An OSMAC Strategy for the Production of Antimicrobial Compounds by the Amazonian Fungi Talaromyces pinophilus CCM-UEA-F0414 and Penicillium paxilli CCM-UEA-F0591
Abstract
1. Introduction
2. Results
2.1. Identification of Fungal Strains
2.2. Antimicrobial Activity
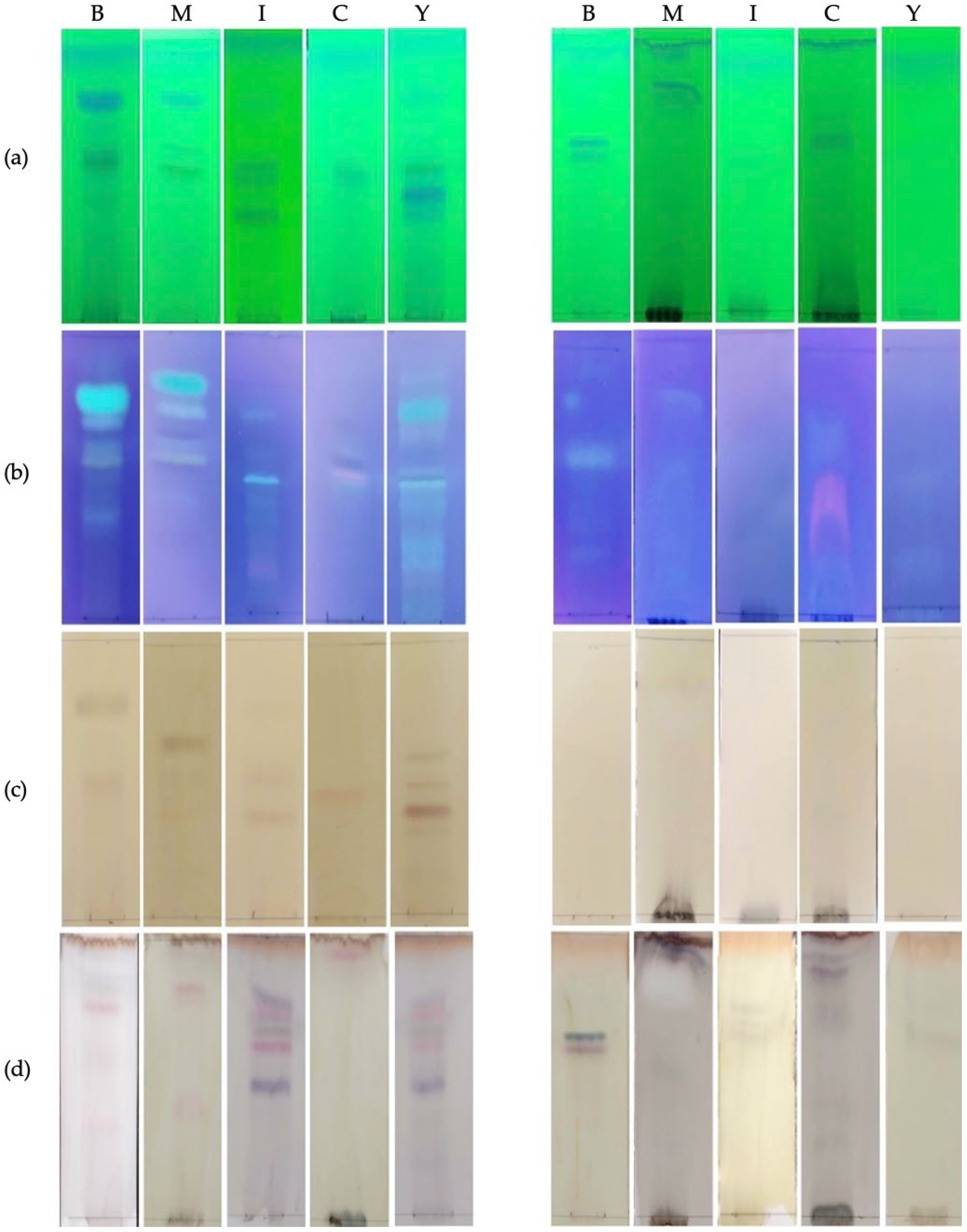
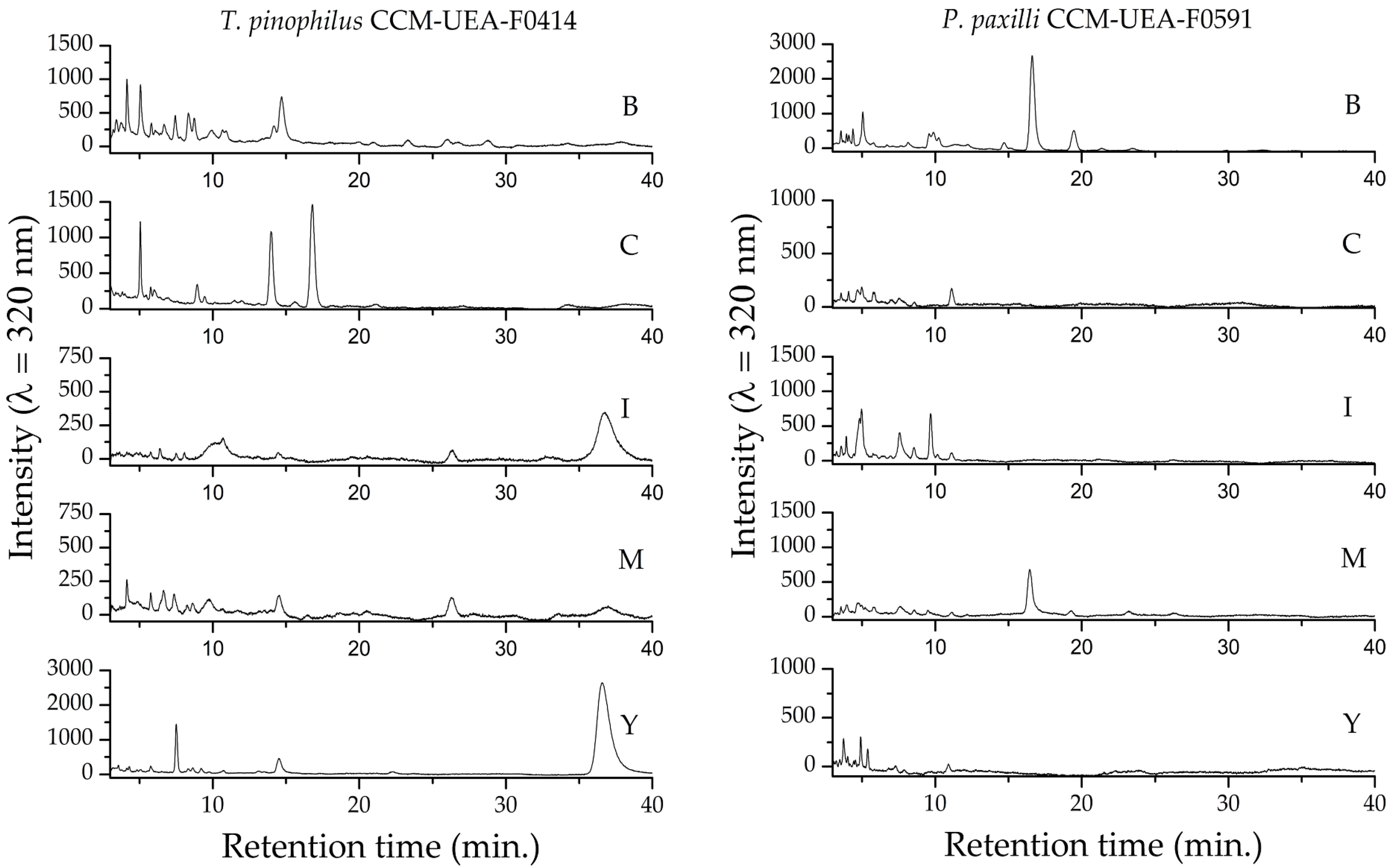
2.3. Chemical Characterization of Fungal Extracts
3. Discussion
4. Materials and Methods
4.1. Fungi
4.2. Fungal Identification
4.2.1. Morphological Characterization
4.2.2. Molecular Identification
4.2.3. Phylogenetic Analysis
4.3. Production and Extraction of Fungal Metabolites
4.4. Antimicrobial Activity Assay
4.5. Thin-Layer Chromatography (TLC)
4.6. Total Phenolic Content
4.7. Fourier Transform Infrared Spectroscopy (FTIR)
4.8. Ultra-High-Performance Liquid Chromatography (uHPLC)
4.9. UV–VIS Spectrophotometric Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de la Fuente-Nunez, C.; Cesaro, A.; Hancock, R.E.W. Antibiotic Failure: Beyond Antimicrobial Resistance. Drug Resist. Updates 2023, 71, 101012. [Google Scholar] [CrossRef]
- Walesch, S.; Birkelbach, J.; Jézéquel, G.; Haeckl, F.P.J.; Hegemann, J.D.; Hesterkamp, T.; Hirsch, A.K.H.; Hammann, P.; Müller, R. Fighting Antibiotic Resistance—Strategies and (Pre)Clinical Developments to Find New Antibacterials. EMBO Rep. 2023, 24, e56033. [Google Scholar] [CrossRef]
- Brüssow, H. The Antibiotic Resistance Crisis and the Development of New Antibiotics. Microb. Biotechnol. 2024, 17, e14510. [Google Scholar] [CrossRef]
- Gurgel, R.S.; Pereira, D.Í.d.M.; Garcia, A.V.F.; Souza, A.T.F.d.; Silva, T.M.d.; Andrade, C.P.d.; Silva, W.L.d.; Nunez, C.V.; Fantin, C.; Procópio, R.E.d.L.; et al. Antimicrobial and Antioxidant Activities of Endophytic Fungi Associated with Arrabidaea chica (Bignoniaceae). J. Fungi 2023, 9, 864. [Google Scholar] [CrossRef]
- Mendieta-Brito, S.; Sayed, M.; Son, E.; Kim, D.-S.; Dávila, M.; Pyo, S.-H. Identification, Characterization, and Antibacterial Evaluation of Five Endophytic Fungi from Psychotria poeppigiana Müll. Arg., an Amazon Plant. Microorganisms 2024, 12, 1590. [Google Scholar] [CrossRef]
