Old and New Glycopeptide Antibiotics: Action and Resistance
Abstract
:1. Natural Glycopeptide Antibiotics
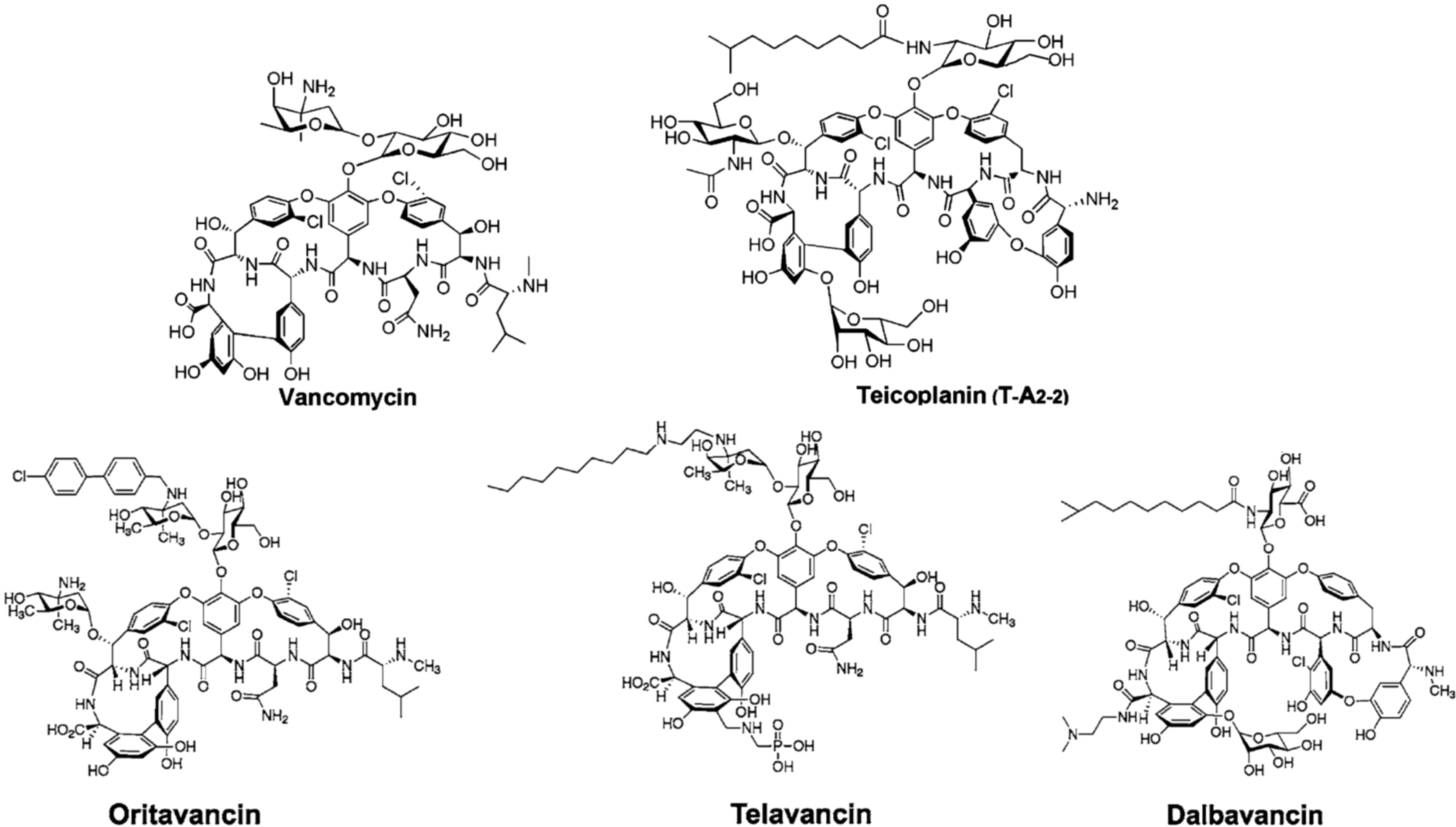
2. Semi-Synthetic Glycopeptide Antibiotics
| Drug | GPA Precursor | Microbiological Spectrum | Main Clinical Indication | Status |
|---|---|---|---|---|
| Oritavancin (Orbactiv) | Chloroeremomycin | MRSA 1, VRSA 1, VRE 1 | ABSSSI 2 | approved by FDA 3 in 2014 |
| Telavancin (Vibativ) | Vancomycin | MRSA 1, MSSA 1, VSE 1, Streptococcus pyogenes | cSSSI 2 | approved by FDA 3 in 2009 |
| Staphylococcus aureus | HABP/VABP 2 | approved by FDA 3 in 2013 | ||
| Dalbavancin (Dalvance) | A40926 | MRSA 1, MSSA 1, Streptococcus pyogenes | ABSSSI 2 | approved by FDA 3 in 2014 |
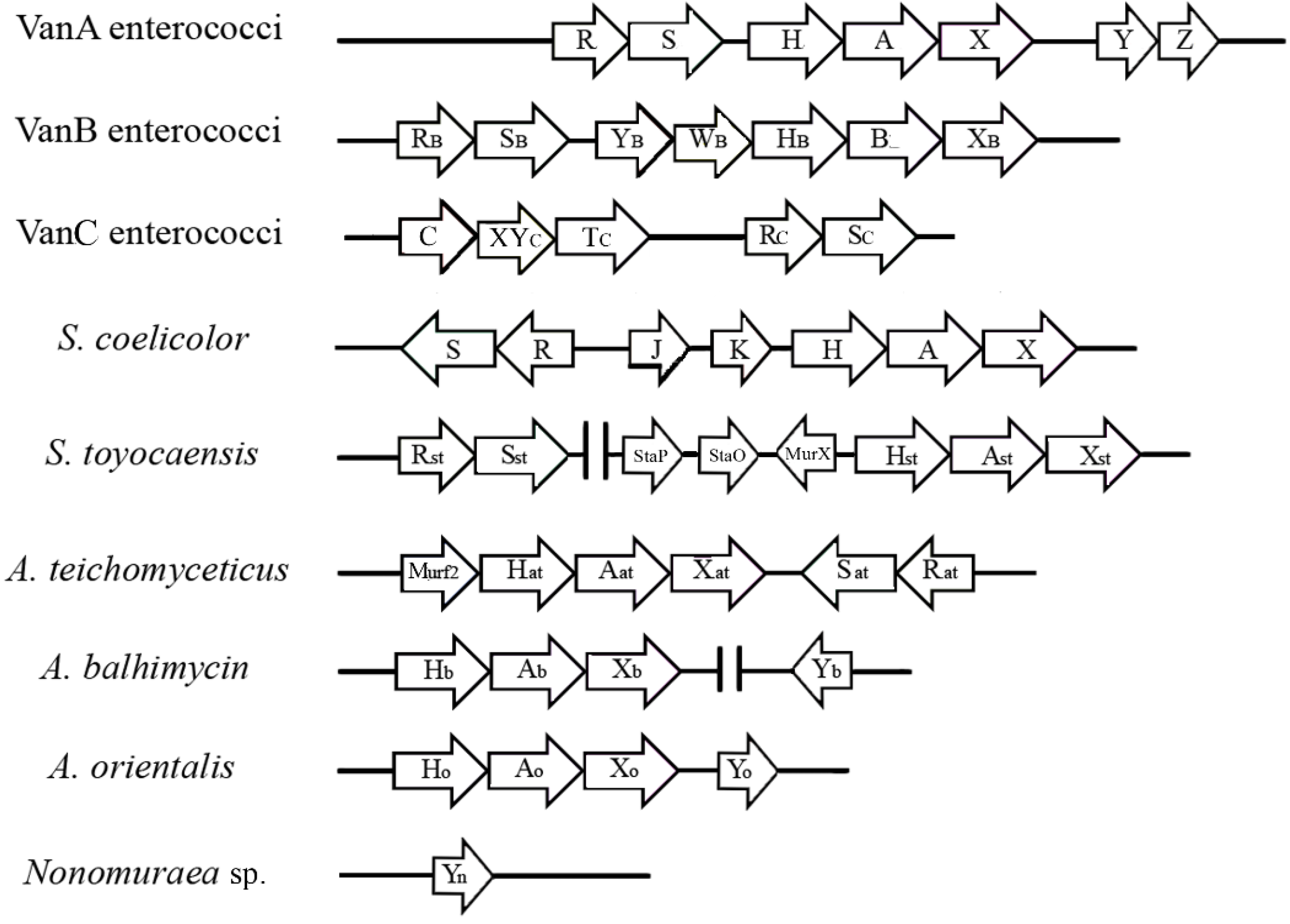
3. The van Gene Clusters in Pathogens
| Microorganisms | GPA Resistance Phenotype | Level of Resistance | MIC (mg/L) of GPAs | Location of van Genes | Transcription of van Genes | C-terminal of Modified PG Target | References |
|---|---|---|---|---|---|---|---|
| E. faecalis E. faecium | VanA | High | Vancomycin 64–100 Teicoplanin 16–512 | Plasmid Chromosome | Inducible | d-Ala-d-Lac | [44,45,46] |
| E. faecalis E. faecium | VanB | Variable | Vancomycin 4–1000 Teicoplanin 0.5–1 | Plasmid Chromosome | Inducible | d-Ala-d-Lac | [33,36,44] |
| E. gallinarum E. casseliflavus E. flavescens | VanC | Intrinsic resistance, low level | Vancomycin 2–32 Teicoplanin 0.5–1 | Chromosome | Constitutive | d-Ala-d-Ser | [33,49,54] |
| E. faecalis E. faecium | VanD | Moderate | Vancomycin 64–128 Teicoplanin 4–64 | Chromosome | Constitutive | d-Ala-d-Lac | [33,53] |
| E. faecalis | VanE | Low | Vancomycin 8–32 Teicoplanin 0.5 | Chromosome | Inducible | d-Ala-d-Ser | [33] |
| E. faecalis E. faecium | VanG | Low | Vancomycin 16 Teicoplanin 0.5 | Chromosome | Inducible | d-Ala-d-Ser | [33] |
| E. faecalis | VanL | Low | Vancomycin 8 Teicoplanin susceptible | Chromosome | Inducible | d-Ala-d-Ser | [54] |
