Oxacillin-Supplemented Mueller-Hinton Agar for In Vitro Inhibition of Ambler Class C β-Lactamases in Enterobacterales
Abstract
1. Introduction
2. Results
2.1. Oxacillin Concentration Determination
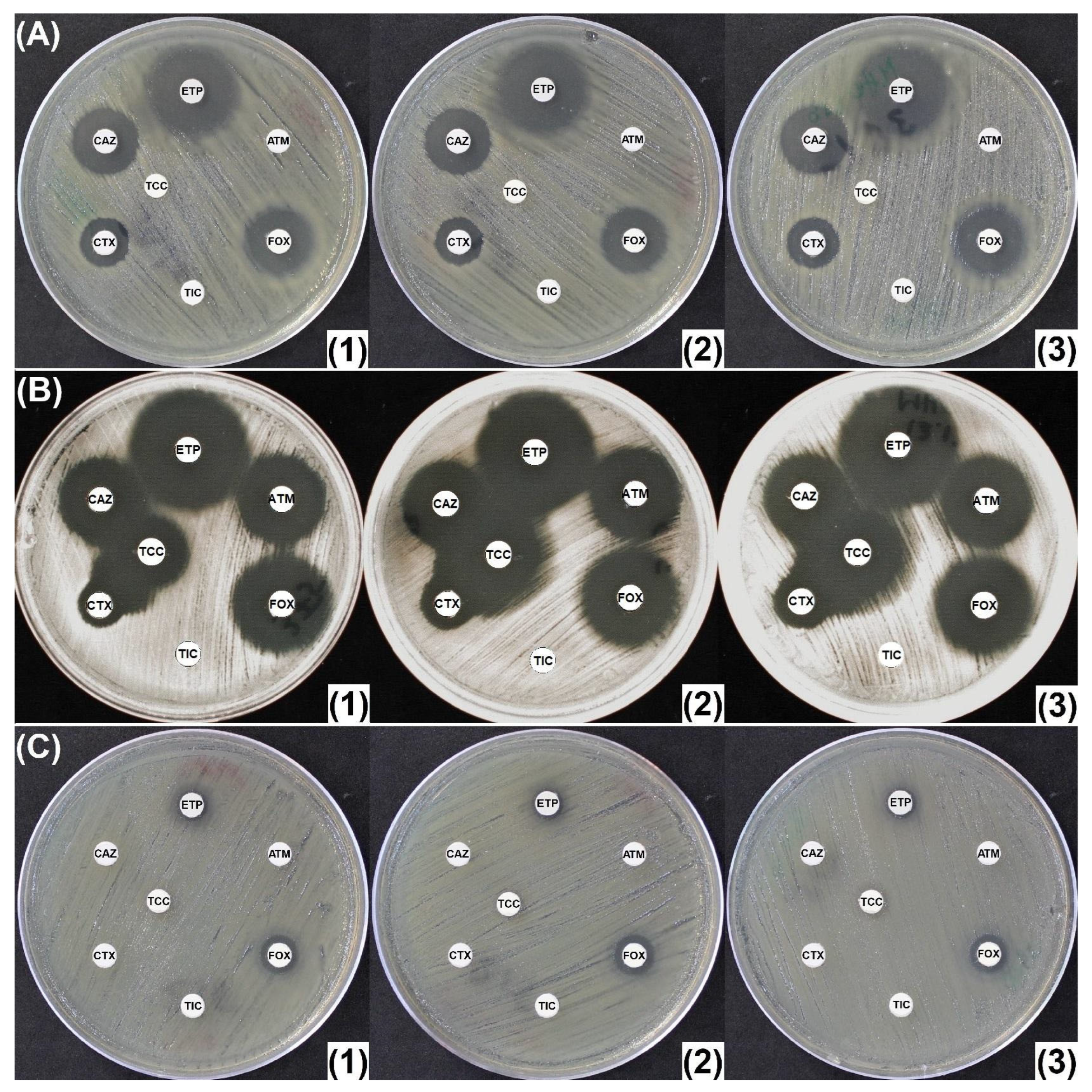
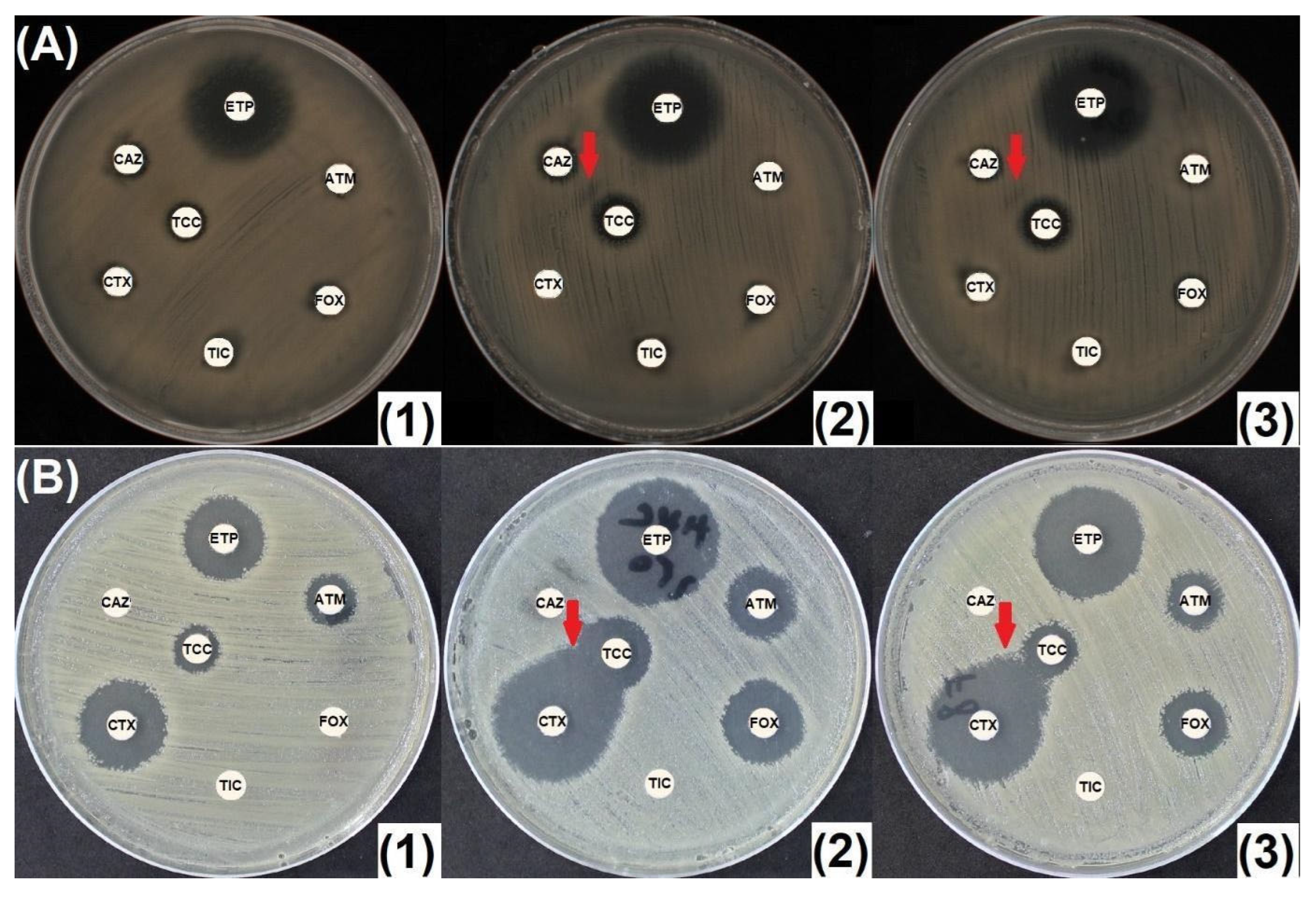
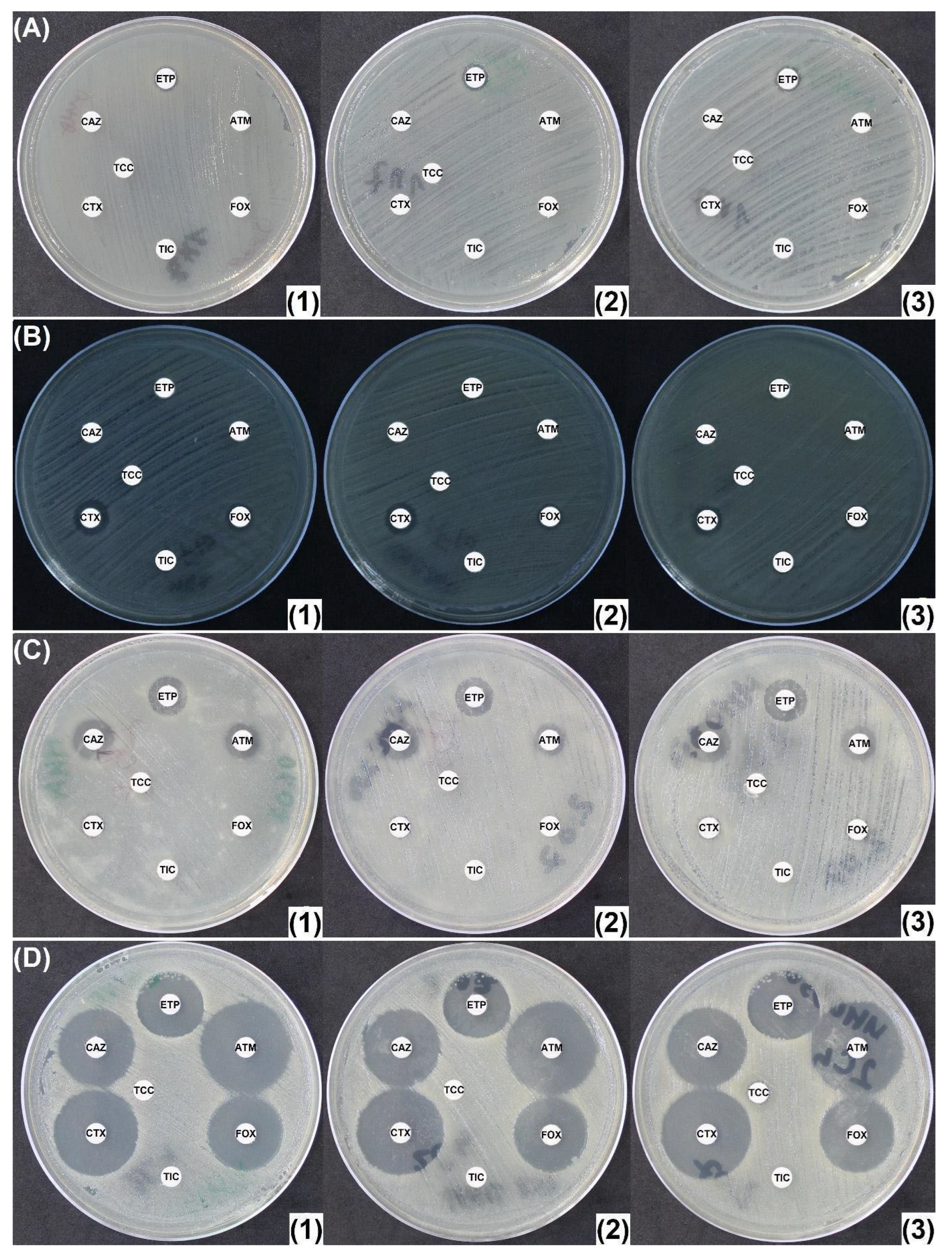
2.2. Penicillinase and ESBL Producers
2.3. CASE and HCASE Producers
2.4. Carbapenemase Producers
3. Discussion
4. Materials and Methods
4.1. Media Preparation
4.2. Susceptibility Testing
4.3. Bacterial Isolates Tested
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AmpC | Chromosomal cephalosporinase |
| AmpC-PE | AmpC-producing Enterobacterales |
| AHE | AmpC-hyperproducing Enterobacterales |
| ATM | Aztreonam |
| ATCC | American Type Culture Collection |
| BSPASE | Broad-spectrum penicillinase |
| CASE | Cephalosporinase |
| CAZ | Ceftazidime |
| CRE | Carbapenem-resistant Enterobacterales |
| CTX | Cefotaxime |
| ESBL | Extended-spectrum β-lactamase |
| ESC | Extended-spectrum cephalorporin |
| ETP | Ertapenem |
| EUCAST | The European Committee on Antimicrobial Susceptibility Testing |
| FOX | Cefoxitin |
| GNB | Gram-negative bacilli |
| HCASE | Hyperproduced cephalosporinase |
| MDR | Multidrug-resistant |
| MHA | Mueller-Hinton agar |
| MHC | Mueller-Hinton agar supplemented with cloxacillin |
| MHC250 | Mueller-Hinton agar supplemented with 250 mg/L cloxacillin |
| MHO | Mueller-Hinton agar supplemented with oxacillin |
| MHO250/300/350/400 | Mueller-Hinton agar supplemented with 250/300/350/400 mg/L oxacillin |
| MBL | Metallo-β-lactamase |
| NA | Not applicable |
| PASE | Penicillinase |
| PBP | Penicillin binding protein |
| PCR | Polymerase chain reaction |
| rCIM | Rapid carbapenem inactivation method |
| RDCK™ | Rosco Diagnostica KPC and MBL confirm kit |
| TIC | Ticarcillin |
| TCC | Ticarcillin/clavulanic acid |
| WT | Wild-type |
| WGS | Whole-genome sequencing |
