Genomic Profiling Reveals Clinically Relevant Antimicrobial Resistance and Virulence Genes in Klebsiella pneumoniae from Hong Kong Wet Markets
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Selection and Genome Sequencing
2.2. Genome Assembly, Annotation, and Quality Control
2.3. Comparative Genomics and Phylogenetic Analysis
2.4. Variant Calling and Functional Analysis
2.5. Antibiotic Resistance and Virulence Gene Detection
2.6. Statistical Analysis
3. Results
3.1. Phylogenetic Analysis
3.2. Principal Component Analysis (PCA)
3.3. Pan-Genome Analysis of 43 K. pneumoniae Genomes
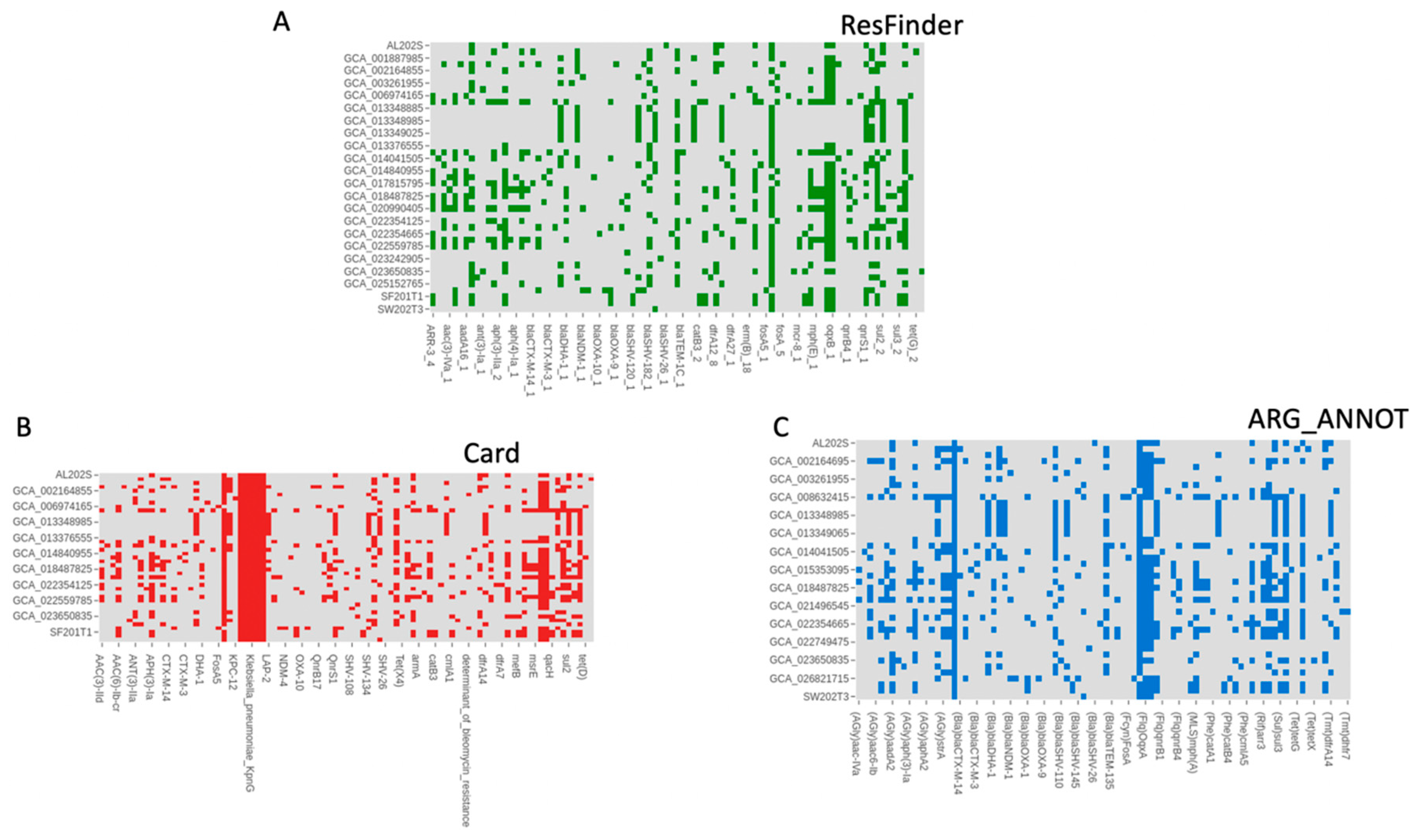
3.4. Antimicrobial Resistance Genes
3.5. Comparative Genomic Analysis of Virulence Factors in Klebsiella pneumoniae Isolates from Wet Market Samples
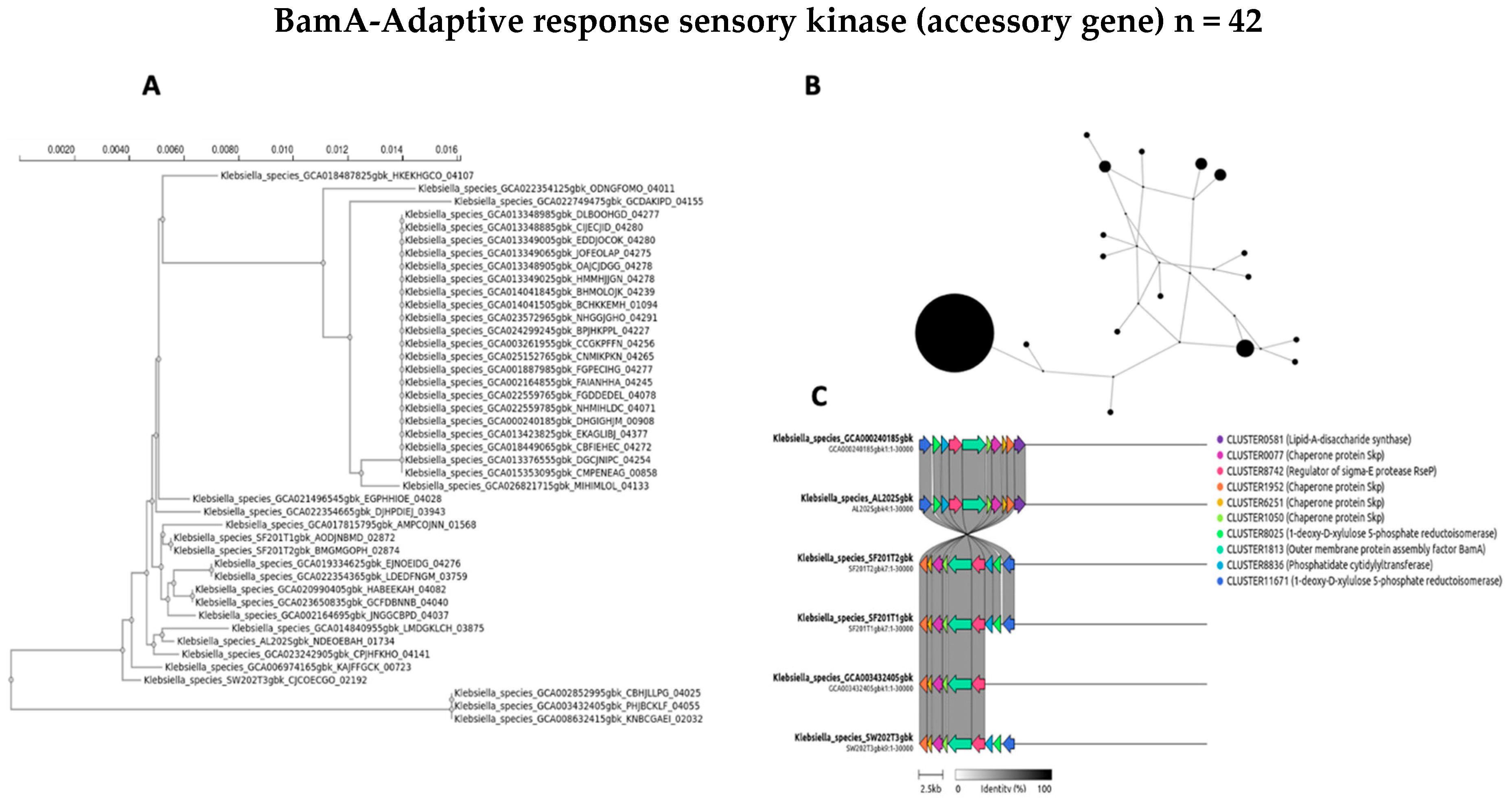
3.6. Phylogenetic and Genomic Contextualization of Selected Accessory Genes in Klebsiella Isolates
4. Discussion
4.1. Virulence Factors in the Wet Market Samples
4.2. Accessory Genes: Evolutionary Dynamics and Horizontal Gene Transfer
4.3. Integrated Results
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riwu, K.H.P.; Effendi, M.H.; Rantam, F.A.; Khairullah, A.R.; Widodo, A. A review: Virulence factors of Klebsiella pneumonia as emerging infection on the food chain. Vet. World 2022, 15, 2172–2179. [Google Scholar] [CrossRef]
- Davis, G.S.; Price, L.B. Recent Research Examining Links Among Klebsiella pneumoniae from Food, Food Animals, and Human Extraintestinal Infections. Curr. Environ. Health Rep. 2016, 3, 128–135. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, N.-l.; Cen, J.-q.; Han, J.-t.; Tang, Y.-x.; Xu, Z.-q.; Zeng, H.; Houf, K.; Yu, Z. Comparative analyses of bacterial contamination and microbiome of broiler carcasses in wet market and industrial processing environments. Int. J. Food Microbiol. 2025, 426, 110937. [Google Scholar] [CrossRef]
