but not orotate relative concentrations were dramatically increased versus the parent VSSA in 13136p−m+V5 and were substantially elevated again in 13136p−m+V20. Orotate levels were comparable in parent and 13136p−m+V5, with substantial accumulation only in 13136p−m+V20.
There were significant decreases in the polyamines putrescine (strain 13136p−m+V20) and spermidine (both strains). Polyamines play important roles in many cellular processes [91,92]. Joshi et al. [93] found polyamines to exhibit a strong antimutagenic effect on several strains of S. aureus. Lower levels of polyamines in VISA strains may facilitate the development of the VISA phenotype by increasing the frequency of mutations. Within the cell polyamines are often complexed with nucleic acids, where they are thought to serve a protective function against stress and possibly participate in the regulation of protein synthesis and DNA-binding protein interactions. Polyamines protect E. coli against oxidative stress and are a component of the acid stress response. On the cell surface polyamines have been shown to influence biofilm formation in Bacillus subtilis and a number of Gram-negative pathogens. Spermidine biosynthesis exerts a regulatory influence on Streptococcus pneumoniae autolytic activity, possibly through cell wall structural interactions [93,94].
Polyamine functions are generally poorly understood [94]. A complication to the assessment of this study’s polyamine data is the unique hypersensitivity of S. aureus to these molecules. It is questionable whether S. aureus is capable of synthesizing these polyamines [93]. Adenine phosphoribosyltransferase, encoded by the apt gene mutated in 13136p−m+V20 but not 13136p−m+V5, is involved in recycling polyamine biosynthesis byproducts in E. coli. Furthermore, in at least some Gram-positives, including S. aureus, the relA gene encoding an alarmone biosynthesis activity that mediates the stringent response is downstream of apt. Streptomyces coelicolor relA is transcribed from transcriptional read-through of apt, linking regulation of the stringent response and that of purine metabolism [31,34,35].
Glutamic acid is known to have roles in the osmoregulatory and stress physiology of S. aureus [89,95]. Alanine and glutamic acid are precursors of the l- and d-alanine, d-glutamic acid, and d-isoglutamine residues found in peptidoglycan. Their presence in increased concentrations in the VISA strains might be related to modifications in peptidoglycan structure and cell wall thickening in VISA strains [7,13,96,97,98]. However, levels of lysine, which is also found in S. aureus peptidoglycan, were decreased in the VISA strains. Possibly also related to peptidoglycan synthesis, glucosamine was undetectable in the parent strain but was present at 6.9/10mg dry weight in 13136p−m+V5 and 129.4/10mg dry weight in 13136p−m+V20. Peptidoglycan biosynthesis has been shown to be activated in some VISA strains [16], with an increased flow of glucose into the cell wall [74]. The levels of glycolytic pathway intermediates generally decreased sequentially from parent to 13136p−m+V5 and again from 13136p−m+V5 to 13136p−m+V20, suggesting an increasing reliance on the high flux of glucose through glycolysis as susceptibility to vancomycin increases during selection.
Mannitol is suspected of playing a role in osmoregulation in S. aureus [99]. Relative concentrations versus parent VSSA of mannitol and mannitol-6-phosphate decreased in 13136p−m+V5 (six- and 4.1-fold, respectively) and 13136p−m+V20 (5.5- and 2.6-fold, respectively). In the parent 13136p−m+, there was almost five-times the relative concentration of the unphosphorylated mannitol as in its VISA derivatives. The lower concentrations in the VISA can be explained by downregulated mtlA expression, which encodes a mannitol phosphotransferase system component, by 2.9- and 4.5-fold in 13136p−m+V5 and 13136p−m+V20, respectively. One reason for the dramatic proline accumulation in the VISA might have been to compensate for the unavailability of mannitol for service in osmoprotection.