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-Producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 2.0. 2017. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 14 March 2025).
- Guillon, H.; Tande, D.; Mammeri, H. Emergence of Ertapenem Resistance in an Escherichia coli Clinical Isolate Producing Extended-Spectrum β-Lactamase AmpC. Antimicrob. Agents Chemother. 2011, 55, 4443–4446. [Google Scholar] [CrossRef]
- AbdelGhani, S.; Thomson, G.K.; Snyder, J.W.; Thomson, K.S. Comparison of the Carba NP, Modified Carba NP, and Updated Rosco Neo-Rapid Carb Kit Tests for Carbapenemase Detection. J. Clin. Microbiol. 2015, 53, 3539–3542. [Google Scholar] [CrossRef]
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic Detection of Extended-Spectrum β-Lactamase Production in Enterobacteriaceae: Review and Bench Guide. Clin. Microbiol. Infect. 2008, 14, 90–103. [Google Scholar] [CrossRef]
- Garrec, H.; Drieux-Rouzet, L.; Golmard, J.-L.; Jarlier, V.; Robert, J. Comparison of Nine Phenotypic Methods for Detection of Extended-Spectrum β-Lactamase Production by Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 1048–1057. [Google Scholar] [CrossRef]
- Bartolini, A.; Frasson, I.; Cavallaro, A.; Richter, S.N.; Palù, G. Comparison of Phenotypic Methods for the Detection of Carbapenem Non-Susceptible Enterobacteriaceae. Gut Pathog. 2014, 6, 13. [Google Scholar] [CrossRef]
- Robert, J.; Pantel, A.; Merens, A.; Meiller, E.; Lavigne, J.-P.; Nicolas-Chanoine, M.-H. Development of an Algorithm for Phenotypic Screening of Carbapenemase-Producing Enterobacteriaceae in the Routine Laboratory. BMC Infect. Dis. 2017, 17, 78. [Google Scholar] [CrossRef]
- Van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Leverstein-Van Hall, M.A.; Stuart, J.W.T.C. A Disc Diffusion Assay for Detection of Class A, B and OXA-48 Carbapenemases in Enterobacteriaceae Using Phenyl Boronic Acid, Dipicolinic Acid and Temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef]
