Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle
Abstract
1. Introduction
2. Results
2.1. Serotype Confirmation
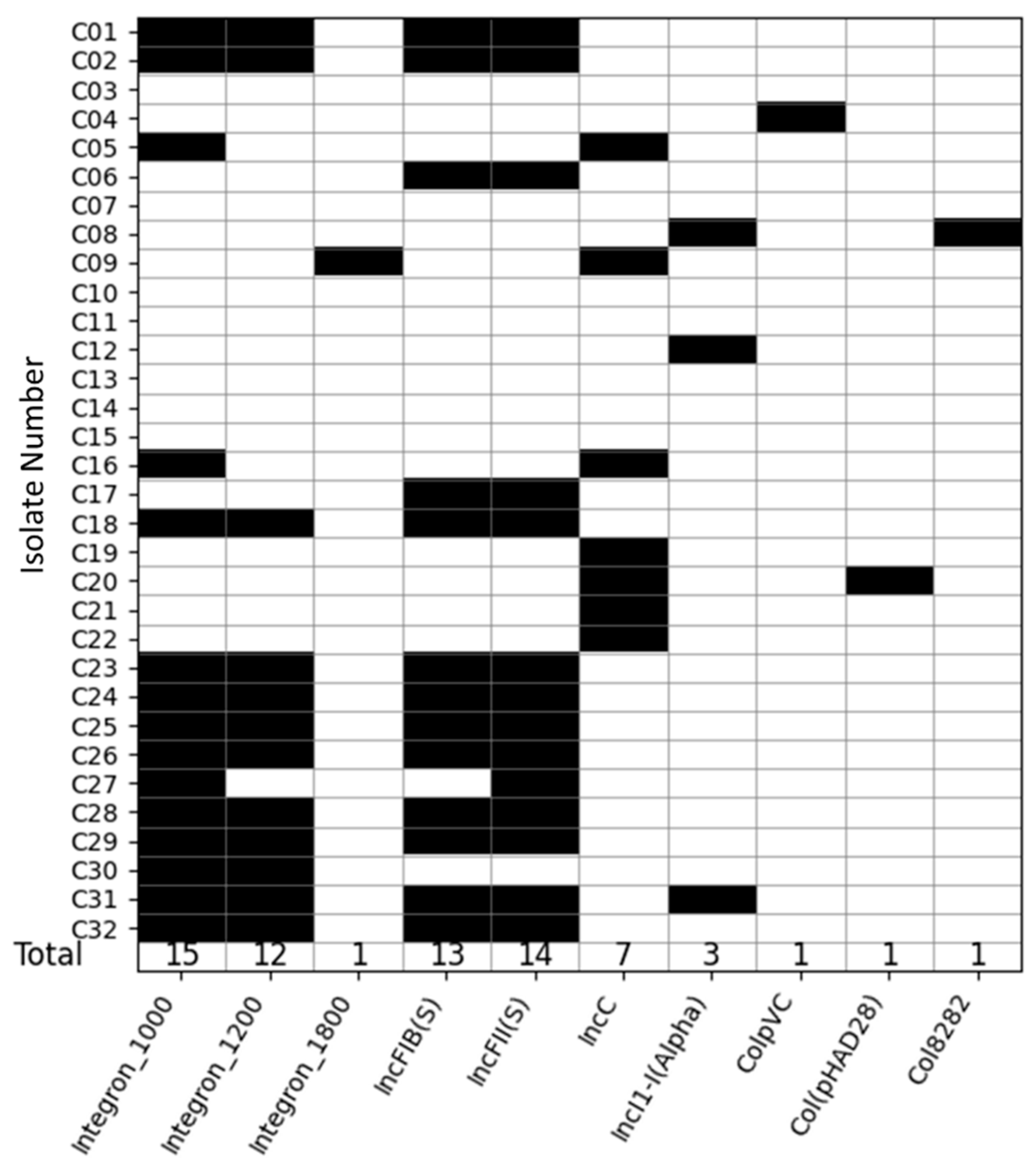
2.2. Integrons and Plasmids Identified
2.3. Identification of Antimicrobial Resistance Genes
2.4. Association of Integrons and AMR Genes
2.5. Location of Integrons and AMR Genes
2.6. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Study Design and Sample Collection
4.2. S. Typhimurium Identification and Antimicrobial Sensitivity Testing
4.3. Antimicrobial Susceptibility Testing
4.4. DNA Extraction
4.5. Integron Identification and Sequencing
4.6. Whole Genome Sample Processing
4.7. Whole Genome Denovo Assembly
4.8. Confirmation of S. Typhimurium and Sequence Typing
4.9. Identification of Plasmids
4.10. Establishing Integron Genomic Locations
4.11. Identification of Antimicrobial Resistance Genes and Their Genomic Location
4.12. Assessment of Genomic Diversity Through Core Genome Phylogeny
4.13. Data Analysis and Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial Resistance |
| ARG | Antimicrobial Resistance Genes |
| WGS | Whole Genome Sequencing |
| BLAST | Basic Local Alignment Search Tool |
| S. Typhimurium | Salmonella enterica serovar Typhimurium |
References
- Kumar, S. Antimicrobial Resistance: A Top Ten Global Public Health Threat. eClinicalMedicine 2021, 41, 101221. [Google Scholar] [CrossRef]
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial Resistance: Impacts, Challenges, and Future Prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Goossens, H.; Ferech, M.; Vander Stichele, R.; Elseviers, M. Outpatient Antibiotic Use in Europe and Association with Resistance: A Cross-National Database Study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef] [PubMed]
- DiMarzio, M.; Shariat, N.; Kariyawasam, S.; Barrangou, R.; Dudley, E.G. Antibiotic Resistance in Salmonella enterica Serovar Typhimurium Associates with CRISPR Sequence Type. Antimicrob. Agents Chemother. 2013, 57, 4282–4289. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Vila, J. Salmonella enterica Serovar Typhimurium Skills To Succeed in the Host: Virulence and Regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Gong, B.; Li, H.; Feng, Y.; Zeng, S.; Zhuo, Z.; Luo, J.; Chen, X.; Li, X. Prevalence, Serotype Distribution and Antimicrobial Resistance of Non-Typhoidal Salmonella in Hospitalized Patients in Conghua District of Guangzhou, China. Front. Cell. Infect. Microbiol. 2022, 12, 805384. [Google Scholar] [CrossRef]
- CDC Salmonella Homepage|CDC. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 30 September 2022).
- Izumiya, H.; Sekizuka, T.; Nakaya, H.; Taguchi, M.; Oguchi, A.; Ichikawa, N.; Nishiko, R.; Yamazaki, S.; Fujita, N.; Watanabe, H.; et al. Whole-Genome Analysis of Salmonella enterica Serovar Typhimurium T000240 Reveals the Acquisition of a Genomic Island Involved in Multidrug Resistance via IS 1 Derivatives on the Chromosome. Antimicrob. Agents Chemother. 2011, 55, 623–630. [Google Scholar] [CrossRef]
- Singh, R.; Mukherjee, S.; Harrision, L.B.; McDermott, P.F.; Ge, B.; Gilbert, J.M.; Li, C.; Whichard, J.M.; Fortenberry, G.Z.; Dessai, U.; et al. Cross-Resistance to 14-, 15- and 16-Membered Ring Macrolides in Salmonella and Campylobacter. J. Antimicrob. Chemother. 2025, 80, 1445–1452. [Google Scholar] [CrossRef]