Ideally, it would be possible to reveal elements of the VISA resistance mechanism by working backwards from phenotypic characteristics through metabolomic, then transcriptomic, and finally genomic data. However, interpretation of the omics data from this study presents several challenges. In-depth knowledge of the function of the gene products of many of the genes with SNPs is lacking, profoundly so for some of them (e.g., ssb1, dinG and stp1). A large proportion of gene expression changes identified by transcriptome analysis encode products in the hypothetical protein, unclassified, and unknown function gene functional groups (27% overall, rising to 40% of the high fold-change subset). This is not a novel observation [45]. As mentioned previously, metabolite relative concentration changes do not always correlate with the expression changes of genes encoding their biosynthetic enzymes and regulatory elements. This may be due in part to the fact that gene expression data do not provide information on gene product activity, and many enzymes and regulatory proteins engage in interactions with other regulatory molecules in ways that are not evident to broad analyses. For example, the alarmone synthetase and hydrolase activities are combined in a single, bifunctional protein in Gram-positive bacteria [45], and gene expression data on relA up- or down-regulation give no guidance on which activities are active at the time of sample processing. Furthermore, the single gene expression change in the mobile and extrachromosomal element functions gene functional group identified by the transcriptional analysis does not reflect the dominant position of the L54a prophage mutations within the genomic analysis.
4. Experimental Section
4.1. Strains and Growth Conditions
S. aureus strains 13136p−m+, 13136p−m+V5 and 13136p−m+V20 were studied [7]. They were grown in Tryptic Soy Broth (Becton Dickinson and Company, Cockeysville, MD, USA) at 37 °C with shaking at 200 rpm. Mid-exponential phase cells (OD600nm 0.5) were used for transcriptional profiling and metabolomics analysis.
4.2. Complete-Genome Comparisons
These were performed with an array-based service provided by Nimblegen Systems Inc. (Madison, WI, USA) [100,101]. The reference genome was strain 13136p−m+ to which the genomes of 13136p−m+V5 and 13136p−m+V20 were compared.
4.3. Transcriptional Profiling
This was carried out as described in previous publications from this laboratory [102,103]. S. aureus genome microarrays version 6.0, provided by the Pathogen Functional Genomics Resource Center of the National Institutes of Allergy and Infectious Diseases, were used.
4.4. Microarray Validation by Real-Time Reverse Transcription-PCR (RT-PCR)
This was carried out as described by Song et al. [103] on selected genes.
4.5. Metabolomic Analysis
This was carried out as described by Singh et al. [104]. Briefly, 500 mL of mid-exponential phase cells growing in a 2-liter flask with shaking (200 rpm) at 37 °C were harvested, washed and extracted with 40% (vol/vol) methanol. The extracts were analyzed by liquid chromatography/mass spectroscopy and gas chromatography/mass spectroscopy. Data from each chromatogram were normalized to an internal standard of 10 mg/mL hentriacontanoic acid to allow dry weight comparisons between samples. Relative concentrations were calculated as metabolite peak area divided by internal standard peak area. These analyses were carried out at the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, Urbana, IL, USA. Three separate batches of each strain were analyzed, and the data were combined.
5. Conclusions
This study added new members to the lists of mutations and transcriptomes that are associated with the VISA phenotype. Overall trends in VISA characteristics were consistent with previous studies, including a great variability in the mutations and gene expression patterns among VISA of different lineages. This variability has been noted previously by Howden et al. [3,4,105]. Those authors and Gardete et al. [106] also confirm that VISA transcriptomes are generally consistent with decreased virulence. Parent VSSA 13136p−m+ in this study had been laboratory-propagated for decades, although for much of that time it was in frozen storage, and its VISA derivatives were selected in vitro and have never been passaged through animals. The extensive modulation of virulence determinants in these and other in vitro-selected and -propagated VISA demonstrates that virulence and central intermediary metabolism are inextricably intertwined. Extended in vitro passage has been shown to change the expression pattern of virulence determinants, but it does not eliminate them [107,108].
