Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria
Abstract
:1. Introduction
2. Extracellular Mechanisms of Resistance: Enzymatic Degradation and AMP Blocking
2.1. Extracellular Proteases
| Name | Mechanism of Action | Antimicrobial Resistance | Organisms | Reference |
|---|---|---|---|---|
| AMP Degradation | ||||
| Aureolysin | Protease | LL-37 | S. aureus | [5,11] |
| Gelatinase | Protease | LL-37 | E. faecalis | [4,10] |
| SepA | Protease | dermcidin | S. epidermidis | [6,16] |
| SpeB | Protease | LL-37 | S. pyogenes | [4,21,22] |
| Sequestration/Competition for AMP target | ||||
| M Protein | Binding at surface | LL-37 | S. pyogenes | [24] |
| PilB | Binding at surface | cathelicidins | S. agalactiae | [25] |
| SIC | Extracellular binding | α-defensins, LL-37, lysozyme | S. pyogenes | [26,27] |
| Staphylokinase | Extracellular binding | Cathelicidin, defensins | S. aureus | [28,29] |
| LciA | Binding at surface | Lactococcin A | L. lactis | [30,31] |
| Capsule | Binding/shielding | Polymyxin B, HNP-1 | S. pneumoniae | [32] |
| Exopolysaccharide | Shielding/ Sequestration | LL-37, hBD-3, dermcidin | S. epidermidis | [33,34,35] |
| LanI lipoproteins | Binding or competition | lantibiotics | L. lactis, B. subtilis, other lantibiotic producers | [36,37,38] |
| Cell Surface Modifications | ||||
| DltABCD | d-alanylation of teichoic acids | daptomycin, vancomycin, nisin, defensins, protegrins | S. aureus, L. monocytogenes, B. cereus, C. difficile, S. pyogenes, S. agalactiae, B. anthracis, S. suis | [2,39,40,41,42,43,44,45] |
| MprF | Lysylation of phoshatidylglycerol | defensins, thrombin-induced platelet microbicidal protein | S. aureus, L. monocytogenes, B. anthracis, M. tuberculosis | [46,47,48,49,50] |
| OatA | Peptidoglycan O-acetylase | lysozyme | S. aureus, S. epidermidis, S. lugdunensis, E. faecalis, L. monocytogenes | [51,52,53,54] |
| PdgA | Peptidoglycan N-acetylglucosamine deacetylase A | lysozyme | S. pneumoniae, E. faecalis, S. suis, L. monocytogenes, B. anthracis | [55,56,57,58] |
| NamH | N-acetylmuramic acid hydroxylase | lysozyme | M. smegmatis | [59] |
| AMP Efflux | ||||
| One-component transporter | ||||
| LmrB | ABC transporter | LsbA/LsbB | L. lactis | [60] |
| QacA | ABC transporter/alteration of membrane structure | thrombin-induced platelet microbicidal protein (tPMP) | S. aureus | [61] |
| BceAB type | ||||
| AnrAB | ABC transporter | nisin, gallidermin, bacitracin, β-lactams | L. monocytogenes | [62,63] |
| BceAB | ABC transporter | Bacitracin a, actagardine, mersacidin, plectasin | B. subtilis a, S. mutans | [64,65,66,67,68] |
| BraAB | ABC transporter | nisin, nukacin ISK-1, bacitracin | S. aureus | [69] |
| PsdAB | ABC transporter | nisin, enduracidin, gallidermin, subtilin | B. subtilis | [66] |
| MbrAB | ABC transporter | bacitracin | S. mutans | [35] |
| SP0812-SP0813 | ABC transporter | bacitracin, vancoresmycin | S. pneumoniae | [70] |
| SP0912-SP0913 | ABC transporter | bacitracin, lincomycin, nisin | S. pneumoniae | [71] |
| VraDE | ABC transporter | bacitracin, nisin, nukacin ISK-1 | S. aureus | [69,72,73,74,75,76] |
| VraFG | ABC transporter | nisin, colistin, bacitracin, vancomycin, indolicidin, LL-37, hBD3 | S. aureus, S. epidermidis | [69,72,75,77,78,79] |
| YsaCB | ABC transporter | nisin | L. lactis | [80] |
| BcrAB type | ||||
| BcrAB(C) | ABC transporter | bacitracin | B. licheniformis | [81] |
| BcrAB(D) | ABC transporter | bacitracin | E. faecalis | [82,83] |
| LanFEG type | ||||
| As-48EFG(H) | ABC transporter | AS-48 | E. faecalis | [84] |
| CprABC | ABC transporter | nisin, galidermin, other lantibiotics | C. difficile | [85,86] |
| EpiFEG(H) | ABC transporter | epidermin, gallidermin | S. epidermidis | [87] |
| LtnFE(I) | ABC transporter | lacticin 3147 | L. lactis | [88,89] |
| McdFEG | ABC transporter | macedocin | S. macedonicus | [90] |
| MrsFGE | ABC transporter | mersacidin | Bacillus sp. HIL Y-84, 54728 | [91,92] |
| MutFEG | ABC transporter | mutacin II | S. mutans | [93] |
| NisFEG(I) | ABC transporter | nisin | L. lactis | [37,94] |
| NukFEG(H) | ABC transporter | nukacin | S. warneri | [95,96] |
| SboFEG | ABC transporter | salivaricin B | S. salivarius | [97] |
| ScnFEG | ABC transporter | streptococcin A-FF22 | S. pyogenes | [98] |
| SmbFT | ABC transporter | Smb, haloduracin | S. mutans | [99] |
| SpaFEG | ABC transporter | subtilin | B. subtilis | [36,100] |
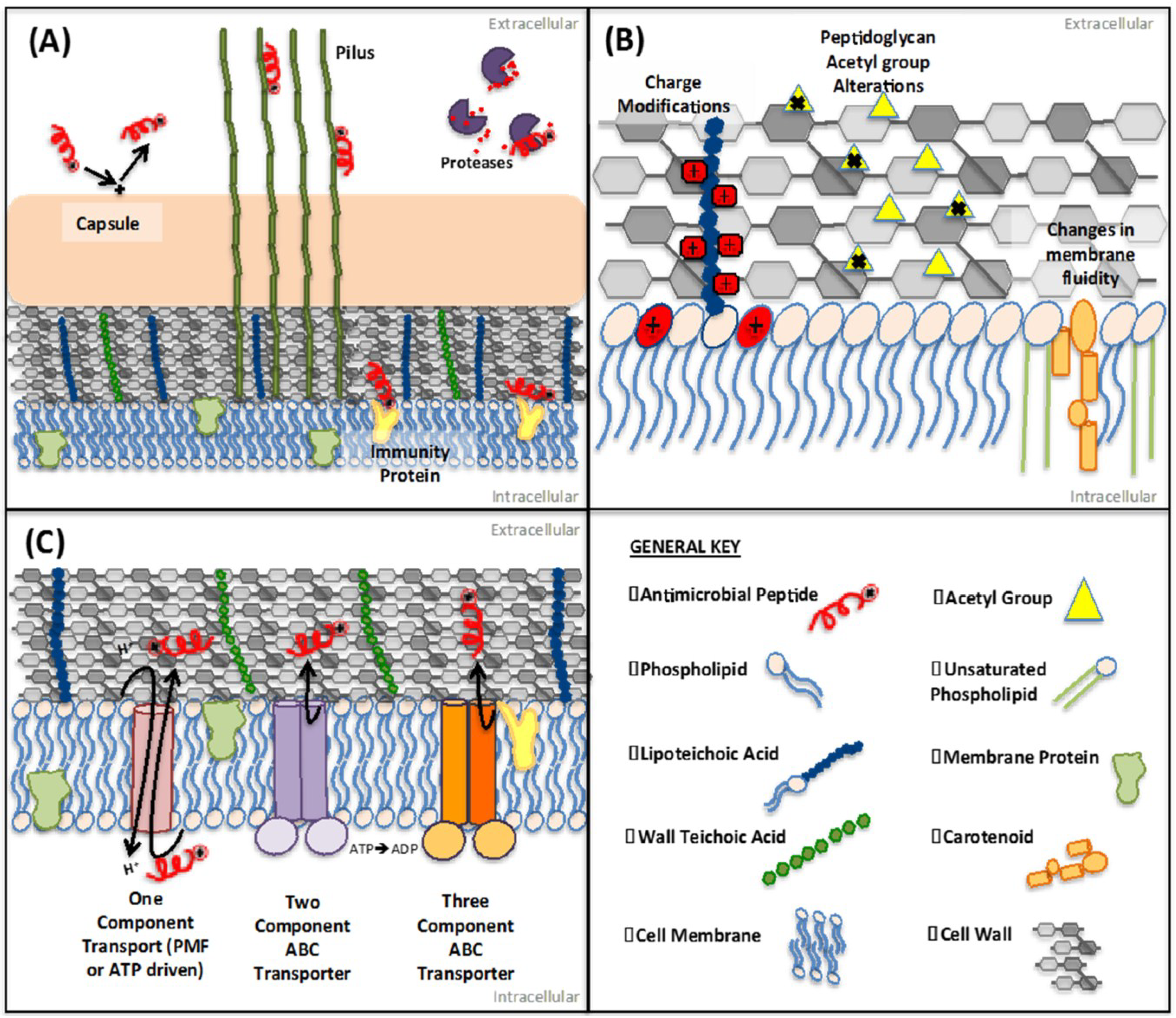
2.2. Protein-Mediated Sequestration
2.3. Inhibition of AMP Activity by Surface-Associated Polysaccharides
3. Membrane and Cell Wall Modifications
3.1. Repulsion of AMPs
3.2. Target Modification
3.3. Alterations to Membrane Order
4. AMP Efflux Mechanisms
4.1. Three-Component (LanFEG) Transporter Systems
4.2. Two-Component ABC-Transporter Systems
4.3. Single Membrane Protein Antimicrobial Transporters
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Koprivnjak, T.; Peschel, A. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci. 2011, 68, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Gotz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [PubMed]
- Staubitz, P.; Neumann, H.; Schneider, T.; Wiedemann, I.; Peschel, A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 2004, 231, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Schmidtchen, A.; Frick, I.M.; Andersson, E.; Tapper, H.; Bjorck, L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 2002, 46, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.; Kosowska, K.; Poulsen, K.; Kasprowicz, A.; Sekowska, A.; van Den Burg, B.; Travis, J.; Potempa, J. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect. Immun. 2000, 68, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Villaruz, A.E.; Li, M.; Cha, D.J.; Sturdevant, D.E.; Otto, M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 2007, 63, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Hase, C.C.; Finkelstein, R.A. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 1993, 57, 823–837. [Google Scholar] [PubMed]
- Del Papa, M.F.; Hancock, L.E.; Thomas, V.C.; Perego, M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacteriol. 2007, 189, 8835–8843. [Google Scholar]
- Engelbert, M.; Mylonakis, E.; Ausubel, F.M.; Calderwood, S.B.; Gilmore, M.S. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 2004, 72, 3628–3633. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef] [PubMed]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wojcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silberring, J.; Reed, M.; Pohl, J.; et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef] [PubMed]