- Hendriksen, S.W.M.; Orsel, K.; Wagenaar, J.A.; Miko, A.; Van Duijkeren, E. Animal-to-Human Transmission of Salmonella Typhimurium DT104A Variant. Emerg. Infect. Dis. 2004, 10, 2225–2227. [Google Scholar] [CrossRef]
- Gutema, F.D.; Agga, G.E.; Abdi, R.D.; De Zutter, L.; Duchateau, L.; Gabriël, S. Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017. Front. Vet. Sci. 2019, 6, 102. [Google Scholar] [CrossRef]
- Habing, G.G.; Lombard, J.E.; Kopral, C.A.; Dargatz, D.A.; Kaneene, J.B. Farm-Level Associations with the Shedding of Salmonella and Antimicrobial-Resistant Salmonella in U.S. Dairy Cattle. Foodborne Pathog. Dis. 2012, 9, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.; Mir, R.A.; Qadri, H.; Dhiman, R.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Integrons in the Development of Antimicrobial Resistance: Critical Review and Perspectives. Front. Microbiol. 2023, 14, 1231938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, B.; Zhang, Y.; Zhang, T.; Yang, L.; Fang, H.H.P.; Ford, T.; Cheng, S. Class 1 Integronase Gene and Tetracycline Resistance Genes tetA and tetC in Different Water Environments of Jiangsu Province, China. Ecotoxicology 2009, 18, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Krishnan, S.; Worden, P.J.; Hardwick, S.A. Recovery of Diverse Genes for Class 1 Integron-Integrases from Environmental DNA Samples. FEMS Microbiol. Lett. 2008, 287, 56–62. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef]
- Rao, S.; Linke, L.; Doster, E.; Hyatt, D.; Burgess, B.A.; Magnuson, R.; Pabilonia, K.L.; Morley, P.S. Genomic Diversity of Class I Integrons from Antimicrobial Resistant Strains of Salmonella Typhimurium Isolated from Livestock, Poultry and Humans. PLoS ONE 2020, 15, e0243477. [Google Scholar] [CrossRef]
- Kim, E.; Nealon, N.J.; Murray, K.A.; Jardine, C.; Magnuson, R.; Rao, S. Integron-Mediated Antimicrobial Resistance and Virulence Factors in Salmonella Typhimurium Isolated from Poultry. Animals 2024, 14, 3483. [Google Scholar] [CrossRef]
- Ali, M.S.; Na, S.-H.; Moon, B.-Y.; Kang, H.Y.; Kang, H.-S.; Kim, S.-J.; Kim, T.-S.; Heo, Y.-E.; Hwang, Y.-J.; Yoon, S.S.; et al. Antimicrobial Resistance Profiles and Molecular Characteristics of Extended-Spectrum β-Lactamase-Producing Salmonella enterica Serovar Typhimurium Isolates from Food Animals During 2010–2021 in South Korea. Foodborne Pathog. Dis. 2024, 21, 634–642. [Google Scholar] [CrossRef]
- Bawn, M.; Alikhan, N.-F.; Thilliez, G.; Kirkwood, M.; Wheeler, N.E.; Petrovska, L.; Dallman, T.J.; Adriaenssens, E.M.; Hall, N.; Kingsley, R.A. Evolution of Salmonella enterica Serotype Typhimurium Driven by Anthropogenic Selection and Niche Adaptation. PLoS Genet. 2020, 16, e1008850. [Google Scholar] [CrossRef]
- Khan, M.; Shamim, S. Understanding the Mechanism of Antimicrobial Resistance and Pathogenesis of Salmonella enterica Serovar Typhi. Microorganisms 2022, 10, 2006. [Google Scholar] [CrossRef]