| E. faecium | VanM | Variable | Vancomycin > 256 Teicoplanin 0.75 | Plasmid Chromosome | Inducible | d-Ala-d-Lac | [56] |
| E. faecium | VanN | Low | Vancomycin 16 Teicoplanin 0.5 | Plasmid | Constitutive | d-Ala-d-Ser | [55,57] |
4. The van Gene Clusters in the Producing Actinomycetes
| Microorganisms | Produced GPA | MIC (mg/L) of GPAs | Location of vanHAX Genes | Transcription of vanHAX Genes | C-terminal 1 of PG Target in Absence of Inducer | C-terminal 1 of PG Target in Presence of Inducer | References |
|---|---|---|---|---|---|---|---|
| Streptomyces coelicolor | none | Vancomycin >100 Teicoplanin <0.5 | Chromosome | Inducible by vancomycin | d-Ala-d-Ala | d-Ala-d-Lac | [47,78,79,80] |
| Streptomyces toyocaensis | A47934 | Vancomycin <0.25 Teicoplanin <0.25 A47934 >5 | Chromosome, A47934 Cluster | Inducible by A47934 | d-Ala-d-Ala | d-Ala-d-Lac | [47,68,72,74] |
| Actinoplanes teichomyceticus | Teicoplanin | Vancomycin 90 Teicoplanin 25 | Chromosome, tei Cluster | Constitutive | d-Ala-d-Lac | d-Ala-d-Lac | [69,70,75,76] |
| Amycolatopsis balhimycin | Balhimycin | Vancomycin n.r.2 Teicoplanin n.r.2 Balhimycin >100 | Chromosome, out of bal Cluster | Constitutive | d-Ala-d-Lac | d-Ala-d-Lac | [71] |
| Nonomuraea sp. ATCC 39727 | A40926 | Vancomycin 30 Teicoplanin 0.9 A40926 4 | n.d. 3 | Inducible by A40926 | d-Ala 4 | d-Ala 4 | [81,82] |
5. The Model System Streptomyces Coelicolor
6. The Case of Nonomuraea sp. ATCC 39727
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rossolini, G.M.; Arena, F.; Pollini, S. Novel infectious diseases and emerging gram-positive multiresistant pathogens in hospital and community acquired infections. In Antimicrobials; Marinelli, F., Genilloud, O., Eds.; Springer Verlag: Berlin Heidelberg, Germany, 2014. [Google Scholar]