- Kubica, M.; Guzik, K.; Koziel, J.; Zarebski, M.; Richter, W.; Gajkowska, B.; Golda, A.; Maciag-Gudowska, A.; Brix, K.; Shaw, L. A potential new pathway for Staphylococcus aureus dissemination: The silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One 2008, 3, e1409. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Santiago, B.; Hernandez-Pando, R.; Carranza, C.; Juarez, E.; Contreras, J.L.; Aguilar-Leon, D.; Torres, M.; Sada, E. Expression of cathelicidin LL-37 during Mycobacterium tuberculosis infection in human alveolar macrophages, monocytes, neutrophils, and epithelial cells. Infect. Immun. 2008, 76, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Schittek, B.; Hipfel, R.; Sauer, B.; Bauer, J.; Kalbacher, H.; Stevanovic, S.; Schirle, M.; Schroeder, K.; Blin, N.; Meier, F.; et al. Dermcidin: A novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2001, 2, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Teufel, P.; Gotz, F. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J. Bacteriol. 1993, 175, 4218–4224. [Google Scholar] [PubMed]
- Cheung, G.Y.; Rigby, K.; Wang, R.; Queck, S.Y.; Braughton, K.R.; Whitney, A.R.; Teintze, M.; DeLeo, F.R.; Otto, M. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 2010, 6, e1001133. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R.; Stevens, D.L.; Kaplan, E.L.; Schlievert, P.M. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes isolates associated with toxic shock-like syndrome. J. Clin. Microbiol. 1991, 29, 1562–1567. [Google Scholar] [PubMed]
- Elliott, S.D. A proteolytic enzyme produced by group A Streptococci with special reference to its effect on the type-specific M antigen. J. Exp. Med. 1945, 81, 573–592. [Google Scholar] [CrossRef] [PubMed]
- Kapur, V.; Majesky, M.W.; Li, L.L.; Black, R.A.; Musser, J.M. Cleavage of interleukin 1 beta (IL-1 beta) precursor to produce active IL-1 beta by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 1993, 90, 7676–7680. [Google Scholar] [CrossRef] [PubMed]
- Kapur, V.; Topouzis, S.; Majesky, M.W.; Li, L.L.; Hamrick, M.R.; Hamill, R.J.; Patti, J.M.; Musser, J.M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 1993, 15, 327–346. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.; Bjorck, L. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 2002, 43, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, P.; Rasmussen, M.; Bjorck, L. Alpha2-Macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J. Biol. Chem. 2004, 279, 52820–52823. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.; Muller, H.P.; Bjorck, L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J. Biol. Chem. 1999, 274, 15336–15344. [Google Scholar] [CrossRef] [PubMed]
- Lauth, X.; von Kockritz-Blickwede, M.; McNamara, C.W.; Myskowski, S.; Zinkernagel, A.S.; Beall, B.; Ghosh, P.; Gallo, R.L.; Nizet, V. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J. Innate. Immun. 2009, 1, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Maisey, H.C.; Quach, D.; Hensler, M.E.; Liu, G.Y.; Gallo, R.L.; Nizet, V.; Doran, K.S. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 2008, 22, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Frick, I.M.; Akesson, P.; Rasmussen, M.; Schmidtchen, A.; Bjorck, L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J. Biol. Chem. 2003, 278, 16561–16566. [Google Scholar] [CrossRef] [PubMed]
- Fernie-King, B.A.; Seilly, D.J.; Davies, A.; Lachmann, P.J. Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: Secretory leukocyte proteinase inhibitor and lysozyme. Infect. Immun. 2002, 70, 4908–4916. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Bokarewa, M.; Foster, T.; Mitchell, J.; Higgins, J.; Tarkowski, A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 2004, 172, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Braff, M.H.; Jones, A.L.; Skerrett, S.J.; Rubens, C.E. Staphylococcus aureus exploits cathelicidin antimicrobial peptides produced during early pneumonia to promote staphylokinase-dependent fibrinolysis. J. Infect. Dis. 2007, 195, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Diep, D.B.; Havarstein, L.S.; Nes, I.F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 1996, 178, 4472–4483. [Google Scholar] [PubMed]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef] [PubMed]
- Llobet, E.; Tomas, J.M.; Bengoechea, J.A. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology 2008, 154, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [PubMed]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [PubMed]
- Tsuda, H.; Yamashita, Y.; Shibata, Y.; Nakano, Y.; Koga, T. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 2002, 46, 3756–3764. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Entian, K.D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 1994, 60, 2793–2801. [Google Scholar] [PubMed]
- Kuipers, O.P.; Beerthuyzen, M.M.; Siezen, R.J.; de Vos, W.M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 1993, 216, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Saris, P.E.; Immonen, T.; Reis, M.; Sahl, H.G. Immunity to lantibiotics. Antonie Van Leeuwenhoek 1996, 69, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Vuong, C.; Otto, M.; Gotz, F. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 2000, 44, 2845–2847. [Google Scholar] [CrossRef] [PubMed]
- Abachin, E.; Poyart, C.; Pellegrini, E.; Milohanic, E.; Fiedler, F.; Berche, P.; Trieu-Cuot, P. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 2002, 43, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Abi Khattar, Z.; Rejasse, A.; Destoumieux-Garzon, D.; Escoubas, J.M.; Sanchis, V.; Lereclus, D.; Givaudan, A.; Kallassy, M.; Nielsen-Leroux, C.; Gaudriault, S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J. Bacteriol. 2009, 191, 7063–7073. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.H.; Ruiz-Bustos, E.; Courtney, H.S.; Dale, J.B.; Pence, M.A.; Nizet, V.; Aziz, R.K.; Gerling, I.; Price, S.M.; Hasty, D.L. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One 2009, 4, e5366. [Google Scholar] [PubMed]