- Ngan, W.Y.; Rao, S.; Chan, L.C.; Sekoai, P.T.; Pu, Y.; Yao, Y.; Fung, A.H.Y.; Habimana, O. Impacts of Wet Market Modernization Levels and Hygiene Practices on the Microbiome and Microbial Safety of Wooden Cutting Boards in Hong Kong. Microorganisms 2020, 8, 1941. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; Srinivas, K.; Milton, A.A.P.; Priya, G.B.; Das, S.; Lindahl, J.F. Limiting the spillover of zoonotic pathogens from traditional food markets in developing countries and a new market design for risk-proofing. Epidemiol. Health 2023, 45, e2023097. [Google Scholar] [CrossRef]
- Sekoai, P.T.; Feng, S.; Zhou, W.; Ngan, W.Y.; Pu, Y.; Yao, Y.; Pan, J.; Habimana, O. Insights into the Microbiological Safety of Wooden Cutting Boards Used for Meat Processing in Hong Kong’s Wet Markets: A Focus on Food-Contact Surfaces, Cross-Contamination and the Efficacy of Traditional Hygiene Practices. Microorganisms 2020, 8, 579. [Google Scholar] [CrossRef]
- Liu, H.Y.; Prentice, E.L.; Webber, M.A. Mechanisms of antimicrobial resistance in biofilms. npj Antimicrob. Resist. 2024, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Olaimat, A.N.; Ababneh, A.M.; Al-Holy, M.; Al-Nabulsi, A.; Osaili, T.; Abughoush, M.; Ayyash, M.; Holley, R.A. A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces. Microbiol. Res. 2024, 15, 1973–1992. [Google Scholar] [CrossRef]
- Lo, M.Y.; Ngan, W.Y.; Tsun, S.M.; Hsing, H.-L.; Lau, K.T.; Hung, H.P.; Chan, S.L.; Lai, Y.Y.; Yao, Y.; Pu, Y.; et al. A Field Study Into Hong Kong’s Wet Markets: Raised Questions Into the Hygienic Maintenance of Meat Contact Surfaces and the Dissemination of Microorganisms Associated With Nosocomial Infections. Front. Microbiol. 2019, 10, 2618. [Google Scholar] [CrossRef]
- Burns, J.P. Administrative reform in Hong Kong: An institutional analysis of food safety. In Comparative Governance Reform in Asia: Democracy, Corruption, and Government Trust; Emerald Publishing Limited: Leeds, UK, 2008; pp. 21–38. [Google Scholar]
- Dong, Y.; Liu, H.; Habimana, O. High risk of Vibrio pathogen and antibiotic resistance transfer in live seafood wet markets of Shantou, China. Int. J. Food Microbiol. 2025, 432, 111098. [Google Scholar] [CrossRef]