The importance of activated elements of the stress response network was reinforced by the results from this study. Stress response modulation in the VISA phenotype was evident across all three omics analyses, from mutations and expression changes with genes known to respond to stresses or encode stress-related products, to modulation of stress-protection metabolites, such as proline and polyamines. It was surprising how many mutations were identified in the two VISA strains that were associated with stress related to DNA damage and the SOS response, especially so for the strain with the higher vancomycin MIC (13136p−m+V20). The pattern of downregulation of central metabolic processes related to protein synthesis and purine and pyrimidine biosynthetic pathways indicates a stringent response-like state for the two VISA, as has been reported for both MRSA and VISA [45,66]. Downregulation of these processes was not simply a component of the slowed growth rates of these VISA versus their VSSA parent, for while central metabolism in VISA 13136p−m+V20 was clearly less active than that in 13136p−m+V5, their doubling times are very similar [7].
The greatest value derived from the metabolomics analysis was in providing physiologic evidence supporting the association of stress response activation and stringent response characteristics with the VISA phenotype. This was evident in progressive changes in metabolite relative concentrations that paralleled the selection of greater vancomycin resistance between VSSA 13136p−m+ and 13136p−m+V5, and then between 13136p−m+V5 and 13136p−m+V20. Numbers of stress response-related mutations consistently changed in this fashion, as did broad expression patterns for genes associated with aspects of metabolism related to the stress response network and central intermediary metabolism. This pattern was also evident in metabolite levels of osmoprotectants, polyamines, and intermediates in nucleotide biosynthesis and central carbon and energy utilization pathways.
Supplementary Tables and Figures
- Supplementary Table S1. Two-fold or greater gene expression changes in VISA 13136p−m+V5 and 13136p−m+V20 versus parent VSSA 13136p−m+.
- Supplementary Table S2. Expression patterns by gene functional group in VISA 13136p−m+V5 and 13136p−m+V20 as number of genes upregulated and downregulated at least two-fold relative to gene expression in VSSA 13136p−m+.
- Supplementary Table S3. Concordance of expression patterns vs. parent VSSA 13136p−m+ between 13136p−m+V5 and 13136p−m+V20 for the 335 genes with expression data for both VISA.
- Supplementary Table S4. The 45 metabolites with at least one five-fold change among comparisons between 13136p−m+, 13136p−m+V5 and 13136p−m+V20.
- Supplementary Figure S1. Number of genes by functional group upregulated and downregulated at least eight-fold in VISA 13136p−m+V5 (A) and 13136p−m+V20 (B) relative to gene expression in VSSA parent 13136p−m+.
- Supplementary Figure S2. Metabolomic profiles by metabolite class (A–D).
- Supplementary File S1. Metabolomic analysis background information.
Acknowledgments
The DNA microarrays were obtained through National Institutes of Allergy and Infectious Diseases (NIAID) Pathogen Functional Genomics Resource Center, Division of Microbiology and Infectious Disease, NIAID, NIH. This work was funded by NIH Grant 1R15AI084006 to BJW. We are grateful to Zhong Li and Alexander Ulanov for the metabolomics results.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hiramatsu, K.; Aritaka, N.; Hanaki, H.; Kawasaki, S.; Hosoda, Y.; Hori, S.; Fukuchi, Y.; Kobayashi, I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997, 350, 1670–1673. [Google Scholar] [CrossRef] [PubMed]
- Pfeltz, R.F.; Wilkinson, B.J. The escalating challenge of vancomycin resistance in Staphylococcus aureus. Curr. Drug Targets Infect. Disord. 2004, 4, 273–294. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Peleg, A.Y.; Stinear, T.P. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect. Genet. Evol. 2013, 13, S1567–S1348. [Google Scholar] [PubMed]