- Fisher, N.; Shetron-Rama, L.; Herring-Palmer, A.; Heffernan, B.; Bergman, N.; Hanna, P. The dltABCD operon of Bacillus anthracis sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 2006, 188, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Sekizaki, T.; Takamatsu, D.; Harel, J.; Dominguez-Punaro Mde, L.; von Aulock, S.; Draing, C.; Marois, C.; Kobisch, M.; Gottschalk, M. d-Alanylation of lipoteichoic acid contributes to the virulence of Streptococcus suis. Infect. Immun. 2008, 76, 3587–3594. [Google Scholar] [CrossRef] [PubMed]
- Poyart, C.; Pellegrini, E.; Marceau, M.; Baptista, M.; Jaubert, F.; Lamy, M.C.; Trieu-Cuot, P. Attenuated virulence of Streptococcus agalactiae deficient in d-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 2003, 49, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Maloney, E.; Stankowska, D.; Zhang, J.; Fol, M.; Cheng, Q.J.; Lun, S.; Bishai, W.R.; Rajagopalan, M.; Chatterjee, D.; Madiraju, M.V. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 2009, 5, e1000534. [Google Scholar] [CrossRef] [PubMed]
- Samant, S.; Hsu, F.F.; Neyfakh, A.A.; Lee, H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 2009, 191, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Thedieck, K.; Hain, T.; Mohamed, W.; Tindall, B.J.; Nimtz, M.; Chakraborty, T.; Wehland, J.; Jansch, L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 2006, 62, 1325–1339. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Jack, R.W.; Otto, M.; Collins, L.V.; Staubitz, P.; Nicholson, G.; Kalbacher, H.; Nieuwenhuizen, W.F.; Jung, G.; Tarkowski, A.; et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 2001, 193, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Oku, Y.; Kurokawa, K.; Ichihashi, N.; Sekimizu, K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 2004, 150, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Gotz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Biswas, R.; Herbert, S.; Gotz, F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 2006, 74, 4598–4604. [Google Scholar] [CrossRef]
- Herbert, S.; Bera, A.; Nerz, C.; Kraus, D.; Peschel, A.; Goerke, C.; Meehl, M.; Cheung, A.; Gotz, F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007, 3, e102. [Google Scholar] [CrossRef] [PubMed]
- Aubry, C.; Goulard, C.; Nahori, M.A.; Cayet, N.; Decalf, J.; Sachse, M.; Boneca, I.G.; Cossart, P.; Dussurget, O. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 2011, 204, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Tomasz, A. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 2000, 275, 20496–20501. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Sekizaki, T.; Takamatsu, D.; de la Cruz Dominguez-Punaro, M.; Harel, J.; Bui, N.K.; Vollmer, W.; Gottschalk, M. Significant contribution of the pgdA gene to the virulence of Streptococcus suis. Mol. Microbiol. 2008, 70, 1120–1135. [Google Scholar] [CrossRef] [PubMed]
- Boneca, I.G.; Dussurget, O.; Cabanes, D.; Nahori, M.A.; Sousa, S.; Lecuit, M.; Psylinakis, E.; Bouriotis, V.; Hugot, J.P.; Giovannini, M.; et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA 2007, 104, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Laaberki, M.H.; Pfeffer, J.; Clarke, A.J.; Dworkin, J. O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J. Biol. Chem. 2011, 286, 5278–5288. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.B.; Mahapatra, S.; Crick, D.C.; Pavelka, M.S., Jr. Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 2005, 280, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Gajic, O.; Buist, G.; Kojic, M.; Topisirovic, L.; Kuipers, O.P.; Kok, J. Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 2003, 278, 34291–34298. [Google Scholar] [CrossRef] [PubMed]
- Kupferwasser, L.I.; Skurray, R.A.; Brown, M.H.; Firth, N.; Yeaman, M.R.; Bayer, A.S. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: Role of the qacA locus. Antimicrob. Agents Chemother. 1999, 43, 2395–2399. [Google Scholar] [PubMed]
- Mandin, P.; Fsihi, H.; Dussurget, O.; Vergassola, M.; Milohanic, E.; Toledo-Arana, A.; Lasa, I.; Johansson, J.; Cossart, P. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 2005, 57, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.; Curtis, N.; Cotter, P.D.; Hill, C.; Ross, R.P. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob. Agents Chemother. 2010, 54, 4416–4423. [Google Scholar] [CrossRef] [PubMed]
- Rietkotter, E.; Hoyer, D.; Mascher, T. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 2008, 68, 768–785. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventos, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Staron, A.; Finkeisen, D.E.; Mascher, T. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 2011, 55, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T.; Margulis, N.G.; Wang, T.; Ye, R.W.; Helmann, J.D. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003, 50, 1591–604. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Giyanto; Tateno, K.; Masuyama, W.; Moriya, S.; Kobayashi, K.; Ogasawara, N. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 2003, 49, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Kawada-Matsuo, M.; Yoshida, Y.; Zendo, T.; Nagao, J.; Oogai, Y.; Nakamura, Y.; Sonomoto, K.; Nakamura, N.; Komatsuzawa, H. Three distinct two-component systems are involved in resistance to the class I bacteriocins, Nukacin ISK-1 and nisin A, in Staphylococcus aureus. PLoS One 2013, 8, e69455. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; Hakenbeck, R.; Henrich, B. An ABC transporter of Streptococcus pneumoniae involved in susceptibility to vancoresmycin and bacitracin. Antimicrob. Agents Chemother. 2009, 53, 2034–2041. [Google Scholar] [CrossRef] [PubMed]
- Majchrzykiewicz, J.A.; Kuipers, O.P.; Bijlsma, J.J. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 2010, 54, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 136–147. [Google Scholar]
- Sass, P.; Jansen, A.; Szekat, C.; Sass, V.; Sahl, H.G.; Bierbaum, G. The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 2008, 8, e186. [Google Scholar] [CrossRef]
- Yoshida, Y.; Matsuo, M.; Oogai, Y.; Kato, F.; Nakamura, N.; Sugai, M.; Komatsuzawa, H. Bacitracin sensing and resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 320, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Hiron, A.; Falord, M.; Valle, J.; Debarbouille, M.; Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef] [PubMed]
- Pietiainen, M.; Francois, P.; Hyyrylainen, H.L.; Tangomo, M.; Sass, V.; Sahl, H.G.; Schrenzel, J.; Kontinen, V.P. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 2009, 10, e429. [Google Scholar] [CrossRef]