- Yim, G.; Thaker, M.N.; Koteva, K.; Wright, G. Glycopeptide antibiotic biosynthesis. J. Antibiot. (Tokyo) 2014, 67, 31–41. [Google Scholar] [CrossRef]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Jovetic, S.; Zhu, Y.; Marcone, G.L.; Marinelli, F.; Tramper, J. β-Lactam and glycopeptide antibiotics: First and last line of defense? Trends Biotechnol. 2010, 28, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Marinelli, F. Glycopeptides: An old but up to date successful antibiotic class. In Antimicrobials; Marinelli, F., Genilloud, O., Eds.; Springer Verlag: Berlin Heidelberg, Germany, 2014. [Google Scholar]
- James, R.C.; Pierce, J.G.; Okano, A.; Xie, J.; Boger, D.L. Redesign of glycopeptide antibiotics: Back to the future. ACS Chem. Biol. 2012, 7, 797–804. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Boddy, C.N.; Bräse, S.; Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. Int. Ed. Engl. 1999, 38, 2096–2152. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Wright, G.D. Opportunities for synthetic biology in antibiotics: Expanding glycopeptide chemical diversity. ACS Synth. Biol. 2012. [Google Scholar] [CrossRef]
- Kalan, L.; Perry, J.; Koteva, K.; Thaker, M.; Wright, G. Glycopeptide sulfation evades resistance. J. Bacteriol. 2013, 195, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Brady, S.F. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. USA 2008, 105, 17273–17277. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Brady, S.F. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 2010, 13, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Reddy, B.V.; Ternei, M.A.; Charlop-Powers, Z.; Calle, P.Y.; Kim, J.H.; Brady, S.F. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 2013, 110, 11797–11802. [Google Scholar] [CrossRef] [PubMed]
- Borghi, A.; Edwards, D.; Zerilli, L.F.; Lancini, G.C. Factors affecting the normal and branched-chain acyl moieties of teicoplanin components produced by Actinoplanes teichomyceticus. J. Gen. Microbiol. 1991, 137, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Taurino, C.; Frattini, L.; Marcone, G.L.; Gastaldo, L.; Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 2011, 10, e82. [Google Scholar] [CrossRef]