- Mueller, J.H.; Hinton, J. A Protein-Free Medium for Primary Isolation of the Gonococcus and Meningococcus. Exp. Biol. Med. 1941, 48, 330–333. [Google Scholar] [CrossRef]
- Ananthan, S.; Subha, A. Cefoxitin Resistance Mediated by Loss of a Porin in Clinical Strains of Klebsiella pneumoniae and Escherichia coli. Indian J. Med. Microbiol. 2005, 23, 20–23. [Google Scholar] [CrossRef]
- Coudron, P.E. Inhibitor-Based Methods for Detection of Plasmid-Mediated AmpC β-Lactamases in Klebsiella Spp., Escherichia coli , and Proteus mirabilis. J. Clin. Microbiol. 2005, 43, 4163–4167. [Google Scholar] [CrossRef]
- Soliman, A.M.; Nariya, H.; Tanaka, D.; Shimamoto, T.; Shimamoto, T. A Novel Single-Tube Eicosaplex/Octaplex PCR System for the Detection of Extended-Spectrum β-Lactamases, Plasmid-Mediated AmpC β-Lactamases, and Integrons in Gram-Negative Bacteria. Antibiotics 2023, 12, 90. [Google Scholar] [CrossRef]
- Perera, V.; Silva, N.D.; Jayatilleke, K.; Silva, S.D.; Corea, E. Performance of Phenotypic Tests to Detect β-Lactamases in a Population of β-Lactamase Coproducing Enterobacteriaceae Isolates. J. Lab. Physicians 2023, 15, 117–125. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 6098, Cloxacillin. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Cloxacillin (accessed on 14 March 2025).
- Michael Marcy, S.; Klein, J.O. The Isoxazolyl Penicillins: Oxacillin, Cloxacillin, and Dicloxacillin. Med. Clin. N. Am. 1970, 54, 1127–1143. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Larsen, D.; Huber, T. Identification of Extended-Spectrum Beta-Lactamases in Clinical Isolates of Enterobacter cloacae: Cloxacillin-Infused Agar Enhances Detection. Am. J. Clin. Pathol. 2012, 138, A059. [Google Scholar] [CrossRef]
- Dortet, L.; Cuzon, G.; Plésiat, P.; Naas, T. Prospective Evaluation of an Algorithm for the Phenotypic Screening of Carbapenemase-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 2016, 71, 135–140. [Google Scholar] [CrossRef]