- Meehl, M.; Herbert, S.; Gotz, F.; Cheung, A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Falord, M.; Karimova, G.; Hiron, A.; Msadek, T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lai, Y.; Villaruz, A.E.; Cha, D.J.; Sturdevant, D.E.; Otto, M. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 2007, 104, 9469–9474. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.E.; van Hijum, S.A.; Knol, J.; Kok, J.; Kuipers, O.P. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 2006, 50, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Podlesek, Z.; Comino, A.; Herzog-Velikonja, B.; Zgur-Bertok, D.; Komel, R.; Grabnar, M. Bacillus licheniformis bacitracin-resistance ABC transporter: Relationship to mammalian multidrug resistance. Mol. Microbiol. 1995, 16, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Manson, J.M.; Keis, S.; Smith, J.M.; Cook, G.M. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 2004, 48, 3743–3748. [Google Scholar] [CrossRef] [PubMed]
- Matos, R.; Pinto, V.V.; Ruivo, M.; Lopes Mde, F. Study on the dissemination of the bcrABDR cluster in Enterococcus spp. reveals that the BcrAB transporter is sufficient to confer high-level bacitracin resistance. Int. J. Antimicrob. Agents 2009, 34, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Valdivia, E.; Martinez-Bueno, M.; Fernandez, M.; Soler-Gonzalez, A.S.; Ramirez-Rodrigo, H.; Maqueda, M. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 2003, 69, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.M.; Sonenshein, A.L. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun. 2011, 79, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Suarez, J.M.; Edwards, A.N.; McBride, S.M. The Clostridium difficile cpr locus is regulated by a non-contiguous two-component system in response to type A and B lantibiotics. J. Bacteriol. 2013, 195, 2621–2631. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Peschel, A.; Gotz, F. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tu3298. FEMS Microbiol. Lett. 1998, 166, 203–211. [Google Scholar] [PubMed]
- Draper, L.A.; Grainger, K.; Deegan, L.H.; Cotter, P.D.; Hill, C.; Ross, R.P. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 2009, 71, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O.; O’Keeffe, T.; Hill, C.; Ross, R.P. Regulation of immunity to the two-component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Mol. Microbiol. 2001, 39, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Papadelli, M.; Karsioti, A.; Anastasiou, R.; Georgalaki, M.; Tsakalidou, E. Characterization of the gene cluster involved in the biosynthesis of macedocin, the lantibiotic produced by Streptococcus macedonicus. FEMS Microbiol. Lett. 2007, 272, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Altena, K.; Guder, A.; Cramer, C.; Bierbaum, G. Biosynthesis of the lantibiotic mersacidin: Organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 2000, 66, 2565–2571. [Google Scholar] [CrossRef] [PubMed]
- Guder, A.; Schmitter, T.; Wiedemann, I.; Sahl, H.G.; Bierbaum, G. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl. Environ. Microbiol. 2002, 68, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Qi, F.; Novak, J.; Caufield, P.W. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 1999, 65, 1356–1360. [Google Scholar] [PubMed]
- Siegers, K.; Entian, K.D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 1995, 61, 1082–1089. [Google Scholar] [PubMed]
- Aso, Y.; Nagao, J.; Koga, H.; Okuda, K.; Kanemasa, Y.; Sashihara, T.; Nakayama, J.; Sonomoto, K. Heterologous expression and functional analysis of the gene cluster for the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. J. Biosci. Bioeng. 2004, 98, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Aso, Y.; Sashihara, T.; Nagao, J.; Kanemasa, Y.; Koga, H.; Hashimoto, T.; Higuchi, T.; Adachi, A.; Nomiyama, H.; Ishizaki, A.; et al. Characterization of a gene cluster of Staphylococcus warneri ISK-1 encoding the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. Biosci. Biotechnol. Biochem. 2004, 68, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Hyink, O.; Wescombe, P.A.; Upton, M.; Ragland, N.; Burton, J.P.; Tagg, J.R. Salivaricin A2 and the novel lantibiotic salivaricin B are encoded at adjacent loci on a 190-kilobase transmissible megaplasmid in the oral probiotic strain Streptococcus salivarius K12. Appl. Environ. Microbiol. 2007, 73, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.E.; Ferretti, J.J.; Hynes, W.L. Nucleotide sequence of the streptococcin A-FF22 lantibiotic regulon: model for production of the lantibiotic SA-FF22 by strains of Streptococcus pyogenes. FEMS Microbiol. Lett. 1999, 175, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Biswas, I. SmbFT, a putative ABC transporter complex, confers protection against the lantibiotic Smb in Streptococci. J. Bacteriol. 2013, 195, 5592–5601. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Heinzmann, S.; Dusterhus, S.; Borchert, S.; Entian, K.D. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 2005, 187, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Rabijns, A.; de Bondt, H.L.; de Ranter, C. Three-dimensional structure of staphylokinase, a plasminogen activator with therapeutic potential. Nat. Struct. Biol. 1997, 4, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Akesson, P.; Sjoholm, A.G.; Bjorck, L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 1996, 271, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Pence, M.A.; Rooijakkers, S.H.; Cogen, A.L.; Cole, J.N.; Hollands, A.; Gallo, R.L.; Nizet, V. Streptococcal inhibitor of complement promotes innate immune resistance phenotypes of invasive M1T1 group A Streptococcus. J. Innate Immun. 2010, 2, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.M.; Spencer, J.; Candlish, D.; Irvine, J.J.; Douce, G.R. Infection of hamsters with the UK Clostridium difficile ribotype 027 outbreak strain R20291. J. Med. Microbiol. 2011, 60, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Van Wamel, W.J.; Rooijakkers, S.H.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.C.; Sullivan, D.J.; Russell, R.J.; Arbuthnott, J.P.; Carey, B.F.; Pomeroy, H.M. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: Molecular mechanism of triple conversion. J. Gen. Microbiol. 1989, 135, 1679–1697. [Google Scholar] [PubMed]