- Cooper, M.A.; Williams, D.H. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem. Biol. 1999, 6, 891–899. [Google Scholar] [CrossRef]
- Van Bambeke, F. Glycopeptides and glycodepsipeptides in clinical development: A comparative review of their antibacterial spectrum, pharmacokinetics and clinical efficacy. Curr. Opin. Investig. Drugs 2006, 7, 740–749. [Google Scholar]
- Jeya, M.; Moon, H.J.; Lee, K.M.; Kim, I.W.; Lee, J.K. Glycopeptide antibiotics and their novel semi-synthetic derivatives. Curr. Pharm. Biotechnol. 2011, 12, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Cynamon, M.H.; Granato, P.A. Comparison of the in vitro activities of teichomycin A2 and vancomycin against staphylococci and enterococci. Antimicrob. Agents Chemother. 1982, 21, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Mackay, J.P.; Gerhard, U.; Beauregard, D.A.; Westwell, M.S.; Searle, M.S.; Williams, D.H. Glycopeptide antibiotic activity and the possible role of dimerization: A model for biological signaling. J. Am. Chem. Soc. 1994, 116, 4581–4590. [Google Scholar] [CrossRef]
- Ashford, P.A.; Bew, S.P. Recent advances in the synthesis of new glycopeptide antibiotics. Chem. Soc. Rev. 2012, 41, 957–978. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Calic, D.; Schweizer, F.; Zelenitsky, S.; Adam, H.; Lagacé-Wiens, P.R.; Rubinstein, E.; Gin, A.S.; Hoban, D.J.; Karlowsky, J.A. New lipoglycopeptides: A comparative review of dalbavancin, oritavancin and telavancin. Drugs 2010, 70, 859–886. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.L.; Chang, R.; Debabov, D.V.; Leung, J.; Wu, T.; Krause, K.M.; Sandvik, E.; Hubbard, J.M.; Kaniga, K.; Schmidt, D.E.; et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 1127–1134. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Available online: http://www.fda.gov/ (accessed on 23 October 2014).