- Takayama, Y.; Adachi, Y.; Nihonyanagi, S.; Okamoto, R. Modified Hodge Test Using Mueller–Hinton Agar Supplemented with Cloxacillin Improves Screening for Carbapenemase-Producing Clinical Isolates of Enterobacteriaceae. J. Med. Microbiol. 2015, 64, 774–777. [Google Scholar] [CrossRef]
- Maurer, F.P.; Castelberg, C.; Quiblier, C.; Bloemberg, G.V.; Hombach, M. Evaluation of Carbapenemase Screening and Confirmation Tests with Enterobacteriaceae and Development of a Practical Diagnostic Algorithm. J. Clin. Microbiol. 2015, 53, 95–104. [Google Scholar] [CrossRef]
- Birgy, A.; Bidet, P.; Genel, N.; Doit, C.; Decré, D.; Arlet, G.; Bingen, E. Phenotypic Screening of Carbapenemases and Associated β-Lactamases in Carbapenem-Resistant Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 1295–1302. [Google Scholar] [CrossRef]
- Fournier, D.; Garnier, P.; Jeannot, K.; Mille, A.; Gomez, A.-S.; Plésiat, P. A Convenient Method To Screen for Carbapenemase-Producing Pseudomonas aeruginosa. J. Clin. Microbiol. 2013, 51, 3846–3848. [Google Scholar] [CrossRef]
- CNR Résistance aux Antibiotiques. Detection des Beta-Lactamases a Spectre Etendu de Classe a et d par la Realisation de Tests de Synergie Chez Pseudomonas aeruginosa, Fiche D’informations. 2017. Available online: https://www.cnr-resistance-antibiotiques.fr/ressources/pages/Tests_synergie_BLSE_V1.pdf (accessed on 14 March 2025).
- Jeannot, K.; Fournier, D.; Müller, E.; Cholley, P.; Plésiat, P. Clonal Dissemination of Pseudomonas aeruginosa Isolates Producing Extended-Spectrum β-Lactamase SHV-2a. J. Clin. Microbiol. 2013, 51, 673–675. [Google Scholar] [CrossRef]
- Fournier, D.; Grisot, E.; Potron, A.; Liapis, E.; Jeannot, K.; Plésiat, P. Nouvelle Stratégie de Dépistage des P. aeruginosa BLSE ou Carbapénèmase Positifs. Available online: https://www.cnr-resistance-antibiotiques.fr/ressources/pages/Poster_RICAI_CT_algorithmev2.pdf (accessed on 14 March 2025).