- Alam, B.; Lǐ, J.; Gě, Q.; Khan, M.A.; Gōng, J.; Mehmood, S.; Yuán, Y.; Gǒng, W. Endophytic Fungi: From Symbiosis to Secondary Metabolite Communications or Vice Versa? Front. Plant. Sci. 2021, 12, 791033. [Google Scholar] [CrossRef]
- Devi, R.; Kaur, T.; Guleria, G.; Rana, K.L.; Kour, D.; Yadav, N.; Yadav, A.N.; Saxena, A.K. Fungal Secondary Metabolites and Their Biotechnological Applications for Human Health. In New and Future Developments in Microbial Biotechnology and Bioengineering; Rastegari, A.A., Yadav, A.N., Yadav, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 147–161. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, J.; Singh, V. Types and Applications of Potential Antibiotics Produced by Fungi. In Nanobiotechnology for Plant Protection, Fungal Secondary Metabolites; Abd-Elsalam, K.A., Mohamed, H.I., Eds.; Elsevier: Amsterdam, The Netherlands, 2024; pp. 493–517. [Google Scholar] [CrossRef]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic Metabolites from Marine-Derived Microorganisms Using OSMAC and Epigenetic Approaches. Mar. Drugs 2022, 20, 84. [Google Scholar] [CrossRef]
- Palma Esposito, F.; Giugliano, R.; Della Sala, G.; Vitale, G.A.; Buonocore, C.; Ausuri, J.; Galasso, C.; Coppola, D.; Franci, G.; Galdiero, M.; et al. Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus. Int. J. Mol. Sci. 2021, 22, 9055. [Google Scholar] [CrossRef]
- Staropoli, A.; Iacomino, G.; De Cicco, P.; Woo, S.L.; Di Costanzo, L.; Vinale, F. Induced Secondary Metabolites of the Beneficial Fungus Trichoderma harzianum M10 through OSMAC Approach. Chem. Biol. Technol. Agric. 2023, 10, 28. [Google Scholar] [CrossRef]
- Barreto, D.L.C.; de Carvalho, C.R.; de Almeida Alves, T.M.; Zani, C.L.; Cantrell, C.L.; Duke, S.O.; Rosa, L.H. Influence of Genetics on the Secondary Metabolism of Fungi. In Natural Secondary Metabolites: From Nature, Through Science, to Industry; Carocho, M., Heleno, S.A., Barros, L., Eds.; Springer: Cham, Switzerland, 2023; pp. 687–704. [Google Scholar] [CrossRef]
