Characterization of Enterobacter cloacae and Citrobacter freundii Species Complex Isolates with Decreased Susceptibility to Cephalosporins from United States Hospitals and Activity of Aztreonam–Avibactam and Comparator Agents (2019–2023)
Abstract
1. Introduction
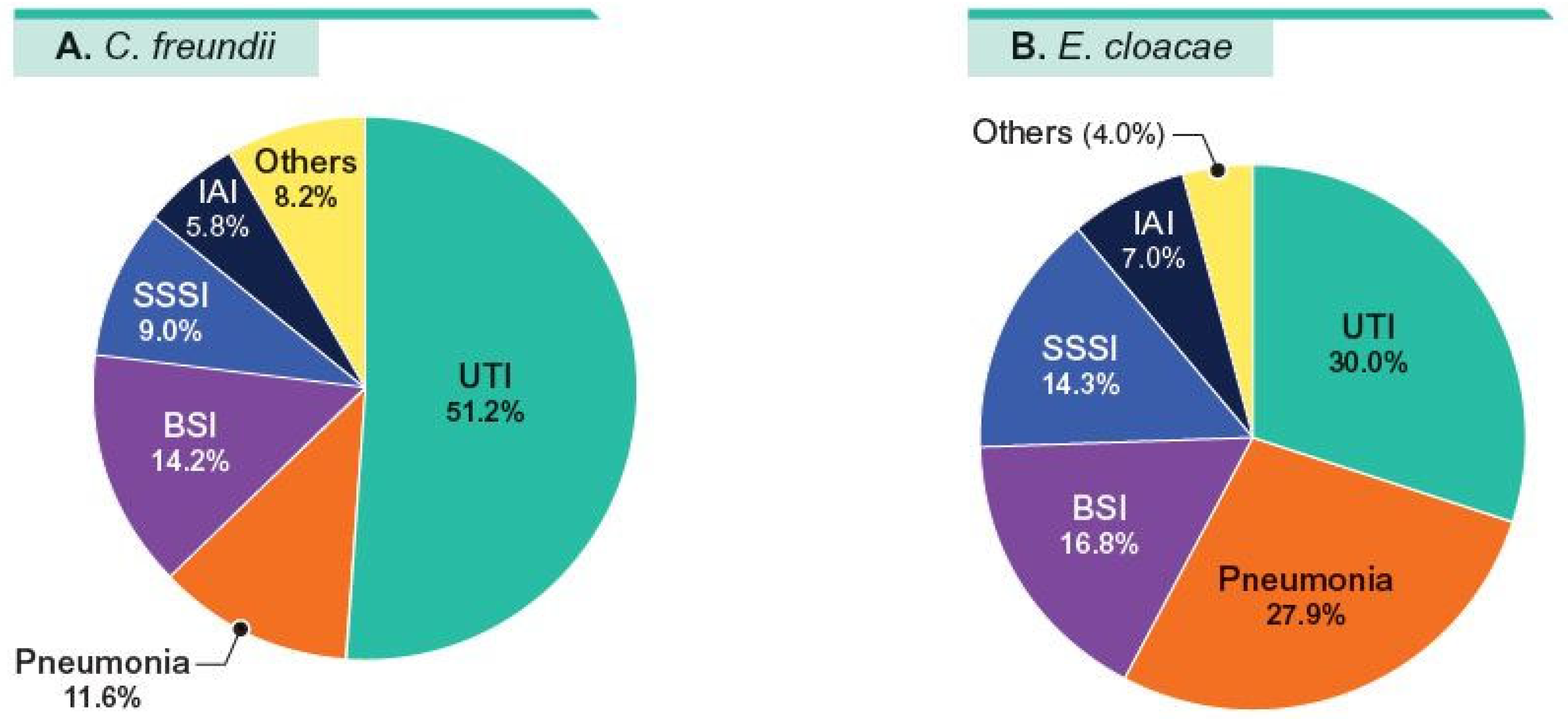
2. Results
3. Discussion
4. Materials and Methods
4.1. Organism Collection
4.2. Susceptibility Testing
4.3. β-Lactamase Screening
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alvarez-Marin, R.; Navarro-Amuedo, D.; Gasch-Blasi, O.; Rodriguez-Martinez, J.M.; Calvo-Montes, J.; Lara-Contreras, R.; Lepe-Jimenez, J.A.; Tubau-Quintano, F.; Cano-Garcia, M.E.; Rodriguez-Lopez, F.; et al. A prospective, multicenter case control study of risk factors for acquisition and mortality in Enterobacter species bacteremia. J. Infect. 2020, 80, 174–181. [Google Scholar] [CrossRef]
- Jabeen, I.; Islam, S.; Hassan, A.; Tasnim, Z.; Shuvo, S.R. A brief insight into Citrobacter species—A growing threat to public health. Front. Antibiot. 2023, 2, 1276982. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Alonso, E.; Bourgeois-Nicolaos, N.; Lepainteur, M.; Derouin, V.; Barreault, S.; Waalkes, A.; Augusto, L.A.; Gera, S.; Gleizes, O.; Tissieres, P.; et al. Contaminated Incubators: Source of a Multispecies Enterobacter Outbreak of Neonatal Sepsis. Microbiol. Spectr. 2022, 10, e0096422. [Google Scholar] [CrossRef] [PubMed]
- Jolivet, S.; Couturier, J.; Le Neindre, K.; Ehmig, M.; Dortet, L.; Emeraud, C.; Barbut, F. Persistence of OXA-48-producing ST-22 Citrobacter freundii in patients and the hospital environment, Paris, France, 2016 to 2022. Eurosurveillance 2024, 29, 2400262. [Google Scholar] [CrossRef] [PubMed]
- Morhart, P.; Gerlach, R.G.; Kunz, C.; Held, J.; Valenza, G.; Wolfle, J.; Reutter, H.; Hanslik, G.J.; Fahlbusch, F.B. Application of Next-Generation Sequencing to Enterobacter Hormaechei Subspecies Analysis During a Neonatal Intensive Care Unit Outbreak. Children 2023, 10, 1696. [Google Scholar] [CrossRef]
- Cai, S.; Quan, J.; Wang, Z.; Hu, H.; Han, X.; Jiang, Y.; Yang, Q.; Yu, Y.; Zhou, Z. High prevalence of carbapenem-resistant Enterobacter cloacae complex in a tertiary hospital over a decade. Microbiol. Spectr. 2024, 12, e0078024. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Lavigne, J.P.; Pages, J.M. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef]
- Gartzonika, K.; Politi, L.; Mavroidi, A.; Tsantes, A.G.; Spanakis, N.; Priavali, E.; Vrioni, G.; Tsakris, A. High prevalence of clonally related ST182 NDM-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int. J. Antimicrob. Agents 2023, 62, 106837. [Google Scholar] [CrossRef]