- CNR Résistance aux Antibiotiques. Etude des Mécanismes de Résistance Chez A. baumannii: Logigramme. Version 5. 2024. Available online: https://www.cnr-resistance-antibiotiques.fr/ressources/pages/v5_logigramme_prise_en_charge_Acinetobacter.pdf (accessed on 14 March 2025).
- CNR Résistance aux Antibiotiques. Etude des Mécanismes de Résistance Chez P. aeruginosa: Logigramme. Version 5. 2024. Available online: https://www.cnr-resistance-antibiotiques.fr/ressources/pages/v5_logigramme_prise_en_charge_Pseudomonas.pdf (accessed on 14 March 2025).
- Poirel, L.; Menuteau, O.; Agoli, N.; Cattoen, C.; Nordmann, P. Outbreak of Extended-Spectrum β-Lactamase VEB-1-Producing Isolates of Acinetobacter baumannii in a French Hospital. J. Clin. Microbiol. 2003, 41, 3542–3547. [Google Scholar] [CrossRef]
- Jean, S.-S.; Harnod, D.; Hsueh, P.-R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Manquat, E.; Le Dorze, M.; Pean De Ponfilly, G.; Benmansour, H.; Amarsy, R.; Cambau, E.; Soyer, B.; Chousterman, B.G.; Jacquier, H. Impact of Systematic Screening for AmpC-Hyperproducing Enterobacterales Intestinal Carriage in Intensive Care Unit Patients. Ann. Intensive Care 2020, 10, 149. [Google Scholar] [CrossRef]
- Duque, M.; Bonnin, R.A.; Dortet, L. Comparison of the French Novel Disc Diffusion-Based Algorithm and the Current EUCAST Guidelines for the Screening of Carbapenemase-Producing Enterobacterales. J. Antimicrob. Chemother. 2024, 79, 1194–1196. [Google Scholar] [CrossRef]
- Ferous, S.; Anastassopoulou, C.; Pitiriga, V.; Vrioni, G.; Tsakris, A. Antimicrobial and Diagnostic Stewardship of the Novel β-Lactam/β-Lactamase Inhibitors for Infections Due to Carbapenem-Resistant Enterobacterales Species and Pseudomonas aeruginosa. Antibiotics 2024, 13, 285. [Google Scholar] [CrossRef]
- De Champs, C.; Poirel, L.; Bonnet, R.; Sirot, D.; Chanal, C.; Sirot, J.; Nordmann, P. Prospective Survey of β-Lactamases Produced by Ceftazidime- Resistant Pseudomonas aeruginosa Isolated in a French Hospital in 2000. Antimicrob. Agents Chemother. 2002, 46, 3031–3034. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. EUCAST Disk Diffusion Test Methodology. 2025. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology (accessed on 14 March 2025).
- Livermore, D.M.; Oakton, K.J.; Carter, M.W.; Warner, M. Activity of Ertapenem (MK-0826) versus Enterobacteriaceae with Potent β-Lactamases. Antimicrob. Agents Chemother. 2001, 45, 2831–2837. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Muntean, M.M.; Muntean, A.-A.; Guerin, F.; Cattoir, V.; Creton, E.; Cotellon, G.; Oueslati, S.; Popa, M.I.; Girlich, D.; Iorga, B.I.; et al. Optimization of the Rapid Carbapenem Inactivation Method for Use with AmpC Hyperproducers. J. Antimicrob. Chemother. 2021, 76, 2294–2301. [Google Scholar] [CrossRef]