- Zhou, J.-T.; Li, B.-C.; Liang, X.-M.; Liang, M.; Li, J.; Xu, W.-F.; Yang, R.-Y. Secondary Metabolites from the Endophytic Fungus Xylaria sp. GDG-102 by Osmac Strategy and Their Antibacterial Activities. Chem. Nat. Compd. 2024, 60, 189–191. [Google Scholar] [CrossRef]
- Takahashi, J.A.; de Queiroz, L.L.; Vidal, D.M. A Close View of the Production of Bioactive Fungal Metabolites Mediated by Chromatin Modifiers. Molecules 2024, 29, 3536. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. Chembiochem. 2002, 3, 619. [Google Scholar] [CrossRef]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of Microbial Secondary Metabolic Pathways: Avenues and Challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef]
- Zhong, M.; Kang, H.; Liu, W.; Ma, L.; Liu, D. Alkaloid Diversity Expansion of a Talent Fungus Penicillium raistrichii through OSMAC-Based Cultivation. Front. Microbiol. 2023, 14, 1279140. [Google Scholar] [CrossRef]
- Smith, H.; Doyle, S.; Murphy, R. Target Directed Identification of Natural Bioactive Compounds from Filamentous Fungi. Food Chem. 2023, 405, 134743. [Google Scholar] [CrossRef]
- Grasel, F.d.S.; Ferrão, M.F.; Wolf, C.R. Development of Methodology for Identification the Nature of the Polyphenolic Extracts by FTIR Associated with Multivariate Analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 153, 94–101. [Google Scholar] [CrossRef]
- Harnly, J.M.; Bhagwat, S.; Lin, L.Z. Profiling Methods for the Determination of Phenolic Compounds in Foods and Dietary Supplements. Anal. Bioanal. Chem. 2007, 389, 47–61. [Google Scholar] [CrossRef]
- Yao, L.H.; Jiang, Y.M.; Shi, J.; Tomas, F.A.; Tomás-Barberán, T.; Barberán, B.; Datta, N.; Singanusong, R.; Chen, S.S. Flavonoids in Food and Their Health Benefits. Plant Foods Hum. Nutr. 2004, 59, 113–122. [Google Scholar] [CrossRef]
- Mishra, R.; Venkateswara Sarma, V. Current Perspectives of Endophytic Fungi in Sustainable Development. In Fungi and their Role in Sustainable Development: Current Perspectives; Gehlot, P., Singh, J., Eds.; Springer: Singapore, 2018; pp. 553–584. [Google Scholar] [CrossRef]