- Miller, W.R.; Arias, C.A. ESKAPE pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- Yao, Y.; Falgenhauer, L.; Falgenhauer, J.; Hauri, A.M.; Heinmuller, P.; Domann, E.; Chakraborty, T.; Imirzalioglu, C. Carbapenem-Resistant Citrobacter spp. as an Emerging Concern in the Hospital-Setting: Results From a Genome-Based Regional Surveillance Study. Front. Cell. Infect. Microbiol. 2021, 11, 744431. [Google Scholar] [CrossRef]
- Sader, H.S.; Mendes, R.E.; Carvalhaes, C.G.; Kimbrough, J.H.; Castanheira, M. Changing Epidemiology of Carbapenemases Among Carbapenem-Resistant Enterobacterales From United States Hospitals and the Activity of Aztreonam-Avibactam Against Contemporary Enterobacterales (2019–2021). Open Forum Infect. Dis. 2023, 10, ofad046. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Mendes, R.E.; Doyle, T.B.; Davis, A.P.; Castanheira, M. Characterization of Enterobacter cloacae and Citrobacter freundii species complex isolates with decreased susceptibility to cephalosporins from United States hospitals and activity of ceftazidime/avibactam and comparator agents. JAC Antimicrob. Resist. 2021, 3, dlab136. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J.; Antibacterial Resistance Leadership Group. A Primer on AmpC beta-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [PubMed]
- Tamma, P.D.; Heil, E.L.; Justo, J.A.; Mathers, A.J.; Satlin, M.J.; Bonomo, R.A. Infectious Diseases Society of America 2024 Guidance on the Treatment of Antimicrobial-Resistant Gram-Negative Infections. Clin. Infect. Dis. 2024, ciae403. [Google Scholar]
- Rodriguez-Villodres, A.; Lepe-Balsalobre, E.; Ortiz De La Rosa, J.M.; Giner Almaraz, S.; Gonzalez De Herrero, E.; Cercenado, E.; Garcia-Fernandez, S.; Benito, R.; Ponz Mir, R.; Canton, R.; et al. Activity of cefepime, carbapenems and new β-lactam/β-lactamase inhibitor combinations on Enterobacter cloacae complex and Klebsiella aerogenes in Spain (SMART 2016–2022). JAC Antimicrob. Resist. 2024, 6, dlae087. [Google Scholar]
- Agergaard, C.N.; Porsbo, L.J.; Sydenham, T.V.; Hansen, S.G.K.; Steinke, K.; Larsen, S.L.; Helgason, K.O.; Hansen, F.; Karstensen, K.T.; Henius, A.E.; et al. Contaminated dicloxacillin capsules as the source of an NDM-5/OXA-48-producing Enterobacter hormaechei ST79 outbreak, Denmark and Iceland, 2022 and 2023. Eurosurveillance 2023, 28, 2300108. [Google Scholar]
- Wendel, A.F.; Meyer, S.; Deenen, R.; Kohrer, K.; Kolbe-Busch, S.; Pfeffer, K.; Willmann, M.; Kaasch, A.J.; MacKenzie, C.R. Long-Term, Low-Frequency Cluster of a German-Imipenemase-1-Producing Enterobacter hormaechei ssp. steigerwaltii ST89 in a Tertiary Care Hospital in Germany. Microb. Drug Resist. 2018, 24, 1305–1315. [Google Scholar]
- Yeh, T.K.; Lin, H.J.; Liu, P.Y.; Wang, J.H.; Hsueh, P.R. Antibiotic resistance in Enterobacter hormaechei. Int. J. Antimicrob. Agents 2022, 60, 106650. [Google Scholar]
- CLSI. M07Ed12; MethMethods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Clinical Laboratory Standards Institute: Berwyn, PA, USA, 2024.