- Bouza, E.; Burillo, A. Oritavancin: A novel lipoglycopeptide active against gram-positive pathogens including multiresistant strains. Int. J. Antimicrob. Agents 2010, 36, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Belley, A.; McKay, G.A.; Arhin, F.F.; Sarmiento, I.; Beaulieu, S.; Fadhil, I.; Parr, T.R.; Moeck, G. Oritavancin disrupts membrane integrity of Staphylococcus aureus and vancomycin-resistant enterococci to effect rapid bacterial killing. Antimicrob. Agents Chemother. 2010, 54, 5369–5371. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.P.; Selva, E.; Gastaldo, L.; Berti, M.; Pallanza, R.; Ripamonti, F.; Ferrari, P.; Denaro, M.; Arioli, V.; Cassani, G. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 1987, 31, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, A.; Ciabatti, R. Glycopeptide derivatives. Curr. Med. Chem. 2001, 8, 1759–1773. [Google Scholar] [CrossRef]
- Malabarba, A.; Goldstein, B.P. Origin, structure, and activity in vitro and in vivo of dalbavancin. J. Antimicrob. Chemother. 2005, 55, 15–20. [Google Scholar]
- Billeter, M.; Zervos, M.J.; Chen, A.Y.; Dalovisio, J.R.; Kurukularatne, C. Dalbavancin: A novel once-weekly lipoglycopeptide antibiotic. Clin. Infect. Dis. 2008, 46, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Treviño, J.; Bayón, C.; Ardá, A.; Marinelli, F.; Gandolfi, R.; Molinari, F.; Jimenez-Barbero, J.; Hernáiz, M.J. New insights into glycopeptide antibiotic binding to cell wall precursors using SPR and NMR spectroscopy. Chemistry 2014, 20, 7363–7372. [Google Scholar] [CrossRef] [PubMed]
- Derlot, R.; Duval, E.J.; Courvalin, P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 1988, 319, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Courvalin, P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006, 42, S25–S34. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.; Wright, G.D.; Dutka-Malen, S.; Arthur, M.; Courvalin, P.; Walsh, C.T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: Biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 1991, 30, 10408–10415. [Google Scholar] [CrossRef] [PubMed]
- Lessard, I.A.; Healy, V.L.; Park, I.S.; Walsh, C.T. Determinants for differential effects on d-ala-d-lactate vs. d-ala-d-ala formation by the VanA ligase from vancomycin-resistant enterococci. Biochemistry 1999, 38, 14006–14022. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Reynolds, P.; Courvalin, P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996, 4, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Molinas, C.; Bugg, T.D.; Wright, G.D.; Walsh, C.T.; Courvalin, P. Evidence for in vivo incorporation of d-lactate into peptidoglycan precursors of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 1992, 36, 867–869. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.E.; Depardieu, F.; Dutka-Malen, S.; Arthur, M.; Courvalin, P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 1994, 13, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wright, G.D.; Walsh, C.T. Overexpression, purification, and characterization of VanX, a d,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 1995, 34, 2455–2463. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Depardieu, F.; Molinas, C.; Reynolds, P.; Courvalin, P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 1995, 154, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Depardieu, F.; Cabanié, L.; Reynolds, P.; Courvalin, P. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 1998, 30, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Depardieu, F.; Gerbaud, G.; Galimand, M.; Leclercq, R.; Courvalin, P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 1997, 179, 97–106. [Google Scholar] [PubMed]