- Kreutz, T.; Duarte, E.O.S.; Quatrin, P.M.; Carneiro, S.B.; Veiga-Junior, V.F.; Fuentefria, A.M.; Koester, L.S. Antifungal Activity of Aniba canelilla (Kunth) Mez Essential Oil and Its Main Compound 1-Nitro-2-Phenylethane against Dermatophytes. Antibiotics 2024, 13, 488. [Google Scholar] [CrossRef]
- Banhos, E.F.d.; Souza, A.Q.L.d.; Andrade, J.C.d.; Souza, A.D.L.d.; Koolen, H.H.F.; Albuquerque, P.M. Endophytic Fungi from Myrcia guianensis at the Brazilian Amazon: Distribution and Bioactivity. Braz. J. Microbiol. 2014, 45, 153–162. [Google Scholar] [CrossRef]
- Matias, R.R.; Sepúlveda, A.M.G.; Batista, B.N.; de Lucena, J.M.V.M.; Albuquerque, P.M. Degradation of Staphylococcus aureus Biofilm Using Hydrolytic Enzymes Produced by Amazonian Endophytic Fungi. Appl. Biochem. Biotechnol. 2021, 193, 2145–2161. [Google Scholar] [CrossRef]
- Lobiuc, A.; Pavăl, N.-E.; Mangalagiu, I.I.; Gheorghiță, R.; Teliban, G.-C.; Oroian, M.; Humelnicu, D.; Goleț, I. Future Antimicrobials: Natural and Functionalized Phenolics. Molecules 2023, 28, 1114. [Google Scholar] [CrossRef]
- De Rossi, L.; Rocchetti, G.; Lucini, L.; Rebecchi, A. Antimicrobial Potential of Polyphenols: Mechanisms of Action and Microbial Responses—A Narrative Review. Antioxidants 2025, 14, 200. [Google Scholar] [CrossRef]
- Daglia, M. Polyphenols as Antimicrobial Agents. Curr. Opin. Biotechnol. 2012, 23, 174–181. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Padhi, S.; Masi, M.; Cimmino, A.; Tuzi, A.; Jena, S.; Tayung, K.; Evidente, A. Funiculosone, a Substituted Dihydroxanthene-1,9-Dione with Two of Its Analogues Produced by an Endolichenic Fungus Talaromyces funiculosus and their Antimicrobial Activity. Phytochem. 2019, 157, 175–183. [Google Scholar] [CrossRef]
- Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A.A.; Khan, G.J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; Modarresi-Ghazani, F.; Xiao, H.; et al. Chlorogenic Acid (CGA): A Pharmacological Review and Call for Further Research. Biomed. Pharmacother. 2018, 97, 67–74. [Google Scholar] [CrossRef]
- Lunardelli, P.; de Carvalho, N.; Silva, E.d.O.; Chagas-Paula, D.A.; Honorata, J.; Luiz, H.; Ikegaki, M. Importance and Implications of the Production of Phenolic Secondary Metabolites by Endophytic Fungi: A Mini-Review. Mini Ver. Med. Chem. 2016, 16, 259–271. [Google Scholar] [CrossRef]