- Zhang, S.; Den Bakker, H.C.; Li, S.; Chen, J.; Dinsmore, B.A.; Lane, C.; Lauer, A.C.; Fields, P.I.; Deng, X. SeqSero2: Rapid and Improved Salmonella Serotype Determination Using Whole-Genome Sequencing Data. Appl. Environ. Microbiol. 2019, 85, e01746-19. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Integron and Its Role in Antimicrobial Resistance: A Literature Review on Some Bacterial Pathogens. Iran. J. Basic Med. Sci. 2021, 24, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Bao, X.; Ji, L.; Chen, L.; Liu, J.; Miao, J.; Chen, D.; Bian, H.; Li, Y.; Yu, G. Resistance Integrons: Class 1, 2 and 3 Integrons. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 45. [Google Scholar] [CrossRef]
- Asgharpour, F.; Rajabnia, R.; Ferdosi Shahandashti, E.; Marashi, M.A.; Khalilian, M.; Moulana, Z. Investigation of Class I Integron in Salmonella infantis and Its Association with Drug Resistance. Jundishapur J. Microbiol. 2014, 7, e10019. [Google Scholar] [CrossRef]
- Antunes, P. Characterization of Antimicrobial Resistance and Class 1 and 2 Integrons in Salmonella enterica Isolates from Different Sources in Portugal. J. Antimicrob. Chemother. 2006, 58, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Majtánová, L.; Majtán, T.; Majtán, V. Detection of the Class 1 Integrons and SGI1 among Salmonella enterica Serovar Typhimurium DT104, U302, DT120, DT193, and Nontypable Human Isolates. Jpn. J. Infect. Dis. 2010, 63, 292–295. [Google Scholar] [CrossRef]
- Monte, D.F.M.; Sellera, F.P.; Lopes, R.; Keelara, S.; Landgraf, M.; Greene, S.; Fedorka-Cray, P.J.; Thakur, S. Class 1 Integron-Borne Cassettes Harboring blaCARB-2 Gene in Multidrug-Resistant and Virulent Salmonella Typhimurium ST19 Strains Recovered from Clinical Human Stool Samples, United States. PLoS ONE 2020, 15, e0240978. [Google Scholar] [CrossRef] [PubMed]
- Pavelquesi, S.L.S.; De Oliveira Ferreira, A.C.A.; Rodrigues, A.R.M.; De Souza Silva, C.M.; Orsi, D.C.; Da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef]
- Glenn, L.M.; Lindsey, R.L.; Frank, J.F.; Meinersmann, R.J.; Englen, M.D.; Fedorka-Cray, P.J.; Frye, J.G. Analysis of Antimicrobial Resistance Genes Detected in Multidrug-Resistant Salmonella enterica Serovar Typhimurium Isolated from Food Animals. Microb. Drug Resist. 2011, 17, 407–418. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of Sulfonamide Resistance Genes (Sul1, Sul2, and Sul3) in Portuguese Salmonella enterica Strains and Relation with Integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Khemtong, S.; Padungtod, P. Occurrence of qacE/qacEDelta1 Genes and Their Correlation with Class 1 Integrons in Salmonella enterica Isolates from Poultry and Swine. Southeast Asian J. Trop. Med. Public Health 2007, 38, 855–862. [Google Scholar] [PubMed]
- Kamolvit, W.; Derrington, P.; Paterson, D.L.; Sidjabat, H.E. A Case of IMP-4-, OXA-421-, OXA-96-, and CARB-2-Producing Acinetobacterpittii Sequence Type 119 in Australia. J. Clin. Microbiol. 2015, 53, 727–730. [Google Scholar] [CrossRef]
- Gallardo, F.; Ruiz, J.; Soto, S.M.; Jimenez de Anta, M.T.; Vila, J. Different antibiotic resistance mechanisms associated with integrons in clinical isolates of Salmonella typhimurium. Rev. Esp. Quimioter. 2003, 16, 398–402. [Google Scholar]