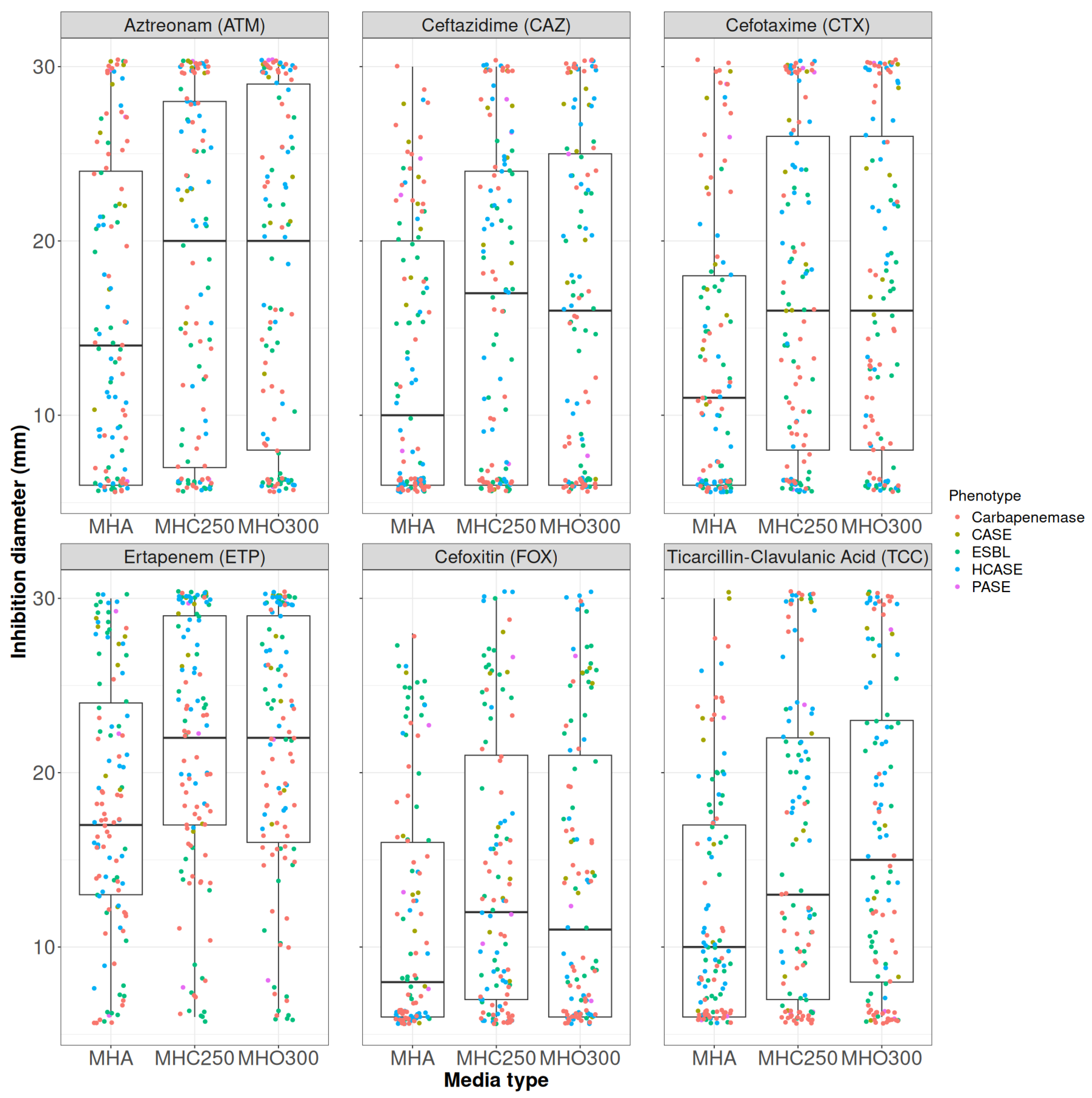

| Kruskal–Wallis Test MHA-MHC250-MHO300 | Wilcoxon Signed-Rank Test MHC250-MHO300 | |
|---|---|---|
| Aztreonam | 0.05355 | 0.7152 |
| Ceftazidime | 0.001271 * | 0.05592 |
| Cefotaxime | 0.00101 * | 0.9244 |
| Cefoxitin | 0.006151 * | 0.1013 |
| Ticarcillin | 0.1358 | 0.04704 * |
| Ticarcillin/clavulanic acid | 0.001322 * | 0.1073 |
| Ertapenem | 0.0004103 * | 0.5564 |
| No. | Species | Phenotype | β-Lactamase Content | Carbapenemase |
|---|---|---|---|---|
| 1 | K. pneumoniae | PASE | SHV-28 | NA |
| 2 | K. pneumoniae | BSPASE | TEM-1 + SHV-28 | NA |
| 3 | E. coli | BSPASE | TEM-1B | NA |
| 4 | K. pneumoniae | ESBL | SHV-11 | NA |
| 5 | E. coli | ESBL | CTX-M-1 | NA |
| 6 | E. coli | ESBL | CTX-M-3 | NA |
| 7 | K. pneumoniae | ESBL | CTX-M-3 | NA |
| 8 | E. coli | ESBL | CTX-M-14 | NA |
| 9 | K. pneumoniae | ESBL | CTX-M-14 | NA |
| 10 | E. coli | ESBL | CTX-M-15 | NA |
| 11 | E. coli | ESBL | CTX-M-15 | NA |
| 12 | K. pneumoniae | ESBL | CTX-M-15 | NA |
| 13 | K. pneumoniae | ESBL | CTX-M-15 | NA |
| 14 | K. pneumoniae | ESBL | CTX-M-15 | NA |
| 15 | E. cloacae | ESBL | VEB-1 | NA |
| 16 | E. coli | ESBL | CTX-M-15 | NA |
| 17 | K. pneumoniae | ESBL | CTX-M-15 + SHV-1 | NA |
| 18 | K. pneumoniae | ESBL | CTX-M-15 + SHV-11 | NA |
| 19 | K. pneumoniae | ESBL | CTX-M-15 + TEM-1 + SHV-11 | NA |
| 20 | K. pneumoniae | ESBL | CTX-M-15 + TEM-1 + SHV-11 | NA |
| 21 | K. pneumoniae | ESBL | CTX-M-15 + TEM-1 + SHV-11 | NA |
| 22 | K. pneumoniae | ESBL | CTX-M-15 + TEM-1 + SHV-12 | NA |
| 23 | K. pneumoniae | ESBL | CTX-M-15 +TEM-1 + SHV-11 | NA |
| 24 | K. pneumoniae | ESBL | CTX-M-15 + TEM-1 + SHV-1 + OXA-1 | NA |
| 25 | K. pneumoniae | ESBL | CTX-M-15, TEM-1B, SHV-28 | NA |
| 26 | K. pneumoniae | ESBL | CTX-M-15, TEM-1B, SHV-83 | NA |
| 27 | K. pneumoniae | ESBL | CTX-M-14 | NA |
| 28 | E. coli | ESBL | CTX-M-2 | NA |
| 29 | K. oxytoca | ESBL | Hyper-K1 (OXY-2-8) | NA |
| 30 | K. pneumoniae | ESBL + BSPASE | CTX-M-15 + TEM-1 + SHV-1 | NA |
| 31 | K. pneumoniae | ESBL + BSPASE | CTX-M-15 + TEM-1 + SHV-1 | NA |
| 32 | K. pneumoniae | ESBL + BSPASE | CTX-M-15 + SHV-28 + TEM-1 | NA |
| 33 | E. coli | Class D ESBL | OXA-1 | NA |
| 34 | K. oxytoca | Class D ESBL | OXA-2-3 | NA |
| 35 * | E. cloacae | CASE | CASE | NA |
| 36 | E. cloacae | CASE | CASE | NA |
| 37 | M. morganii | CASE | CASE | NA |
| 38 | E. coli | CASE plasmidic | CMY-2 | NA |
| 39 * | E. cloacae | CASE + ESBL | CASE + ESBL | NA |
| 40 | E. cloacae | CASE + ESBL | CASE + CTX-M-15 | NA |
| 41 | K. aerogenes | CASE + ESBL | CASE + TEM-24 | NA |
| 42 | E. cloacae | CASE + Class D ESBL | CASE + OXA-35 + ACT-6 | NA |
| 43 * | E. coli | HCASE | HCASE | NA |