- Singh, A.K.; Kim, J.Y.; Lee, Y.S. Phenolic Compounds in Active Packaging and Edible Films/Coatings: Natural Bioactive Molecules and Novel Packaging Ingredients. Molecules 2022, 27, 7513. [Google Scholar] [CrossRef]
- Albuquerque, B.R.; Heleno, S.A.; Oliveira, M.B.P.P.; Barros, L.; Ferreira, I.C.F.R. Phenolic Compounds: Current Industrial Applications, Limitations and Future Challenges. Food Funct. 2021, 12, 14–29. [Google Scholar] [CrossRef]
- Anh, N.M.; Huyen, V.T.T.; Quyen, V.T.; Dao, P.T.; Quynh, D.T.; Huong, D.T.M.; Van Cuong, P.; Dat, T.T.H.; Minh, L.T.H. Antimicrobial and Cytotoxic Secondary Metabolites from a Marine-Derived Fungus Penicillium citrinum VM6. Curr. Microbiol. 2024, 81, 32. [Google Scholar] [CrossRef]
- Borkunov, G.V.; Leshchenko, E.V.; Berdyshev, D.V.; Popov, R.S.; Chingizova, E.A.; Shlyk, N.P.; Gerasimenko, A.V.; Kirichuk, N.N.; Khudyakova, Y.V.; Chausova, V.E.; et al. New Piperazine Derivatives Helvamides B–C from the Marine-Derived Fungus Penicillium velutinum ZK-14 Uncovered by OSMAC (One Strain Many Compounds) Strategy. Nat. Prod. Bioprospect. 2024, 14, 32. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K.M. Strategies for Fermentation Medium Optimization: An In-Depth Review. Front. Microbiol. 2017, 7, 2087. [Google Scholar] [CrossRef]
- Palomino Garcia, L.R.; Biasetto, C.R.; Araujo, A.R.; Bianchi, V.L.D. Enhanced Extraction of Phenolic Compounds from Coffee Industry’s Residues through Solid State Fermentation by Penicillium purpurogenum. Food Sci. Technol. 2015, 35, 704–711. [Google Scholar] [CrossRef]
- Song, Z.; Chen, D.; Dai, J. Isolation of Four New Phenolic Compounds from the Fungus Talaromyces cellulolyticus. Phytochem. Let. 2024, 64, 19–24. [Google Scholar] [CrossRef]
- Li, L.; Li, C.B.; Wu, Y.H.; Li, B.; Wu, Y.R.; Yan, Z.Y. Combining Metabolomics and Metagenomics to Analyze the Biosynthetic Potential of Culturable Endophytic Fungi Isolated from Salvia miltiorrhiza. Appl. Biochem. Microbiol. 2023, 59, 659–672. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Crozier, A.; Jaganath, I.B.; Clifford, M.N. Phenols, Polyphenols and Tannins: An Overview. In Plant Secondary Metabolites: Occurrence, Structure and Role in the Human Diet; Crozier, A., Clifford, M.N., Ashihara, H., Eds.; Blackwell Publishing: Oxford, UK, 2006; pp. 1–24. [Google Scholar]