- Utaida, S.; Dunman, P.M.; Macapagal, D.; Murphy, E.; Projan, S.J.; Singh, V.K.; Jayaswal, R.K.; Wilkinson, B.J. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 2003, 149, 2719–2732. [Google Scholar] [CrossRef] [PubMed]
- McAleese, F.; Wu, S.W.; Sieradzki, K.; Dunman, P.; Murphy, E.; Projan, S.; Tomasz, A. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus Exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 2006, 188, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Pfeltz, R.F.; Singh, V.K.; Schmidt, J.L.; Batten, M.A.; Baranyk, C.S.; Nadakavukaren, M.J.; Jayaswal, R.K.; Wilkinson, B.J. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 2000, 44, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Jevons, M.P. “Celbenin”—Resistant Staphylococci. Br. Med. J. 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Dyke, K.G. Penicillinase production and intrinsic resistance to penicillins in methicillin-resistant cultures of Staphylococcus aureus. J. Med. Microbiol. 1969, 3, 261–278. [Google Scholar] [CrossRef]
- Lee, C.Y.; Iandolo, J.J. Integration of staphylococcal phage L54a occurs by site-specific recombination: Structural analysis of the attachment sites. Proc. Natl. Acad. Sci. USA 1986, 83, 5474–5478. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; Deboy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.T.; Jevons, M.P. A survey of methicillin resistance in Staphylococcus. Postgrad. Med. J. 1964, 40 (Suppl), 170–178. [Google Scholar] [CrossRef] [PubMed]
- Koehl, J.L.; Muthaiyan, A.; Jayaswal, R.K.; Ehlert, K.; Labischinski, H.; Wilkinson, B.J. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 3749–3757. [Google Scholar] [CrossRef] [PubMed]
- Oshida, T.; Sugai, M.; Komatsuzawa, H.; Hong, Y.M.; Suginaka, H.; Tomasz, A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain. Cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 1995, 92, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant. Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Hanaki, H.; Labischinski, H.; Inaba, Y.; Kondo, N.; Murakami, H.; Hiramatsu, K. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 1998, 42, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.L.; Rice, K.C.; Slater, S.R.; Fox, P.M.; Archer, G.L.; Bayles, K.W.; Fey, P.D.; Kreiswirth, B.N.; Somerville, G.A. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: Implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 2007, 51, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Alexander, E.L.; Gardete, S.; Bar, H.Y.; Wells, M.T.; Tomasz, A.; Rhee, K.Y. Intermediate-type vancomycin resistance (VISA) in genetically-distinct Staphylococcus aureus isolates is linked to specific, reversible metabolic alterations. PLOS ONE 2014, 9, e97137. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, K.; Stephenson, S.; Perego, M. Overexpression of the PepF oligopeptidase inhibits sporulation initiation in Bacillus subtilis. J. Bacteriol. 2002, 184, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.F.; Goss, L.; Dworkin, J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 192–212. [Google Scholar]
- Ogura, M.; Tanaka, T. Bacillus subtilis comZ (yjzA) negatively affects expression of comG but not comK. J. Bacteriol. 2000, 182, 4992–4994. [Google Scholar] [CrossRef] [PubMed]
- Suematsu, T.; Yokobori, S.; Morita, H.; Yoshinari, S.; Ueda, T.; Kita, K.; Takeuchi, N.; Watanabe, Y. A bacterial elongation factor G homologue exclusively functions in ribosome recycling in the spirochaete Borrelia burgdorferi. Mol. Microbiol. 2010, 75, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Tsai, T.Y.; Pan, T.M. Physiological response and protein expression under acid stress of Escherichia coli O157:H7 TWC01 isolated from Taiwan. J. Agric. Food Chem. 2007, 5, 7182–7191. [Google Scholar] [CrossRef]
- Kapralou, S.; Fabbretti, A.; Garulli, C.; Spurio, R.; Gualerzi, C.O.; Dahlberg, A.E.; Pon, C.L. Translation initiation factor IF1 of Bacillus stearothermophilus and Thermus thermophilus substitute for Escherichia coli IF1 in vivo and in vitro without a direct IF1-IF2 interaction. Mol. Microbiol. 2008, 70, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 2006, 188, 1648–1659. [Google Scholar] [CrossRef] [PubMed]