- Jin, T.; Bokarewa, M.; McIntyre, L.; Tarkowski, A.; Corey, G.R.; Reller, L.B.; Fowler, V.G., Jr. Fatal outcome of bacteraemic patients caused by infection with staphylokinase-deficient Staphylococcus aureus strains. J. Med. Microbiol. 2003, 52, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Bisno, A.L.; Brito, M.O.; Collins, C.M. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 2003, 3, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Madzivhandila, M.; Adrian, P.V.; Cutland, C.L.; Kuwanda, L.; Madhi, S.A. Distribution of pilus islands of group B streptococcus associated with maternal colonization and invasive disease in South Africa. J. Med. Microbiol. 2013, 62, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Maisey, H.C.; Hensler, M.; Nizet, V.; Doran, K.S. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 2007, 189, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; Paul, M.; Xie, L.; van der Donk, W.A. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 2005, 105, 633–684. [Google Scholar] [CrossRef] [PubMed]
- Alkhatib, Z.; Abts, A.; Mavaro, A.; Schmitt, L.; Smits, S.H. Lantibiotics: How do producers become self-protected? J. Biotechnol. 2012, 159, 145–154. [Google Scholar]
- Halami, P.M.; Stein, T.; Chandrashekar, A.; Entian, K.D. Maturation and processing of SpaI, the lipoprotein involved in subtilin immunity in Bacillus subtilis ATCC 6633. Microbiol. Res. 2010, 165, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Schneider, T.; Pag, U.; Sahl, H.G. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 2004, 70, 3263–3271. [Google Scholar] [CrossRef] [PubMed]
- Christ, N.A.; Bochmann, S.; Gottstein, D.; Duchardt-Ferner, E.; Hellmich, U.A.; Dusterhus, S.; Kotter, P.; Guntert, P.; Entian, K.D.; Wohnert, J. The First structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J. Biol. Chem. 2012, 287, 35286–35298. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Immonen, T.; Koponen, O.; Saris, P.E. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 1995, 131, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Takala, T.M.; Saris, P.E. C terminus of NisI provides specificity to nisin. Microbiology 2006, 152, 3543–3549. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.; Eschbach-Bludau, M.; Iglesias-Wind, M.I.; Kupke, T.; Sahl, H.G. Producer immunity towards the lantibiotic Pep5: Identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl. Environ. Microbiol. 1994, 60, 2876–2883. [Google Scholar] [PubMed]
- Skaugen, M.; Abildgaard, C.I.; Nes, I.F. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol. Gen. Genet. 1997, 253, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, C.; Pag, U.; Josten, M.; Metzger, J.; Jack, R.W.; Bierbaum, G.; Jung, G.; Sahl, H.G. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 1998, 64, 3140–3146. [Google Scholar] [PubMed]
- Twomey, D.; Ross, R.P.; Ryan, M.; Meaney, B.; Hill, C. Lantibiotics produced by lactic acid bacteria: structure, function and applications. Antonie Van Leeuwenhoek 2002, 82, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Peterson, P.K.; Wilkinson, B.J.; Kim, Y.; Schmeling, D.; Quie, P.G. Influence of encapsulation on staphylococcal opsonization and phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 1978, 19, 943–949. [Google Scholar] [PubMed]
- Nelson, A.L.; Roche, A.M.; Gould, J.M.; Chim, K.; Ratner, A.J.; Weiser, J.N. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 2007, 75, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Ashbaugh, C.D.; Warren, H.B.; Carey, V.J.; Wessels, M.R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 1998, 102, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Kogan, G.; Uhrin, D.; Brisson, J.R.; Paoletti, L.C.; Blodgett, A.E.; Kasper, D.L.; Jennings, H.J. Structural and immunochemical characterization of the type VIII group B Streptococcus capsular polysaccharide. J. Biol. Chem. 1996, 271, 8786–8790. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Aanensen, D.M.; Mavroidi, A.; Saunders, D.; Rabbinowitsch, E.; Collins, M.; Donohoe, K.; Harris, D.; Murphy, L.; Quail, M.A.; et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006, 2, e31. [Google Scholar] [CrossRef] [PubMed]
- Candela, T.; Fouet, A. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol. Microbiol. 2005, 57, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Kasper, D.L.; Krick, T.P.; Wessels, M.R. Characterization of the linkage between the type III capsular polysaccharide and the bacterial cell wall of group B Streptococcus. J. Biol. Chem. 2000, 275, 7497–7504. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [PubMed]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Alberti, S.; Bengoechea, J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef] [PubMed]
- Rupp, M.E.; Fey, P.D.; Heilmann, C.; Gotz, F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 2001, 183, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Bartscht, K.; Mack, D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 1999, 67, 2627–2632. [Google Scholar] [PubMed]
- Beiter, K.; Wartha, F.; Hurwitz, R.; Normark, S.; Zychlinsky, A.; Henriques-Normark, B. The capsule sensitizes Streptococcus pneumoniae to alpha-defensins human neutrophil proteins 1 to 3. Infect. Immun. 2008, 76, 3710–3716. [Google Scholar] [CrossRef] [PubMed]
- Wartha, F.; Beiter, K.; Albiger, B.; Fernebro, J.; Zychlinsky, A.; Normark, S.; Henriques-Normark, B. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 2007, 9, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.; Szekat, C.; Schroder, W.; Wolz, C.; Goerke, C.; Lee, J.C.; Turck, M.; Bierbaum, G. Production of capsular polysaccharide does not influence Staphylococcus aureus vancomycin susceptibility. BMC Microbiol. 2013, 13, e65. [Google Scholar] [CrossRef]
- Boman, H.G. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1995, 13, 61–92. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.; Hancock, R.E. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Nizet, V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues. Mol. Biol. 2006, 8, 11–26. [Google Scholar] [PubMed]