- Arthur, M.; Depardieu, F.; Courvalin, P. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 1999, 145, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Quintiliani, R. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 2001, 45, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D.; Holman, T.R.; Walsh, C.T. Purification and characterization of VanR and the cytosolic domain of VanS: A two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 1993, 32, 5057–5063. [Google Scholar] [CrossRef] [PubMed]
- Healy, V.L.; Lessard, I.A.; Roper, D.I.; Knox, J.R.; Walsh, C.T. Vancomycin resistance in enterococci: Reprogramming of the d-ala-d-ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 2000, 7, R109–R119. [Google Scholar] [CrossRef] [PubMed]
- Koteva, K.; Hong, H.J.; Wang, X.D.; Nazi, I.; Hughes, D.; Naldrett, M.J.; Buttner, M.J.; Wright, G.D. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 2010, 6, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.J.; Hutchings, M.I.; Buttner, M.J.; Biotechnology and Biological Sciences Research Council, U.K. Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 2008, 631, 200–213. [Google Scholar] [PubMed]
- Depardieu, F.; Podglajen, I.; Leclercq, R.; Collatz, E.; Courvalin, P. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 2007, 20, 79–114. [Google Scholar] [CrossRef] [PubMed]
- Billot-Klein, D.; Blanot, D.; Gutmann, L.; van Heijenoort, J. Association constants for the binding of vancomycin and teicoplanin to N-acetyl-d-alanyl-d-alanine and N-acetyl-d-alanyl-d-serine. Biochem. J. 1994, 304, 1021–1022. [Google Scholar] [PubMed]
- Reynolds, P.E.; Courvalin, P. Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob. Agents Chemother. 2005, 49, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Baptista, M.; Depardieu, F.; Reynolds, P.; Courvalin, P.; Arthur, M. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 1997, 25, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Depardieu, F.; Kolbert, M.; Pruul, H.; Bell, J.; Courvalin, P. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2004, 48, 3892–3904. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.A.; Willey, B.M.; Fawcett, D.; Gillani, N.; Mulvey, M.R. Molecular characterization of Enterococcus faecalis N06–0364 with low-level vancomycin resistance harboring a novel d-ala-d-ser gene cluster, VanL. Antimicrob. Agents Chemother. 2008, 52, 2667–2672. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Depardieu, F.; Bourdon, N.; Fines-Guyon, M.; Berger, P.; Camiade, S.; Leclercq, R.; Courvalin, P.; Cattoir, V. d-Ala-d-ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2011, 55, 4606–4612. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lin, D.; Yan, G.; Ye, X.; Wu, S.; Guo, Y.; Zhu, D.; Hu, F.; Zhang, Y.; Wang, F.; et al. VanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4643–4647. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Tanimoto, K.; Shibayama, K.; Arakawa, Y.; Fujimoto, S.; Ike, Y.; Tomita, H. Identification of VanN-type vancomycin resistance in an Enterococcus faecium isolate from chicken meat in Japan. Antimicrob. Agents Chemother. 2012, 56, 6389–6392. [Google Scholar] [CrossRef] [PubMed]
- Bartley, J. First case of VRSA identified in Michigan. Infect. Control Hosp. Epidemiol. 2002, 23, 480. [Google Scholar] [PubMed]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; McDougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Moravvej, Z.; Estaji, F.; Askari, E.; Solhjou, K.; Naderi Nasab, M.; Saadat, S. Update on the global number of vancomycin-resistant Staphylococcus aureus (VRSA) strains. Int. J. Antimicrob. Agents 2013, 42, 370–371. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Diaz, L.; Wollam, A.; Panesso, D.; Zhou, Y.; Rincon, S.; Narechania, A.; Xing, G.; di Gioia, T.S.; Doi, A.; et al. Transferable vancomycin resistance in a community-associated MRSA lineage. N. Engl. J. Med. 2014, 370, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Foucault, M.L.; Courvalin, P.; Grillot-Courvalin, C. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 2354–2359. [Google Scholar] [CrossRef] [PubMed]