- Maul, R.W.; Sanders, L.H.; Lim, J.B.; Benitez, R.; Sutton, M.D. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J. Bacteriol. 2007, 189, 4688–4695. [Google Scholar] [CrossRef] [PubMed]
- Sobral, R.G.; Jones, A.E.; Des Etages, S.G.; Dougherty, T.J.; Peitzsch, R.M.; Gaasterland, T.; Ludovice, A.M.; de Lencastre, H.; Tomasz, A. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 2007, 189, 2376–2391. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.H.; Lee, C.Y. Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J. Bacteriol. 1989, 171, 4146–4153. [Google Scholar] [PubMed]
- Campbell, J.; Singh, A.K.; Santa Maria, J.P., Jr.; Kim, Y.; Brown, S.; Swoboda, J.G.; Mylonakis, E.; Wilkinson, B.J.; Walker, S. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 2011, 6, 106–116. [Google Scholar] [CrossRef] [PubMed]
- McRobbie, A.M.; Meyer, B.; Rouillon, C.; Petrovic-Stojanovska, B.; Liu, H.; White, M.F. Staphylococcus aureus DinG, a helicase that has evolved into a nuclease. Biochem. J. 2012, 442, 77–84. [Google Scholar] [PubMed]
- Singh, V.; Evans, G.B.; Lenz, D.H.; Mason, J.M.; Clinch, K.; Mee, S.; Painter, G.F.; Tyler, P.C.; Furneaux, R.H.; Lee, J.E.; et al. Femtomolar transition state analogue inhibitors of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J. Biol. Chem. 2005, 280, 18265–18273. [Google Scholar] [CrossRef] [PubMed]
- Kappes, R.M.; Kempf, B.; Kneip, S.; Boch, J.; Gade, J.; Meier-Wagner, J.; Bremer, E. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 1999, 32, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Wehmeier, L.; Schäfer, A.; Burkovski, A.; Krämer, R.; Mechold, U.; Malke, H.; Pühler, A.; Kalinowski, J. The role of the Corynebacterium glutamicum rel gene in (p)ppGpp metabolism. Microbiology 1998, 144, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Wendrich, T.M.; Marahiel, M.A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 1997, 26, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Chakraburtty, R.; White, J.; Takano, E.; Bibb, M. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2). Mol. Microbiol. 1996, 19, 357–368. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Lindsay, J.A. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: Implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010, 10, e173. [Google Scholar] [CrossRef]
- Ythier, M.; Resch, G.; Waridel, P.; Panchaud, A.; Gfeller, A.; Majcherczyk, P.; Quadroni, M.; Moreillon, P. Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: Correlation with agr genotypes and adherence phenotypes. Mol. Cell Proteomics 2012, 11, 1123–1139. [Google Scholar] [CrossRef] [PubMed]
- Komatsuzawa, H.; Ohta, K.; Sugai, M.; Fujiwara, T.; Glanzmann, P.; Berger-Bachi, B.; Suginaka, H. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Ohlsen, K.; Donat, S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Beltramini, A.M.; Mukhopadhyay, C.D.; Pancholi, V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 2009, 77, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Renzoni, A.; Andrey, D.O.; Jousselin, A.; Barras, C.; Monod, A.; Vaudaux, P.; Lew, D.; Kelley, W.L. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLOS ONE 2011, 6, e21577. [Google Scholar] [CrossRef] [PubMed]
- Passalacqua, K.D.; Satola, S.W.; Crispell, E.K.; Read, T.D. A mutation in the PP2C phosphatase gene in a Staphylococcus aureus USA300 clinical isolate with reduced susceptibility to vancomycin and daptomycin. Antimicrob. Agents Chemother. 2012, 13, 5212–5223. [Google Scholar] [CrossRef]
- Cameron, D.R.; Ward, D.V.; Kostoulias, X.; Howden, B.P.; Moellering, R.C.J.; Eliopoulos, G.; Peleg, A.Y. The serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J. Infect. Dis. 2012, 205, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Hatzios, S.K.; Baer, C.E.; Rustad, T.R.; Siegrist, M.S.; Pang, J.M.; Ortega, C.; Alber, T.; Grundner, C.; Sherman, D.R.; Bertozzi, C.R. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. USA 2013, 110, E5069–E5077. [Google Scholar] [CrossRef] [PubMed]