- Shende, V.V.; Bauman, K.D.; Moore, B.S. The Shikimate Pathway: Gateway to Metabolic Diversity. Nat. Prod. Rep. 2024, 41, 604–648. [Google Scholar] [CrossRef]
- Liu, C.; Dong, H.; Zhao, M.; Zhang, L. Polyketide Biosynthesis in Fungi: Insights from Gene Clusters and Beyond. Fungal Genet. Biol. 2019, 132, 103256. [Google Scholar] [CrossRef]
- Castellani, A. Viability of Some Pathogenic Fungi in Distilled Water. J. Trop. Med. Hyg. 1939, 42, 225–226. [Google Scholar]
- Frisvad, J.C.; Samson, R.A. Polyphasic Taxonomy of Penicillium subgenus Penicillium. A Guide to Identification of Food and Air-Borne Terverticillate penicillia and Their Mycotoxins. Stud. Mycol. 2004, 49, 1–174. [Google Scholar]
- Procop, G.W.; Church, D.L.; Hall, G.S.; Janda, W.M.; Koneman, E.W.; Schreckenberger, P.C.; Woods, G.L. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 7th ed.; Jones & Bartlett Learning: Burlington, MA, USA, 2016. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Pereira, D.Í.d.M.; Gurgel, R.S.; Souza, A.T.F.d.; Matias, R.R.; Falcão, L.d.S.; Chaves, F.C.M.; Silva, G.F.d.; Martínez, J.G.; Procópio, R.E.d.L.; Fantin, C.; et al. Isolation and Identification of Pigment-Producing Endophytic Fungi from the Amazonian Species Fridericia chica. J. Fungi 2024, 10, 77. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- CLSI M27; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017.
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
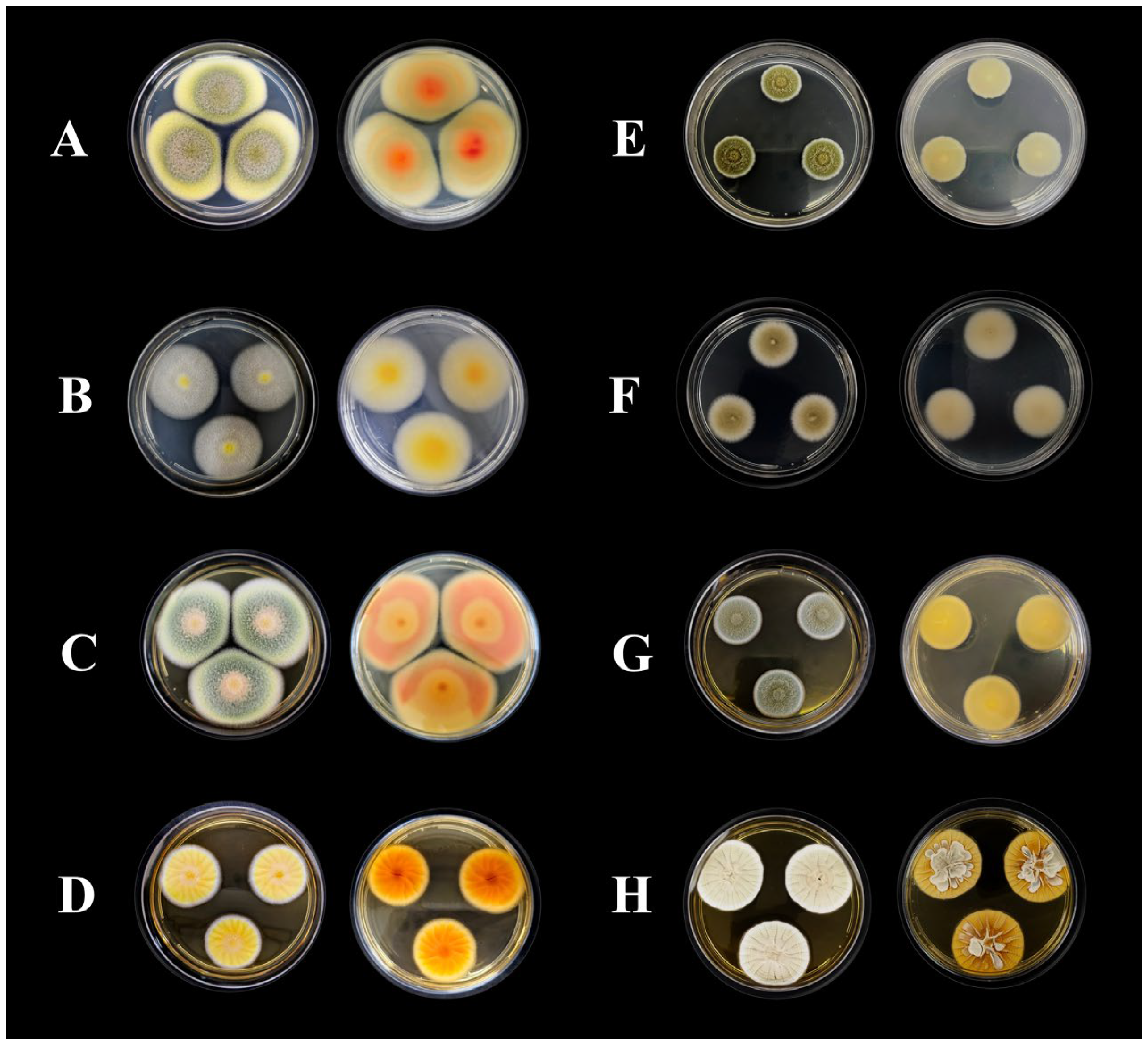





| Isolate | Species | Source | GenBank Accession Numbers | ||
|---|---|---|---|---|---|
| ITS | tub2 | rpb2 | |||
| CCM-UEA-F0414 | Talaromyces pinophilus | Myrcia guianensis | PQ336018 | - | PQ349268 |
| CCM-UEA-F0591 | Penicillium paxilli | Aniba canelilla | PQ336019 | PQ349270 | PQ349269 |