- Périchon, B.; Courvalin, P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 4580–4587. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, F. Chapter 2. From microbial products to novel drugs that target a multitude of disease indications. Methods Enzymol. 2009, 458, 29–58. [Google Scholar] [PubMed]
- Marinelli, F.; Marcone, G. Small molecules, microbial secondary metabolites. In Comprehensive Biotechnology, 2nd ed.; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, pp. 285–297. [Google Scholar]
- Cundliffe, E.; Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 2010, 37, 643–672. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.G.; Broadhead, G.; Leskiw, B.K.; Wright, G.D. d-Ala-d-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc. Natl. Acad. Sci. USA 1997, 94, 6480–6483. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.G.; Lessard, I.A.; Park, I.; Wright, G.D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 1998, 42, 2215–2220. [Google Scholar] [PubMed]
- Sosio, M.; Kloosterman, H.; Bianchi, A.; de Vreugd, P.; Dijkhuizen, L.; Donadio, S. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 2004, 150, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Li, T.L.; Huang, F.; Haydock, S.F.; Mironenko, T.; Leadlay, P.F.; Spencer, J.B. Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: Characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 2004, 11, 107–119. [Google Scholar] [PubMed]
- Schäberle, T.F.; Vollmer, W.; Frasch, H.J.; Hüttel, S.; Kulik, A.; Röttgen, M.; von Thaler, A.K.; Wohlleben, W.; Stegmann, E. Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob. Agents Chemother. 2011, 55, 4283–4289. [Google Scholar] [CrossRef] [PubMed]
- Pootoolal, J.; Thomas, M.G.; Marshall, C.G.; Neu, J.M.; Hubbard, B.K.; Walsh, C.T.; Wright, G.D. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 2002, 99, 8962–8967. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.S.; Shrader, T.E. Femabx family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J. Biol. Chem. 2001, 276, 6998–7003. [Google Scholar] [CrossRef] [PubMed]
- Neu, J.M.; Wright, G.D. Inhibition of sporulation, glycopeptide antibiotic production and resistance in Streptomyces toyocaensis NRRL15009 by protein kinase inhibitors. FEMS Microbiol. Lett. 2001, 199, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Serina, S.; Radice, F.; Maffioli, S.; Donadio, S.; Sosio, M. Glycopeptide resistance determinants from the teicoplanin producer Actinoplanes teichomyceticus. FEMS Microbiol. Lett. 2004, 240, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Beltrametti, F.; Consolandi, A.; Carrano, L.; Bagatin, F.; Rossi, R.; Leoni, L.; Zennaro, E.; Selva, E.; Marinelli, F. Resistance to glycopeptide antibiotics in the teicoplanin producer is mediated by van gene homologue expression directing the synthesis of a modified cell wall peptidoglycan. Antimicrob. Agents Chemother. 2007, 51, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Hong, H.J.; Buttner, M.J. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 2006, 59, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.J.; Hutchings, M.I.; Neu, J.M.; Wright, G.D.; Paget, M.S.; Buttner, M.J. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 2004, 52, 1107–1121. [Google Scholar] [CrossRef] [PubMed]
- Novotna, G.; Hill, C.; Vincent, K.; Liu, C.; Hong, H.J. A novel membrane protein, VanJ, conferring resistance to teicoplanin. Antimicrob. Agents Chemother. 2012, 56, 1784–1796. [Google Scholar] [CrossRef] [PubMed]