- Reiss, S.; Pané-Farré, J.; Fuchs, S.; François, P.; Liebeke, M.; Schrenzel, J.; Lindequist, U.; Lalk, M.; Wolz, C.; Hecker, M.; et al. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 2012, 56, 787–804. [Google Scholar] [CrossRef] [PubMed]
- Miyafusa, T.; Caaveiro, J.M.; Tanaka, Y.; Tsumoto, K. Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains. Biochem. J. 2012, 443, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Lamamy, C.; Berthelot, A.; Bertrand, X.; Valentin, A.S.; Dos Santos, S.; Thiais, S.; Morange, V.; Girard, N.; Donnio, P.Y.; Quentin, R.; et al. Bloodstream Infection Study Group of the Réseau des Hygiénistes du Centre. CC9 livestock-associated Staphylococcus aureus emerges in bloodstream infections in French patients unconnected with animal farming. Clin. Infect. Dis. 2013, 56, e83–e86. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.; Månsson, M.; Bojer, M.S.; Gram, L.; Larsen, T.O.; Novick, R.P.; Frees, D.; Frøkiær, H.; Ingmer, H. Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLOS ONE 2014, 9, e84992. [Google Scholar] [CrossRef] [PubMed]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Shrihari, R.Y.; Singh, N.P. Multiplex reverse transcription polymerase chain reaction to study the expression of virulence and stress response genes in Staphylococcus aureus. J. Food Sci. 2012, 77, M95–M101. [Google Scholar] [CrossRef] [PubMed]
- Banbula, A.; Potempa, J.; Travis, J.; Fernandez-Catalán, C.; Mann, K.; Huber, R.; Bode, W.; Medrano, F. Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution. Structure 1998, 6, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Arêde, P.; Botelho, T.; Guevara, T.; Usón, I.; Oliveira, D.C.; Gomis-Rüth, F.X. Structure-function studies of the staphylococcal methicillin resistance antirepressor MecR2. J. Biol. Chem. 2013, 288, 21267–21278. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.; Zhao, C.; Xiao, D.; Zhang, J.; Zhang, F.; Chen, M.; Wang, H. Comparative proteomics-based identification of genes associated with glycopeptide resistance in clinically derived heterogeneous vancomycin-intermediate Staphylococcus aureus strains. PLOS ONE 2013, 8, e66880. [Google Scholar] [CrossRef] [PubMed]
- Haemig, H.A.; Moen, P.J.; Brooker, R.J. Evidence that highly conserved residues of transmembrane segment 6 of Escherichia coli MntH are important for transport activity. Biochemistry 2010, 49, 4662–4671. [Google Scholar] [CrossRef] [PubMed]
- Mongodin, E.; Finan, J.; Climo, M.W.; Rosato, A.; Gill, S.; Archer, G.L. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 2003, 185, 4638–4643. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Hall, J.W.; Yang, J.; Ji, Y. The essential yhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus. PLOS ONE 2012, 7, e50608. [Google Scholar] [CrossRef] [PubMed]
- Napier, B.A.; Meyer, L.; Bina, J.E.; Miller, M.A.; Sjöstedt, A.; Weiss, D.S. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc. Natl. Acad. Sci. USA 2012, 109, 18084–18089. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Gantert, C.; Voss, M.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Studies on the mechanism of ring hydrolysis in phenylacetate degradation: A metabolic branching point. J. Biol. Chem. 2011, 286, 11021–11034. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bachhawat, A.K. Pyroglutamic acid: Throwing light on a lightly studied metabolite. Curr. Sci. 2012, 102, 288–297. [Google Scholar]
- Ledala, N.; Zhang, B.; Seravalli, J.; Powers, R.; Somerville, G.A. The influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. J. Bacteriol. 2014, 196, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Liebeke, M.; Meyer, H.; Donat, S.; Ohlsen, K.; Lalk, M. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: involvement in cell wall biosynthesis. Chem. Biol. 2010, 17, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Nandakumar, R.; Sadykov, M.R.; Madayiputhiya, N.; Luong, T.T.; Gaupp, R.; Lee, C.Y. RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 2011, 193, 6187–6196. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane-Khadka, R.; Cantore, S.A.; Riordan, J.T.; Delgado, A.; Norman, A.E.; Dueñas, S.; Zaman, S.; Horan, S.; Wilkinson, B.J.; Gustafson, J.E. sarA inactivation reduces vancomycin-intermediate and ciprofloxacin resistance expression by Staphylococcus aureus. Int. J. Antimicrob. Agents 2009, 34, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Lithgow, J.K.; Ingham, E.; Foster, S.J. Role of the hprT-ftsH locus in Staphylococcus aureus. Microbiology 2004, 150, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Hishinuma, T.; Katayama, Y.; Cui, L.; Kapi, M.; Hiramatsu, K. Mutation of RNApolymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob. Agents Chemother. 2011, 55, 4188–4195. [Google Scholar] [CrossRef] [PubMed]
- Keaton, M.A.; Rosato, R.R.; Plata, K.B.; Singh, C.R.; Rosato, A.E. Exposure of clinical MRSA heterogeneous strains to β-lactams redirects metabolism to optimize energy production through the TCA cycle. PLOS ONE 2013, 8, e71025. [Google Scholar] [CrossRef] [PubMed]
- Dordel, J.; Kim, C.; Chung, M.; Pardos de la Gándara, M.; Holden, M.T.; Parkhill, J.; de Lencastre, H.; Bentley, S.D.; Tomasz, A. Novel determinants of antibiotic resistance: Identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Mwangi, M.; Chung, M.; Milheiriço, C.; de Lencastre, H.; Tomasz, A. The mechanism of heterogeneous beta-lactam resistance in MRSA: Key role of the stringent stress response. PLOS ONE 2013, 8, e82814. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp Conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Moreira, B.; Boyle-Vavra, S.; deJonge, B.L.; Daum, R.S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1997, 41, 1788–1793. [Google Scholar] [PubMed]
- Delaune, A.; Poupel, O.; Mallet, A.; Coic, Y.M.; Msadek, T.; Dubrac, S. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLOS ONE 2011, 6, e17054. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Kuroda, H.; Oshima, T.; Takeuchi, F.; Mori, H.; Hiramatsu, K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 2003, 49, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lian, J.Q.; Neoh, H.M.; Reyes, E.; Hiramatsu, K. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 3404–3413. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Cui, L.; Kim, J.; Hiramatsu, K. Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains derived from hVISA clinical strain Mu3. Antimicrob. Agents Chemother. 2013, 12, 5843–5853. [Google Scholar] [CrossRef]
- Ye, Z.H.; Buranen, S.L.; Lee, C.Y. Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J. Bacteriol. 1990, 172, 2568–2575. [Google Scholar] [PubMed]
- Novick, R.P.; Christie, G.E.; Penadés, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Banerjee, R.; Pani, B.; Sen, U.; Sen, R. The moonlighting function of bacteriophage P4 capsid protein, Psu, as a transcription antiterminator. Bacteriophage 2013, 3, e25657. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Davis, E.; Brown, D.; Campbell, E.A.; Wigneshweraraj, S.; Darst, S.A. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of σ70 domain 1.1. Proc. Natl. Acad. Sci. USA 2013, 110, 19772–19777. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, J.E.; Philipson, L. Role of the phi 11 phage genome in competence of Staphylococcus aureus. J. Bacteriol. 1974, 119, 19–32. [Google Scholar] [PubMed]
- Kuroda, H.; Kuroda, M.; Cui, L.; Hiramatsu, K. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 2007, 268, 98–105. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, J.O.; Langevin, M.J.; Price, C.T.; Blevins, J.S.; Smeltzer, M.S.; Gustafson, J.E. Effects of sarA inactivation on the intrinsic multidrug resistance mechanism of Staphylococcus aureus. FEMS Microbiol. Lett. 2004, 237, 297–302. [Google Scholar] [PubMed]
- Tang, J.; Chen, J.; Li, H.; Zeng, P.; Li, J. Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among Staphylococcus aureus strains isolated from different sources. Foodborne Pathog. Dis. 2013, 10, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Laarman, A.J.; Ruyken, M.; Malone, C.L.; van Strijp, J.A.; Horswill, A.R.; Rooijakkers, S.H. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 2011, 186, 6445–6453. [Google Scholar] [CrossRef] [PubMed]