- CLSI. M100Ed35; Performance Standards for Antimicrobial Susceptability Testing. 35th Informational Supplement; Clinical Laboratory Standards Institute: Berwyn, PA, USA, 2025.
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.J.C.M.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Castanheira, M.; Kimbrough, J.H.; DeVries, S.; Mendes, R.E.; Sader, H.S. Trends of beta-Lactamase Occurrence Among Escherichia coli and Klebsiella pneumoniae in United States Hospitals During a 5-Year Period and Activity of Antimicrobial Agents Against Isolates Stratified by beta-Lactamase Type. Open Forum Infect. Dis. 2023, 10, ofad038. [Google Scholar] [CrossRef] [PubMed]
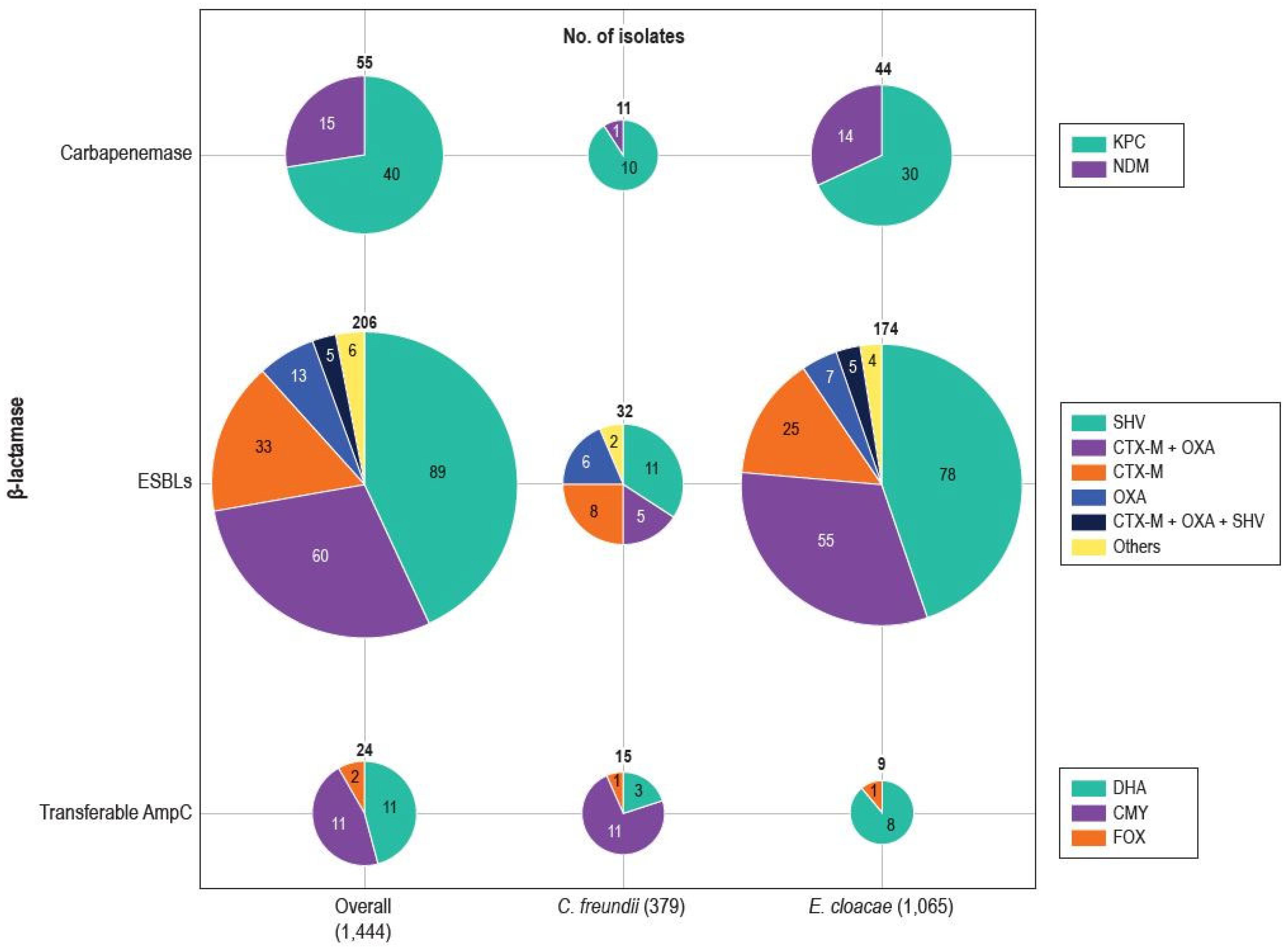
| Beta-Lactamase/Organism | No. of Isolates |
|---|---|
| Carbapenemases | 55 |
| C. freundii | 11 |
| KPC-18 | 1 |
| KPC-2 | 7 |
| KPC-3 | 2 |
| NDM-1 | 1 |
| E. cloacae | 44 |
| KPC-2 | 14 |
| KPC-3 | 12 |
| KPC-4 | 1 |
| KPC-6 | 2 |
| KPC-10, KPC-6 | 1 |
| NDM-1 | 14 |
| ESBL | 206 |
| C. freundii | 32 |
| CTX-M-1 | 1 |
| CTX-M-3 | 2 |
| CTX-M-3, SHV-12 | 1 |
| CTX-M-13 | 1 |
| CTX-M-15 | 4 |
| CTX-M-15, OXA-1/30 | 5 |
| OXA-1/30 | 5 |
| OXA-4, GES-7 | 1 |
| OXA-45 | 1 |
| SHV-5 | 1 |
| SHV-7 | 3 |
| SHV-12 | 7 |
| E. cloacae | 174 |
| CTX-M-3 | 7 |
| CTX-M-9 | 3 |
| CTX-M-14 | 1 |
| CTX-M-15 | 13 |