- Jing, G.; Sun, Z.; Wang, H.; Gong, Y.; Huang, S.; Ning, K.; Xu, J.; Su, X. Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 2017, 7, 40371. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Dereeper, A.; Summo, M.; Meyer, D.F.; Marschall, T. PanExplorer: A web-based tool for exploratory analysis and visualization of bacterial pan-genomes. Bioinformatics 2022, 38, 4412–4414. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M.; Valencia, A. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2014, 6, 80–92. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- De Koster, S.; Rodriguez Ruiz, J.P.; Rajakani, S.G.; Lammens, C.; Glupczynski, Y.; Goossens, H.; Xavier, B.B. Diversity in the Characteristics of Klebsiella pneumoniae ST101 of Human, Environmental, and Animal Origin. Front. Microbiol. 2022, 13, 838207. [Google Scholar] [CrossRef]
- Pajand, O.; Rahimi, H.; Badmasti, F.; Gholami, F.; Alipour, T.; Darabi, N.; Aarestrup, F.M.; Leekitcharoenphon, P. Various arrangements of mobile genetic elements among CC147 subpopulations of Klebsiella pneumoniae harboring blaNDM-1: A comparative genomic analysis of carbapenem resistant strains. J. Biomed. Sci. 2023, 30, 73. [Google Scholar] [CrossRef]
- Rocha, J.; Henriques, I.; Gomila, M.; Manaia, C.M. Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings. Sci. Rep. 2022, 12, 10441. [Google Scholar] [CrossRef]
- Ramos, P.I.P.; Picão, R.C.; de Almeida, L.G.P.; Lima, N.C.B.; Girardello, R.; Vivan, A.C.P.; Xavier, D.E.; Barcellos, F.G.; Pelisson, M.; Vespero, E.C.; et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genom. 2014, 15, 54. [Google Scholar] [CrossRef]
- Dawson, C.J.; Bartczak, A.; Hassan, K.A. Mutations in the efflux regulator gene oqxR provide a simple genetic switch for antimicrobial resistance in Klebsiella pneumoniae. Microbiology 2024, 170, 001499. [Google Scholar] [CrossRef]
- Mihailovskaya, V.S.; Selivanova, P.A.; Kuznetsova, M.V. Prevalence of qacEΔ1, qacE, oqxA, oqxB, acrA, cepA and zitB genes among multidrug-resistant Klebsiella pneumoniae isolated in a cardiac hospital. J. Microbiol. Epidemiol. Immunobiol. 2024, 101, 502–511. [Google Scholar] [CrossRef]
- Joseph, B.J.; Mathew, M.; Rachel, R.; Mathew, J.; Radhakrishnan, E.K. Klebsiella pneumoniae Virulence Factors and Biofilm Components: Synthesis, Structure, Function, and Inhibitors. In ESKAPE Pathogens; Springer: Berlin/Heidelberg, Germany, 2024; pp. 271–295. [Google Scholar]