- Ingavale, S.S.; van Wamel, W.; Cheung, A.L. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Komatsuzawa, H.; Ohta, K.; Fujiwara, T.; Choi, G.H.; Labischinski, H.; Sugai, M. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2001, 203, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Koprivnjak, T.; Mlakar, V.; Swanson, L.; Fournier, B.; Peschel, A.; Weiss, J.P. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 2006, 188, 3622–3630. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.E.; Wilkinson, B.J. Proline transport in Staphylococcus aureus: A high affinity system and a low affinity system involved in osmoregulation. J. Bacteriol. 1992, 174, 2702–2710. [Google Scholar] [PubMed]
- Li, C.; Sun, F.; Cho, H.; Yelavarthi, V.; Sohn, C.; He, C.; Schneewind, O.; Bae, T. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 2010, 192, 3883–3892. [Google Scholar] [CrossRef] [PubMed]
- Bouchereau, A.; Aziz, A.; Larher, F.; Martin-Tanguy, J. Polyamines and environmental challenges: Recent developments. Plant Sci. 1999, 140, 103–125. [Google Scholar] [CrossRef]
- Thomas, T.; Thomas, T.J. Polyamine in cell growth and cell death: Molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.S.; Spontak, J.S.; Klapper, D.G.; Richardson, A.R. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 2011, 82, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Potter, A.J.; Paton, J.C. Spermidine biosynthesis and transport modulates pneumococcal autolysis. J. Bacteriol. 2014. [Google Scholar] [CrossRef]
- Graham, J.E.; Wilkinson, B.J. Staphylococcus aureus osmoregulation: Roles for choline, glycine betaine, proline, and taurine. J. Bacteriol. 1992, 174, 2711–2716. [Google Scholar] [PubMed]
- Hiramatsu, K. Vancomycin-resistant Staphylococcus aureus: A new model of antibiotic resistance. Lancet Infect. Dis. 2001, 1, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Sieradzki, K.; Tomasz, A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 1997, 179, 2557–2566. [Google Scholar] [PubMed]
- Cui, L.; Murakami, H.; Kuwahara-Arai, K.; Hanaki, H.; Hiramatsu, K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 2000, 44, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of oxygen on glucose metabolism: Utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLOS ONE 2013, 8, e58277. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Albert, T.J.; Dailidiene, D.; Dailide, G.; Norton, J.E.; Kalia, A.; Richmond, T.A.; Molla, M.; Singh, J.; Green, R.D.; Berg, D.E. Mutation discovery in bacterial genomes: Metronidazole resistance in Helicobacter pylori. Nat. Meth. 2005, 2, 951–953. [Google Scholar] [CrossRef]
- Muthaiyan, A.; Silverman, J.A.; Jayaswal, R.K.; Wilkinson, B.J. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 2008, 52, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Rubio, A.; Jayaswal, R.K.; Silverman, J.A.; Wilkinson, B.J. Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLOS ONE 2013, 8, e58469. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Ulanov, A.V.; Li, Z.; Jayaswal, R.K.; Wilkinson, B.J. Metabolomes of the psychrotolerant bacterium Listeria monocytogenes 10403S grown at 37 °C and 8 °C. Int. J. Food Microbiol. 2011, 148, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; McEvoy, C.E.; Allen, D.L.; Chua, K.; Gao, W.; Harrison, P.F.; Bell, J.; Coombs, G.; Wood, V.B.; Porter, J.L.; et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLOS Pathog. 2011, 7, e1002359. [Google Scholar] [CrossRef] [PubMed]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLOS Pathog. 2012, 13, e1002505. [Google Scholar] [CrossRef]
- Pandya, U.; Sinha, M.; Luxon, B.A.; Watson, D.A.; Niesel, D.W. Global transcription profiling and virulence potential of Streptococcus pneumoniae after serial passage. Gene 2009, 443, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Somerville, G.A.; Beres, S.B.; Fitzgerald, J.R.; DeLeo, F.R.; Cole, R.L.; Hoff, J.S.; Musser, J.M. In vitro serial passage of Staphylococcus aureus: Changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 2002, 184, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).