| Microorganisms | Fungal Extracts (MIC, µg/mL) | Positive Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. pinophilus CCM-UEA-F0414 | P. paxilli CCM-UEA-F0591 | |||||||||||
| B | I | Y | C | M | B | I | Y | C | M | Lev | Terb | |
| E. coli | - | 5000 | - | - | 5000 | 5000 | - | - | 5000 | 5000 | 0.061 | NT |
| B. subtilis | - | 1250 | 5000 | 5000 | 5000 | 5000 | - | 2500 | - | 2500 | 0.003 | NT |
| E. faecalis | - | - | - | 5000 | - | 5000 | - | - | - | 2500 | 1.953 | NT |
| K. pneumoniae | - | - | - | 5000 | - | - | - | - | - | 5000 | 250 | NT |
| P. aeruginosa | 5000 | 5000 | 5000 | - | - | 5000 | 2500 | - | 5000 | 5000 | 0.001 | NT |
| S. aureus | - | 5000 | 5000 | 156 | - | 2500 | 2.50 | - | - | - | 0.122 | NT |
| S.epidermidis | - | 5000 | 5000 | 1250 | - | 2500 | 313 | - | - | 5000 | 250 | NT |
| S. mutans | - | - | - | 2500 | - | - | 5000 | 5000 | - | 5000 | 0.0007 | NT |
| S. pyogenes | 1250 | - | 625 | 2500 | 1250 | - | 625 | 2500 | - | - | 0.003 | NT |
| S. enterica | - | - | - | 2500 | - | 5000 | - | - | - | 5000 | 0.0007 | NT |
| C. albicans | 156 | 156 | 78 | 313 | 156 | 78 | - | 5000 | - | 625 | NT | 50 |
| C. parapsilosis | - | - | - | 5000 | - | - | - | 2500 | 5000 | 5000 | NT | 400 |
| C. tropicalis | 2500 | 5000 | 5000 | 2500 | 5000 | 2500 | 625 | 1250 | 5000 | 2500 | NT | 25 |
| A. brasiliensis | 2500 | - | 2500 | 2500 | - | - | - | - | - | - | NT | 9.5 × 10−7 |
| Isolate | Medium | Concentration (mg/g of Extract) | |
|---|---|---|---|
| Caffeic Acid | Chlorogenic Acid | ||
| T. pinophilus CCM-UEA-F0414 | B | 1.7 | 0.43 |
| M | - | - | |
| I | - | - | |
| C | 1.6 | - | |
| Y | - | - | |
| P. paxilli CCM-UEA-F0591 | B | 2.2 | - |
| M | - | - | |
| I | 1.6 | - | |
| C | 0.6 | - | |
| Y | 0.6 | - | |
| Locus 1 | Primer | Sequence 5′–3′ | T (°C) 2 | Reference |
|---|---|---|---|---|
| ITS | Its1 | TCCGTAGGTGAACCTGCGG | 62 | [51] |
| Its4 | TCCTCCGCTTATTGATATGC | |||
| tub2 | Bt-2a | GGT AAC CAA ATC GGT GCT GCT TTC | 58 | [52] |
| Bt-2b | ACC CTC AGT GTA GTG ACC CTT GGC | |||
| rpb2 | RPB2-6F | TGG GGK WTG GTY TGY CCT GC | 58 | [53] |
| fRPB2-7cR | CCC ATR GCT TGY TTR CCC AT |
| Culture Medium | Composition (g/L) |
|---|---|