- Peschel, A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002, 10, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Rozek, A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 2002, 206, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Weidenmaier, C.; Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 2008, 6, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Goldfine, H. Bacterial membranes and lipid packing theory. J. Lipid. Res. 1984, 25, 1501–1507. [Google Scholar] [PubMed]
- Wiese, A.; Munstermann, M.; Gutsmann, T.; Lindner, B.; Kawahara, K.; Zahringer, U.; Seydel, U. Molecular mechanisms of polymyxin B-membrane interactions: Direct correlation between surface charge density and self-promoted transport. J. Membr. Biol. 1998, 162, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Peschel, A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 2011, 80, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef] [PubMed]
- Kristian, S.A.; Durr, M.; van Strijp, J.A.; Neumeister, B.; Peschel, A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect. Immun. 2003, 71, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Sakinc, T.; Laverde, D.; Wobser, D.; Benachour, A.; Theilacker, C.; Hartke, A.; Huebner, J. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 2012, 7, e38458. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, A.B.; Angert, E.R.; Helmann, J.D. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 2009, 53, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Maloney, E.; Lun, S.; Stankowska, D.; Guo, H.; Rajagoapalan, M.; Bishai, W.R.; Madiraju, M.V. Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front. Microbiol. 2011, 2, e19. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Peschel, A.; Kempf, V.A.; Lucindo, N.; Yeaman, M.R.; Bayer, A.S. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect. Immun. 2005, 73, 8033–8038. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, K.; Whitmire, W.; Xiong, Y.Q.; Molden, J.; Jones, T.; Peschel, A.; Staubitz, P.; Adler-Moore, J.; McNamara, P.J.; Proctor, R.A.; et al. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 2007, 153, 1187–1197. [Google Scholar]
- Ruzin, A.; Severin, A.; Moghazeh, S.L.; Etienne, J.; Bradford, P.A.; Projan, S.J.; Shlaes, D.M. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta 2003, 1621, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Komatsuzawa, H.; Fujiwara, T.; McCallum, N.; Sugai, M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 4800–4807. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.J.; Proctor, R.A.; Sahl, H.G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Mishra, N.N.; Rubio, A.; Bayer, A.S. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 2013, 57, 5658–5664. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, L.I.; Helmann, J.D. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 2008, 190, 7797–7807. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.; Ibba, M. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J. Biol. Chem. 2009, 284, 29677–29683. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Yang, S.J.; Chen, L.; Muller, C.; Saleh-Mghir, A.; Kuhn, S.; Peschel, A.; Yeaman, M.R.; Nast, C.C.; Kreiswirth, B.N.; et al. Emergence of daptomycin resistance in daptomycin-naive rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS One 2013, 8, e71151. [Google Scholar] [CrossRef] [PubMed]
- Slavetinsky, C.J.; Peschel, A.; Ernst, C.M. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 3492–3497. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.M.; Sonenshein, A.L. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 2011, 157, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Loach, D.M.; Alqumber, M.; Rockel, C.; Hermann, C.; Pfitzenmaier, M.; Tannock, G.W. d-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 2007, 9, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Koprivnjak, T.; Mlakar, V.; Swanson, L.; Fournier, B.; Peschel, A.; Weiss, J.P. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 2006, 188, 3622–3630. [Google Scholar] [CrossRef] [PubMed]
- Le Jeune, A.; Torelli, R.; Sanguinetti, M.; Giard, J.C.; Hartke, A.; Auffray, Y.; Benachour, A. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One 2010, 5, e9658. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, F.C.; Heaton, M.P.; Debabov, D.V.; Zhang, Q. The dlt operon in the biosynthesis of d-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 1996, 2, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Perego, M.; Glaser, P.; Minutello, A.; Strauch, M.A.; Leopold, K.; Fischer, W. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J. Biol. Chem. 1995, 270, 15598–15606. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, F.C.; Baddiley, J. A continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Kreiswirth, B.N.; Sakoulas, G.; Yeaman, M.R.; Xiong, Y.Q.; Sawa, A.; Bayer, A.S. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 2009, 200, 1916–1920. [Google Scholar] [CrossRef] [PubMed]
- Guariglia-Oropeza, V.; Helmann, J.D. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and d-alanylation of teichoic acids. J. Bacteriol. 2011, 193, 6223–6232. [Google Scholar] [CrossRef] [PubMed]
- Jann, N.J.; Schmaler, M.; Kristian, S.A.; Radek, K.A.; Gallo, R.L.; Nizet, V.; Peschelm, A.; Landmann, R. Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J. Leukoc. Biol. 2009, 86, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Saar-Dover, R.; Bitler, A.; Nezer, R.; Shmuel-Galia, L.; Firon, A.; Shimoni, E.; Trieu-Cuot, P.; Shai, Y. d-Alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog. 2012, 8, e1002891. [Google Scholar] [CrossRef] [PubMed]
- Kristian, S.A.; Lauth, X.; Nizet, V.; Goetz, F.; Neumeister, B.; Peschel, A.; Landmann, R. Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J. Infect. Dis. 2003, 188, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.V.; Kristian, S.A.; Weidenmaier, C.; Faigle, M.; van Kessel, K.P.; van Strijp, J.A.; Gotz, F.; Neumeister, B.; Peschel, A. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 2002, 186, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Thompson, R.; Palmer, J.W.; Khorazo, D. The nature of lysozyme action. Science 1934, 79, 61. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Palmer, J.W.; Thompson, R.; Khorazo, D. On the mechanism of lysozyme action. J. Biol. Chem. 1936, 113, 479–486. [Google Scholar]