- Grubwieser, P.; Hilbe, R.; Gehrer, C.M.; Grander, M.; Brigo, N.; Hoffmann, A.; Seifert, M.; Berger, S.; Theurl, I.; Nairz, M.; et al. Klebsiella pneumoniae manipulates human macrophages to acquire iron. Front. Microbiol. 2023, 14, 1223113. [Google Scholar] [CrossRef]
- Lan, Y.; Zhou, M.; Li, X.; Liu, X.; Li, J.; Liu, W. Preliminary Investigation of Iron Acquisition in Hypervirulent Klebsiella pneumoniae Mediated by Outer Membrane Vesicles. Infect. Drug Resist. 2022, 15, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Tan, S.; Ye, W.; Hou, L.; Fang, B. Low-concentration iron promotes Klebsiella pneumoniae biofilm formation by suppressing succinic acid. BMC Microbiol. 2022, 22, 95. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, X.; Chan, E.W.-C.; Zhang, R.; Chen, S.; Zhou, X. Functional Characterization of Plasmid-Borne rmpADC Homologues in Klebsiella pneumoniae. Microbiol. Spectr. 2023, 11, 03081-22. [Google Scholar] [CrossRef]
- Walker, K.A.; Miner, T.A.; Palacios, M.; Trzilova, D.; Frederick, D.R.; Broberg, C.A.; Sepúlveda, V.E.; Quinn, J.D.; Miller, V.L.; Goldberg, J.B. A Klebsiella pneumoniae Regulatory Mutant Has Reduced Capsule Expression but Retains Hypermucoviscosity. mBio 2019, 10, e00089-19. [Google Scholar] [CrossRef] [PubMed]
- Mike, L.A.; Stark, A.J.; Forsyth, V.S.; Vornhagen, J.; Smith, S.N.; Bachman, M.A.; Mobley, H.L.T. A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 2020, 17, e1009376. [Google Scholar] [CrossRef]
- Samoilova, A.A.; Kraeva, L.A.; Mikhailov, N.V.; Saitova, A.T.; Polev, D.E.; Vashukova, M.A.; Gordeeva, S.A.; Smirnova, E.V.; Beljatich, L.I.; Dolgova, A.S.; et al. Genomic analysis of Klebsiella pneumoniae strains virulence and antibiotic resistance. Russ. J. Infect. Immun. 2024, 14, 339–350. [Google Scholar] [CrossRef]
- Isogai, M.; Kawamura, K.; Yagi, T.; Kayama, S.; Sugai, M.; Doi, Y.; Suzuki, M. Evaluation of Klebsiella pneumoniae pathogenicity through holistic gene content analysis. Microb. Genom. 2024, 10, 001295. [Google Scholar] [CrossRef]