| BDL (B) | Potato (200), dextrose (20), and yeast extract (2) |
| Malt (M) | Malt extract (20) |
| Modified ISP2 (I) | Corn Starch (4), yeast extract (4), and malt extract (10) |
| Modified CZAPEK (C) | D-glucose (20), Fe2(SO4)3 (10), K2HPO4 (1); MgSO4 (0.5), KCl (0.5), and yeast extract (2) |
| YES (Y) | Sucrose (150), yeast extract (20), and MgSO4 (0.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Andrade, C.P.; Lacerda, C.D.; Valente, R.A.; Rocha, L.S.d.H.; de Souza, A.T.F.; Pereira, D.Í.d.M.; Barbosa, L.K.; Fantin, C.; Duvoisin Junior, S.; Albuquerque, P.M. An OSMAC Strategy for the Production of Antimicrobial Compounds by the Amazonian Fungi Talaromyces pinophilus CCM-UEA-F0414 and Penicillium paxilli CCM-UEA-F0591. Antibiotics 2025, 14, 756. https://doi.org/10.3390/antibiotics14080756
de Andrade CP, Lacerda CD, Valente RA, Rocha LSdH, de Souza ATF, Pereira DÍdM, Barbosa LK, Fantin C, Duvoisin Junior S, Albuquerque PM. An OSMAC Strategy for the Production of Antimicrobial Compounds by the Amazonian Fungi Talaromyces pinophilus CCM-UEA-F0414 and Penicillium paxilli CCM-UEA-F0591. Antibiotics. 2025; 14(8):756. https://doi.org/10.3390/antibiotics14080756
Chicago/Turabian Stylede Andrade, Cleudiane Pereira, Caroline Dutra Lacerda, Raíssa Assímen Valente, Liss Stone de Holanda Rocha, Anne Terezinha Fernandes de Souza, Dorothy Ívila de Melo Pereira, Larissa Kirsch Barbosa, Cleiton Fantin, Sergio Duvoisin Junior, and Patrícia Melchionna Albuquerque. 2025. "An OSMAC Strategy for the Production of Antimicrobial Compounds by the Amazonian Fungi Talaromyces pinophilus CCM-UEA-F0414 and Penicillium paxilli CCM-UEA-F0591" Antibiotics 14, no. 8: 756. https://doi.org/10.3390/antibiotics14080756
APA Stylede Andrade, C. P., Lacerda, C. D., Valente, R. A., Rocha, L. S. d. H., de Souza, A. T. F., Pereira, D. Í. d. M., Barbosa, L. K., Fantin, C., Duvoisin Junior, S., & Albuquerque, P. M. (2025). An OSMAC Strategy for the Production of Antimicrobial Compounds by the Amazonian Fungi Talaromyces pinophilus CCM-UEA-F0414 and Penicillium paxilli CCM-UEA-F0591. Antibiotics, 14(8), 756. https://doi.org/10.3390/antibiotics14080756