| 44 * | E. cloacae | HCASE | HCASE | NA |
| 45 * | E. cloacae | HCASE | HCASE | NA |
| 46 * | E. cloacae | HCASE | HCASE | NA |
| 47 | E. cloacae | HCASE | HCASE | NA |
| 48 | E. cloacae | HCASE | HCASE | NA |
| 49 | E. cloacae | HCASE | HCASE | NA |
| 50 | E. cloacae | HCASE | HCASE | NA |
| 51 | E. cloacae | HCASE | HCASE | NA |
| 52 | E. cloacae | HCASE | HCASE | NA |
| 53 | K. aerogenes | HCASE | HCASE | NA |
| 54 | E. aerogenes | HCASE | HCASE | NA |
| 55 * | H. alvei | HCASE | HCASE | NA |
| 56 * | S. marcescens | HCASE | HCASE | NA |
| 57 * | E. cloacae | HCASE | HCASE | NA |
| 58 | E. coli | HCASE plasmidic | DHA-1 | NA |
| 59 * | E. coli | HCASE plasmidic | ACC-1 | NA |
| 60 * | K. pneumoniae | HCASE plasmidic | DHA-2 | NA |
| 61 * | P. mirabilis | HCASE plasmidic | ACC-1 | NA |
| 62 | K. pneumoniae | HCASE plasmidic | SHV + DHA-1 | NA |
| 63 | K. pneumoniae | HCASE + BSPASE | HCASE (CMY-16), TEM-1B, SHV-11 | NA |
| 64 | E. coli | HCASE + ESBL | CTX-M-14 | NA |
| 65 | K. pneumoniae | HCASE + ESBL | LEN16 (99.65%), CTX-M-1 | NA |
| 66 * | E. cloacae | HCASE + ESBL | HCASE + ESBL | NA |
| 67 * | E. cloacae | HCASE + ESBL | HCASE + CTX-M-15 | NA |
| 68 * | E. cloacae | HCASE + ESBL | HCASE + CTX-M-15 | NA |
| 69 * | E. cloacae | HCASE + ESBL | HCASE + CTX-M-15 | NA |
| 70 | C. freundii | HCASE + ESBL | HCASE + TEM-3 | NA |
| 71 | E. cloacae | HCASE + ESBL | HCASE + ESBL | NA |
| 72 | E. cloacae | HCASE + ESBL + BSPASE | HCASE (ACT-7 like), TEM-1B, CTX-M-15 | NA |
| 73 | E. coli | Carbapenemase | NDM-1 + OXA-1 + OXA-10 + CMY-16 + TEM-1 | NDM-1 |
| 74 | E. coli | Carbapenemase | NDM-1 + OXA-1 + TEM-1 | NDM-1 |
| 75 | E. coli | Carbapenemase | NDM-4 + CTX-M-15 + CMY-6 | NDM-4 |
| 76 | K. pneumoniae | Carbapenemase | NDM-1 + OXA-1 + SHV-11 | NDM-1 |
| 77 | K. pneumoniae | Carbapenemase | NDM-1 + OXA-1 + CTX-M-15 + TEM-1 + SHV-28 + OXA-9 + CMY-6 | NDM-1 |
| 78 | P. stuartii | Carbapenemase | NDM-1 + OXA-1 + CMY-6 + TEM-1 | NDM-1 |
| 79 | P. rettgeri | Carbapenemase | NDM-1 + CTX-M-15 | NDM-1 |
| 80 | K. pneumoniae | Carbapenemase | VIM-1 + SHV-5 | VIM-1 |
| 81 | E. cloacae | Carbapenemase | VIM-1 + SHV-70 | VIM-1 |
| 82 | E. cloacae | Carbapenemase | VIM-4 + CTX-M-15 + TEM-1 + SHV-31 | VIM-4 |
| 83 | K. pneumoniae | Carbapenemase | IMP-8 + SHV-12 | IMP-8 |
| 84 | E. cloacae | Carbapenemase | IMP-8 | IMP-8 |
| 85 | E. cloacae | Carbapenemase | GIM-1 | GIM-1 |
| 86 | E. coli | Carbapenemase | KPC-2 | KPC-2 |
| 87 | E. coli | Carbapenemase | KPC-2 + TEM-1 + OXA-9 | KPC-2 |
| 88 | K. pneumoniae | Carbapenemase | KPC-2 + SHV-11 | KPC-2 |
| 89 | E. cloacae | Carbapenemase | KPC-2 + TEM-1 + OXA-1 | KPC-2 |
| 90 | E. cloacae | Carbapenemase | KPC-2 + TEM-1 + SHV-11 | KPC-2 |
| 91 | E. cloacae | Carbapenemase | KPC-2 + TEM-3 | KPC-2 |