- Dey, S.; Gaur, M.; Sykes, E.M.E.; Prusty, M.; Elangovan, S.; Dixit, S.; Pati, S.; Kumar, A.; Subudhi, E. Unravelling the Evolutionary Dynamics of High-Risk Klebsiella pneumoniae ST147 Clones: Insights from Comparative Pangenome Analysis. Genes 2023, 14, 1037. [Google Scholar] [CrossRef] [PubMed]
- Morgado, S.; Fonseca, E.; Vicente, A.C. Genomics of Klebsiella pneumoniae Species Complex Reveals the Circulation of High-Risk Multidrug-Resistant Pandemic Clones in Human, Animal, and Environmental Sources. Microorganisms 2022, 10, 2281. [Google Scholar] [CrossRef] [PubMed]








| Sample | Host | Type | BioSampleID |
|---|---|---|---|
| AL202S | Food_proc_surface | Wet market | SAMN45810452 |
| SF201T1 | Food_proc_surface | Wet market | SAMN45810453 |
| SF201T2 | Food_proc_surface | Wet market | SAMN45810454 |
| SW202T3 | Food_proc_surface | Wet market | SAMN45810455 |
| GCA_013349065 | Surface | Environmental | SAMN15186438 |
| GCA_022354665 | Sewage | Environmental | SAMN13925598 |
| GCA_000240185 | Homo-Sapiens | Clinical | SAMN02602959 |
| GCA_013376555 | Water | Environmental | SAMN14548317 |
| GCA_022559765 | Poultry | Animal-associated | SAMN23139062 |
| GCA_001887985 | Homo-Sapiens | Clinical | SAMN06019522 |
| GCA_013423825 | Sewage | Environmental | SAMN15511588 |
| GCA_022559785 | Poultry | Animal-associated | SAMN23139063 |
| GCA_002164695 | Homo-Sapiens | Clinical | SAMN06909168 |
| GCA_014041505 | Surface | Environmental | SAMN15649164 |
| GCA_022749475 | Swine | Animal-associated | SAMN26814915 |
| GCA_002164855 | Homo-Sapiens | Clinical | SAMN06909161 |
| GCA_014041845 | Homo-Sapiens | Clinical | SAMN15649146 |
| GCA_023242905 | Homo-Sapiens | Clinical | SAMN27593784 |
| GCA_002852995 | Homo-Sapiens | Clinical | SAMN07609113 |
| GCA_014840955 | Poultry | Animal-associated | SAMN16222891 |
| GCA_023572965 | Homo-Sapiens | Clinical | SAMN28546263 |
| GCA_003261955 | Homo-Sapiens | Clinical | SAMN08932863 |
| GCA_015353095 | Swine | Animal-associated | SAMN16659219 |
| GCA_023650835 | Sewage | Environmental | SAMN23799605 |
| GCA_003432405 | Swine | Animal-associated | SAMN09691066 |
| GCA_017815795 | Red_Kangaroo | Animal-associated | SAMN14590646 |
| GCA_024299245 | Homo-Sapiens | Clinical | SAMN26245098 |