- Kwun, M.J.; Novotna, G.; Hesketh, A.R.; Hill, L.; Hong, H.J. In vivo studies suggest that induction of VanS-dependent vancomycin resistance requires binding of the drug to d-Ala-d-Ala termini in the peptidoglycan cell wall. Antimicrob. Agents Chemother. 2013, 57, 4470–4480. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Beltrametti, F.; Binda, E.; Carrano, L.; Foulston, L.; Hesketh, A.; Bibb, M.; Marinelli, F. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2010, 54, 2465–2472. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Binda, E.; Carrano, L.; Bibb, M.; Marinelli, F. The relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2014, 58. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.J.; Hutchings, M.I.; Hill, L.M.; Buttner, M.J. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 2005, 280, 13055–13061. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Sosio, M.; Stinchi, S.; Beltrametti, F.; Lazzarini, A.; Donadio, S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 2003, 10, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Hugonnet, J.E.; Haddache, N.; Veckerlé, C.; Dubost, L.; Marie, A.; Shikura, N.; Mainardi, J.L.; Rice, L.B.; Arthur, M. Peptidoglycan cross-linking in glycopeptide-resistant actinomycetales. Antimicrob. Agents Chemother. 2014, 58, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Binda, E.; Marcone, G.L.; Pollegioni, L.; Marinelli, F. Characterization of VanY(n), a novel d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance in Nonomuraea sp. ATCC 39727. FEBS J. 2012, 279, 3203–3213. [Google Scholar] [CrossRef] [PubMed]
- Binda, E.; Marcone, G.L.; Berini, F.; Pollegioni, L.; Marinelli, F. Streptomyces spp. as efficient expression system for a d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance. BMC Biotechnol. 2013, 13, e24. [Google Scholar] [CrossRef]
- Wright, G.D.; Molinas, C.; Arthur, M.; Courvalin, P.; Walsh, C.T. Characterization of VanY, a d,d-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob. Agents Chemother. 1992, 36, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.E.; Arias, C.A.; Courvalin, P. Gene vanxyc encodes d,d-dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 1999, 34, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Mainardi, J.L.; Villet, R.; Bugg, T.D.; Mayer, C.; Arthur, M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in gram-positive bacteria. FEMS Microbiol. Rev. 2008, 32, 386–408. [Google Scholar] [CrossRef] [PubMed]
- Meziane-Cherif, D.; Stogios, P.J.; Evdokimova, E.; Savchenko, A.; Courvalin, P. Structural basis for the evolution of vancomycin resistance d,d-peptidases. Proc. Natl. Acad. Sci. USA 2014, 111, 5872–5877. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Carrano, L.; Marinelli, F.; Beltrametti, F. Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. (Tokyo) 2010, 63, 83–88. [Google Scholar] [CrossRef]
- Marcone, G.L.; Foulston, L.; Binda, E.; Marinelli, F.; Bibb, M.; Beltrametti, F. Methods for the genetic manipulation of Nonomuraea sp. ATCC 39727. J. Ind. Microbiol. Biotechnol. 2010, 37, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Sim, Y.M.; Kim, H.J.; Lee, D.W.; Lim, S.K.; Lee, S.J. Genome sequence of the vancomycin-producing Amycolatopsis orientalis subsp. Orientalis strain KCTC 9412T. Genome Announc. 2013, 1. [Google Scholar] [CrossRef]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G.D. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binda, E.; Marinelli, F.; Marcone, G.L. Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics 2014, 3, 572-594. https://doi.org/10.3390/antibiotics3040572
Binda E, Marinelli F, Marcone GL. Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics. 2014; 3(4):572-594. https://doi.org/10.3390/antibiotics3040572
Chicago/Turabian StyleBinda, Elisa, Flavia Marinelli, and Giorgia Letizia Marcone. 2014. "Old and New Glycopeptide Antibiotics: Action and Resistance" Antibiotics 3, no. 4: 572-594. https://doi.org/10.3390/antibiotics3040572
APA StyleBinda, E., Marinelli, F., & Marcone, G. L. (2014). Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics, 3(4), 572-594. https://doi.org/10.3390/antibiotics3040572