- Giles, W.P.; Benson, A.K.; Olson, M.E.; Hutkins, R.W.; Whichard, J.M.; Winokur, P.L.; Fey, P.D. DNA Sequence Analysis of Regions Surrounding BlaCMY-2 from Multiple Salmonella Plasmid Backbones. Antimicrob. Agents Chemother. 2004, 48, 2845–2852. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carattoli, A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the Wild: Antibiotic Resistance Genes in Natural Environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Ruzante, J.M.; Lombard, J.E.; Wagner, B.; Fossler, C.P.; Karns, J.S.; Van Kessel, J.A.S.; Gardner, I.A. Factors Associated with Salmonella Presence in Environmental Samples and Bulk Tank Milk from US Dairies. Zoonoses Public Health 2010, 57, e217–e225. [Google Scholar] [CrossRef]
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Thakur, S. Genetic Relatedness of Multidrug Resistant Escherichia coli Isolated from Humans, Chickens and Poultry Environments. Antimicrob. Resist. Infect. Control 2021, 10, 58. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; CLSI Document M100-S24; CLSI: Wayne, PA, USA, 2014. [Google Scholar]
- Lucey, B.; Crowley, D.; Moloney, P.; Cryan, B.; Daly, M.; O’Halloran, F.; Threlfall, E.J.; Fanning, S. Integronlike Structures in Campylobacter spp. of Human and Animal Origin. Emerg. Infect. Dis. 2000, 6, 50–55. [Google Scholar] [CrossRef]
- Rao, S.; Maddox, C.W.; Hoien-Dalen, P.; Lanka, S.; Weigel, R.M. Diagnostic Accuracy of Class 1 Integron PCR Method in Detection of Antibiotic Resistance in Salmonella Isolates from Swine Production Systems. J. Clin. Microbiol. 2008, 46, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Fenske, G.J.; Antony, L.; Ghimire, S.; Welsh, R.; Ramachandran, A.; Scaria, J. Whole Genome Sequencing-Based Detection of Antimicrobial Resistance and Virulence in Non-Typhoidal Salmonella enterica Isolated from Wildlife. Gut Pathog. 2017, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBDuk Plugin for Geneious Prime, Version 1.0. Biomatters Ltd.: Auckland, New Zealand, 2015. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Worley, J.; Meng, J.; Allard, M.W.; Brown, E.W.; Timme, R.E. Salmonella enterica Phylogeny Based on Whole-Genome Sequencing Reveals Two New Clades and Novel Patterns of Horizontally Acquired Genetic Elements. mBio 2018, 9, e02303-18. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahadur, S.U.K.; Nealon, N.J.; Daniels, J.B.; Zaheer, M.U.; Salman, M.; Rao, S. Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle. Antibiotics 2025, 14, 633. https://doi.org/10.3390/antibiotics14070633
Bahadur SUK, Nealon NJ, Daniels JB, Zaheer MU, Salman M, Rao S. Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle. Antibiotics. 2025; 14(7):633. https://doi.org/10.3390/antibiotics14070633
Chicago/Turabian StyleBahadur, Sami Ullah Khan, Nora Jean Nealon, Joshua B. Daniels, Muhammad Usman Zaheer, Mo Salman, and Sangeeta Rao. 2025. "Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle" Antibiotics 14, no. 7: 633. https://doi.org/10.3390/antibiotics14070633
APA StyleBahadur, S. U. K., Nealon, N. J., Daniels, J. B., Zaheer, M. U., Salman, M., & Rao, S. (2025). Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle. Antibiotics, 14(7), 633. https://doi.org/10.3390/antibiotics14070633