| GCA_006974165 | Rabbit | Animal-associated | SAMN11650121 |
| GCA_018449065 | Swine | Animal-associated | SAMN19107302 |
| GCA_025152765 | Homo-Sapiens | Clinical | SAMN30672525 |
| GCA_008632415 | Poultry | Animal-associated | SAMN12741260 |
| GCA_018487825 | Swine | Animal-associated | SAMN12741260 |
| GCA_026821715 | Homo-Sapiens | Clinical | SAMN30914361 |
| GCA_013348885 | Homo-Sapiens | Clinical | SAMN15186440 |
| GCA_019334625 | Swine | Animal-associated | SAMN20245095 |
| GCA_013348905 | Surface | Environmental | SAMN15186447 |
| GCA_020990405 | Housefly | Animal-associated | SAMN23139350 |
| GCA_013348985 | Surface | Environmental | SAMN15186444 |
| GCA_021496545 | Swine | Animal-associated | SAMN15097741 |
| GCA_013349005 | Surface | Environmental | SAMN15186443 |
| GCA_022354125 | Sewage | Environmental | SAMN13925581 |
| GCA_013349025 | Surface | Environmental | SAMN15186442 |
| GCA_022354365 | Sewage | Environmental | SAMN13925526 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngan, W.Y.; Rao, S.; Fung, A.H.Y.; Habimana, O. Genomic Profiling Reveals Clinically Relevant Antimicrobial Resistance and Virulence Genes in Klebsiella pneumoniae from Hong Kong Wet Markets. Antibiotics 2025, 14, 922. https://doi.org/10.3390/antibiotics14090922
Ngan WY, Rao S, Fung AHY, Habimana O. Genomic Profiling Reveals Clinically Relevant Antimicrobial Resistance and Virulence Genes in Klebsiella pneumoniae from Hong Kong Wet Markets. Antibiotics. 2025; 14(9):922. https://doi.org/10.3390/antibiotics14090922
Chicago/Turabian StyleNgan, Wing Yui, Subramanya Rao, Aster Hei Yiu Fung, and Olivier Habimana. 2025. "Genomic Profiling Reveals Clinically Relevant Antimicrobial Resistance and Virulence Genes in Klebsiella pneumoniae from Hong Kong Wet Markets" Antibiotics 14, no. 9: 922. https://doi.org/10.3390/antibiotics14090922
APA StyleNgan, W. Y., Rao, S., Fung, A. H. Y., & Habimana, O. (2025). Genomic Profiling Reveals Clinically Relevant Antimicrobial Resistance and Virulence Genes in Klebsiella pneumoniae from Hong Kong Wet Markets. Antibiotics, 14(9), 922. https://doi.org/10.3390/antibiotics14090922