- Chipman, D.M.; Sharon, N. Mechanism of lysozyme action. Science 1969, 165, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Nash, J.A.; Ballard, T.N.; Weaver, T.E.; Akinbi, H.T. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J. Immunol. 2006, 177, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Hebert, L.; Courtin, P.; Torelli, R.; Sanguinetti, M.; Chapot-Chartier, M.P.; Auffray, Y.; Benachour, A. Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 2007, 75, 5390–5398. [Google Scholar] [CrossRef] [PubMed]
- Amano, K.; Araki, Y.; Ito, E. Effect of N-acyl substitution at glucosamine residues on lysozyme-catalyzed hydrolysis of cell-wall peptidoglycan and its oligosaccharides. Eur. J. Biochem. 1980, 107, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Amano, K.; Hayashi, H.; Araki, Y.; Ito, E. The action of lysozyme on peptidoglycan with N-unsubstituted glucosamine residues. Isolation of glycan fragments and their susceptibility to lysozyme. Eur. J. Biochem. 1977, 76, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Psylinakis, E.; Boneca, I.G.; Mavromatis, K.; Deli, A.; Hayhurst, E.; Foster, S.J.; Varum, K.M.; Bouriotis, V. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 2005, 280, 30856–30863. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Schuttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Tomasz, A. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 2002, 70, 7176–7178. [Google Scholar] [CrossRef] [PubMed]
- Benachour, A.; Ladjouzi, R.; le Jeune, A.; Hebert, L.; Thorpe, S.; Courtin, P.; Chapot-Chartier, M.P.; Prajsnar, T.K.; Foster, S.J.; Mesnage, S. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. J. Bacteriol. 2012, 194, 6066–6073. [Google Scholar] [CrossRef] [PubMed]
- Rae, C.S.; Geissler, A.; Adamson, P.C.; Portnoy, D.A. Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect. Immun. 2011, 79, 3596–3606. [Google Scholar] [CrossRef] [PubMed]
- Crisostomo, M.I.; Vollmer, W.; Kharat, A.S.; Inhulsen, S.; Gehre, F.; Buckenmaier, S.; Tomasz, A. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol. Microbiol. 2006, 61, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Biswas, R.; Herbert, S.; Kulauzovic, E.; Weidenmaier, C.; Peschel, A.; Gotz, F. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 2007, 189, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Akinbi, H.T.; Standish, A.J.; Weiser, J.N. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog. 2008, 4, e1000241. [Google Scholar] [CrossRef] [PubMed]
- Veiga, P.; Bulbarela-Sampieri, C.; Furlan, S.; Maisons, A.; Chapot-Chartier, M.P.; Erkelenz, M.; Mervelet, P.; Noirot, P.; Frees, D.; Kuipers, O.P.; et al. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 2007, 282, 19342–19354. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Park, B.G.; Wolf, A.J.; Brikos, C.; Goodridge, H.S.; Becker, C.A.; Reyes, C.N.; Miao, E.A.; Aderem, A.; Gotz, F.; et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 2010, 7, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.J.; Nikaido, H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, A.S.; Montville, T.J. Nisin induces changes in membrane fatty acid composition of Listeria monocytogenes nisin-resistant strains at 10 degrees C and 30 degrees C. J. Appl. Microbiol. 1997, 82, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Verheul, A.; Russell, N.J.; van’t Hof, R.; Rombouts, F.M.; Abee, T. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 1997, 63, 3451–3457. [Google Scholar] [PubMed]
- Ming, X.T.; Daeschel, M.A. Nisin resistance of foodborne bacteria and the specific resistance responses of Listeria monocytogenes Scott A. J. Food Protect. 1993, 56, 944–948. [Google Scholar]
- Crandall, A.D.; Montville, T.J. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl. Environ. Microbiol. 1998, 64, 231–237. [Google Scholar] [PubMed]
- Mishra, N.N.; McKinnell, J.; Yeaman, M.R.; Rubio, A.; Nast, C.C.; Chen, L.; Kreiswirth, B.N.; Bayer, A.S. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011, 55, 4012–4018. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Liu, G.Y.; Yeaman, M.R.; Nast, C.C.; Proctor, R.A.; McKinnell, J.; Bayer, A.S. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 2011, 55, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Rubio, A.; Nast, C.C.; Bayer, A.S. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: Evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int. J. Microbiol. 2012, 2012, e683450. [Google Scholar] [CrossRef]
- Britton, G. Structure and properties of carotenoids in relation to function. FASEB J. 1995, 9, 1551–1558. [Google Scholar] [PubMed]
- Pelz, A.; Wieland, K.P.; Putzbach, K.; Hentschel, P.; Albert, K.; Gotz, F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 2005, 280, 32493–32498. [Google Scholar] [CrossRef] [PubMed]
- Katzif, S.; Lee, E.H.; Law, A.B.; Tzeng, Y.L.; Shafer, W.M. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J. Bacteriol. 2005, 187, 8181–8184. [Google Scholar] [CrossRef] [PubMed]
- Subczynski, W.K.; Wisniewska, A. Physical properties of lipid bilayer membranes: Relevance to membrane biological functions. Acta Biochim. Pol. 2000, 47, 613–625. [Google Scholar] [PubMed]
- Wisniewska, A.; Subczynski, W.K. Effects of polar carotenoids on the shape of the hydrophobic barrier of phospholipid bilayers. Biochim. Biophys. Acta 1998, 1368, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.S.; Prasad, R.; Chandra, J.; Koul, A.; Smriti, M.; Varma, A.; Skurray, R.A.; Firth, N.; Brown, M.H.; Koo, S.P.; et al. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 2000, 68, 3548–3553. [Google Scholar] [CrossRef] [PubMed]
- Van Blitterswijk, W.J.; van der Meer, B.W.; Hilkmann, H. Quantitative contributions of cholesterol and the individual classes of phospholipids and their degree of fatty acyl (un)saturation to membrane fluidity measured by fluorescence polarization. Biochemistry 1987, 26, 1746–1756. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Chen, J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004, 73, 241–268. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [PubMed]
- Reizer, J.; Reizer, A.; Saier, M.H., Jr. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1992, 1, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Heinzmann, S.; Kiesau, P.; Himmel, B.; Entian, K.D. The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 2003, 47, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Heinzmann, S.; Solovieva, I.; Entian, K.D. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 2003, 278, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Immonen, T.; Saris, P.E. Characterization of the nisFEG operon of the nisin Z producing Lactococcus lactis subsp. lactis N8 strain. DNA Seq. 1998, 9, 263–274. [Google Scholar]