| 92 | E. cloacae | Carbapenemase | IMI-1 | IMI-1 |
| 93 | E. asburiae | Carbapenemase | IMI-2 | IMI-2 |
| 94 | E. asburiae | Carbapenemase | IMI-2 | IMI-2 |
| 95 | E. cloacae | Carbapenemase | NmcA | NmcA |
| 96 | E. cloacae | Carbapenemase | GES-5 | GES-5 |
| 97 | E. cloacae | Carbapenemase | FRI-1 | FRI-1 |
| 98 | K. pneumoniae | Carbapenemase | OXA-48 | OXA-48 |
| 99 | E. cloacae | Carbapenemase | OXA-48 + TEM-1 + CTX-M-15 + OXA-1 | OXA-48 |
| 100 | E. cloacae | Carbapenemase | OXA-48 + SHV-5 | OXA-48 |
| 101 | E. coli | Carbapenemase | OXA-181 | OXA-181 |
| 102 | K. pneumoniae | Carbapenemase | OXA-181 + SHV-11 + CTXM-15 + OXA-1 | OXA-181 |
| 103 | E. coli | Carbapenemase | OXA-204 + CMY-4 + CTX-M-15 + OXA-1 | OXA-204 |
| 104 | K. pneumoniae | Carbapenemase | OXA-204 + SHV-28 + TEM-1 + CTX-M-15 | OXA-204 |
| 105 | K. pneumoniae | Carbapenemase | OXA-232 + SHV-1 + TEM-1 + CTX-M-15 + OXA-1 | OXA-232 |
| 106 | E. coli | Carbapenemase | OXA-244 + TEM-1 + CMY-2 | OXA-244 |
| 107 | E. coli | Carbapenemase | OXA-244 + TEM-1 + CMY-2 | OXA-244 |
| 108 | P. mirabilis | Carbapenemase | OXA-23 | OXA-23 |
| 109 | P. mirabilis | Carbapenemase | OXA-58 | OXA-58 |
| 110 | E. cloacae | Carbapenemase | NmcA | NmcA |
| 111 | E. cloacae | Carbapenemase | IMI-17 | IMI-17 |
| 112 | E. cloacae | Carbapenemase | IMI-17 | IMI-17 |
| 113 | E. cloacae | Carbapenemase | IMI-17 | IMI-17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chelaru, E.-C.; Muntean, A.-A.; Muntean, M.-M.; Hogea, M.-O.; Caracoti, C.-Ș.; Ciomaga, B.-F.; Naas, T.; Popa, M.I. Oxacillin-Supplemented Mueller-Hinton Agar for In Vitro Inhibition of Ambler Class C β-Lactamases in Enterobacterales. Antibiotics 2025, 14, 616. https://doi.org/10.3390/antibiotics14060616
Chelaru E-C, Muntean A-A, Muntean M-M, Hogea M-O, Caracoti C-Ș, Ciomaga B-F, Naas T, Popa MI. Oxacillin-Supplemented Mueller-Hinton Agar for In Vitro Inhibition of Ambler Class C β-Lactamases in Enterobacterales. Antibiotics. 2025; 14(6):616. https://doi.org/10.3390/antibiotics14060616
Chicago/Turabian StyleChelaru, Edgar-Costin, Andrei-Alexandru Muntean, Mădălina-Maria Muntean, Mihai-Octav Hogea, Costin-Ștefan Caracoti, Bogdan-Florin Ciomaga, Thierry Naas, and Mircea Ioan Popa. 2025. "Oxacillin-Supplemented Mueller-Hinton Agar for In Vitro Inhibition of Ambler Class C β-Lactamases in Enterobacterales" Antibiotics 14, no. 6: 616. https://doi.org/10.3390/antibiotics14060616
APA StyleChelaru, E.-C., Muntean, A.-A., Muntean, M.-M., Hogea, M.-O., Caracoti, C.-Ș., Ciomaga, B.-F., Naas, T., & Popa, M. I. (2025). Oxacillin-Supplemented Mueller-Hinton Agar for In Vitro Inhibition of Ambler Class C β-Lactamases in Enterobacterales. Antibiotics, 14(6), 616. https://doi.org/10.3390/antibiotics14060616