- Ra, S.R.; Qiao, M.; Immonen, T.; Pujana, I.; Saris, E.J. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactoccocus lactis N8. Microbiology 1996, 142, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Aso, Y.; Okuda, K.; Nagao, J.; Kanemasa, Y.; Thi Bich Phuong, N.; Koga, H.; Shioya, K.; Sashihara, T.; Nakayama, J.; Sonomoto, K. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 2005, 69, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Yanagihara, S.; Shioya, K.; Harada, Y.; Nagao, J.; Aso, Y.; Zendo, T.; Nakayama, J.; Sonomoto, K. Binding specificity of the lantibiotic-binding immunity protein NukH. Appl. Environ. Microbiol. 2008, 74, 7613–7619. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S. ABC transporters of antimicrobial peptides in Firmicutes bacteria—Phylogeny, function and regulation. Mol. Microbiol. 2012, 86, 1295–1317. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. ABC transporters: Physiology, structure and mechanism—An overview. Res. Microbiol. 2001, 152, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Dintner, S.; Staron, A.; Berchtold, E.; Petri, T.; Mascher, T.; Gebhard, S. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 2011, 193, 3851–3862. [Google Scholar] [CrossRef] [PubMed]
- Revilla-Guarinos, A.; Gebhard, S.; Mascher, T.; Zuniga, M. Defence against antimicrobial peptides: Different strategies in Firmicutes. Environ. Microbiol. 2014, 16, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Bernard, R.; El Ghachi, M.; Mengin-Lecreulx, D.; Chippaux, M.; Denizot, F. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J. Biol. Chem. 2005, 280, 28852–28857. [Google Scholar] [CrossRef] [PubMed]
- Shaaly, A.; Kalamorz, F.; Gebhard, S.; Cook, G.M. Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. J. Antimicrob. Chemother. 2013, 68, 1583–1593. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, A.; Jalbert, L.A.; Harel, J.; Masson, L.; Archambault, M. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One 2012, 7, e44449. [Google Scholar] [CrossRef] [PubMed]
- Butaye, P.; Cloeckaert, A.; Schwarz, S. Mobile genes coding for efflux-mediated antimicrobial resistance in Gram-positive and Gram-negative bacteria. Int. J. Antimicrob. Agents 2003, 22, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Van Veen, H.W.; Venema, K.; Bolhuis, H.; Oussenko, I.; Kok, J.; Poolman, B.; Driessen, A.J.; Konings, W.N. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. USA 1996, 93, 10668–10672. [Google Scholar] [CrossRef] [PubMed]
- Saidijam, M.; Benedetti, G.; Ren, Q.; Xu, Z.; Hoyle, C.J.; Palmer, S.L.; Ward, A.; Bettaney, K.E.; Szakonyi, G.; Meuller, J.; et al. Microbial drug efflux proteins of the major facilitator superfamily. Curr. Drug Targets 2006, 7, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, T.G.; Paulsen, I.T.; Gillespie, M.T.; Tennent, J.M.; Midgley, M.; Jones, I.G.; Purewal, A.S.; Skurray, R.A. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 1992, 74, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Leelaporn, A.; Paulsen, I.T.; Tennent, J.M.; Littlejohn, T.G.; Skurray, R.A. Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J. Med. Microbiol. 1994, 40, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.S.; Cheng, D.; Yeaman, M.R.; Corey, G.R.; McClelland, R.S.; Harrel, L.J.; Fowler, V.G., Jr. In vitro resistance to thrombin-induced platelet microbicidal protein among clinical bacteremic isolates of Staphylococcus aureus correlates with an endovascular infectious source. Antimicrob. Agents Chemother. 1998, 42, 3169–3172. [Google Scholar] [PubMed]
- Solheim, M.; Aakra, A.; Vebo, H.; Snipen, L.; Nes, I.F. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl. Environ. Microbiol. 2007, 73, 5767–5774. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fuentes, M.A.; Abriouel, H.; Ortega Morente, E.; Perez Pulido, R.; Galvez, A. Genetic determinants of antimicrobial resistance in Gram positive bacteria from organic foods. Int. J. Food Microbiol. 2014, 172, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gay, K.; Stephens, D.S. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 2001, 184, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R. Road to clinical efficacy: Challenges and novel strategies for antimicrobial peptide development. Future Microbiol. 2011, 6, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; Mu, D.; Montalban-Lopez, M.; Hendriks, D.; Kuipers, O.P. Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth. Biol. 2013, 2, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.J.; Mabood, F.; Souleimanov, A.; Zhou, X.; Jaoua, S.; Kamoun, F.; Smith, D.L. Stability and antibacterial activity of bacteriocins produced by Bacillus thuringiensis and Bacillus thuringiensis ssp. kurstaki. J. Microbiol. Biotechnol. 2008, 18, 1836–1840. [Google Scholar]
- Chehimi, S.; Delalande, F.; Sable, S.; Hajlaoui, M.R.; van Dorsselaer, A.; Limam, F.; Pons, A.M. Purification and partial amino acid sequence of thuricin S, a new anti-Listeria bacteriocin from Bacillus thuringiensis. Can. J. Microbiol. 2007, 53, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; McDougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug. Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Napier, B.A.; Band, V.; Burd, E.M.; Weiss, D.S. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob. Agents Chemother. 2014, 58, 5594–5597. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawrocki, K.L.; Crispell, E.K.; McBride, S.M. Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria. Antibiotics 2014, 3, 461-492. https://doi.org/10.3390/antibiotics3040461
Nawrocki KL, Crispell EK, McBride SM. Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria. Antibiotics. 2014; 3(4):461-492. https://doi.org/10.3390/antibiotics3040461
Chicago/Turabian StyleNawrocki, Kathryn L., Emily K. Crispell, and Shonna M. McBride. 2014. "Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria" Antibiotics 3, no. 4: 461-492. https://doi.org/10.3390/antibiotics3040461
APA StyleNawrocki, K. L., Crispell, E. K., & McBride, S. M. (2014). Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria. Antibiotics, 3(4), 461-492. https://doi.org/10.3390/antibiotics3040461