| CTX-M-15, OXA-1/30 | 55 |
| CTX-M-15, OXA-1/30, SHV-12 | 5 |
| CTX-M-15, SHV-12 | 1 |
| CTX-M-88 | 1 |
| OXA-1/30 | 6 |
| OXA-1/30, SHV-12 | 1 |
| OXA-1/30, SHV-7 | 1 |
| OXA-17 | 1 |
| SHV-5 | 1 |
| SHV-7 | 8 |
| SHV-7-like | 1 |
| SHV-12 | 65 |
| SHV-30 | 3 |
| TEM-19 | 1 |
| Transferable AmpC | 24 |
| C. freundii | 15 |
| CMY-159 | 2 |
| CMY-179 | 3 |
| CMY-180 | 4 |
| CMY-181 | 2 |
| DHA-1 | 3 |
| FOX-5-like | 1 |
| E. cloacae | 9 |
| DHA-1 | 8 |
| FOX-5 | 1 |
| Antimicrobial | C. freundii (No.) | E. cloacae (No.) | All (No.) | |||
|---|---|---|---|---|---|---|
| MIC50/90 | %S a | MIC50/90 | %S a | MIC50/90 | %S a | |
| All organisms | (n = 1374) | (n = 3732) | (n = 5106) | |||
| Aztreonam–avibactam a | 0.06/0.25 | 100.0 | 0.06/0.5 | 99.8 | 0.06/0.5 | 99.9 |
| Ceftazidime–avibactam | 0.12/0.5 | 99.9 | 0.25/0.5 | 99.4 | 0.25/0.5 | 99.5 |
| Meropenem–vaborbactam | 0.03/0.03 | 99.9 | 0.03/0.03 | 99.5 | 0.03/0.03 | 99.6 |
| Ceftolozane–tazobactam | 0.25/16 | 77.1 | 0.25/16 | 77.6 | 0.25/16 | 77.5 |
| Piperacillin–tazobactam | 4/128 | 72.1 | 2/128 | 71.9 | 2/128 | 72.0 |
| Aztreonam | 0.25/>16 | 72.4 | 0.12/>16 | 71.4 | 0.12/>16 | 71.7 |
| Ceftriaxone | 0.25/>8 | 70.2 | 0.25/>8 | 66.4 | 0.25/>8 | 67.4 |
| Ceftazidime | 0.5/>32 | 72.5 | 0.5/>32 | 70.6 | 0.5/>32 | 71.1 |
| Cefepime | 0.06/2 | 93.2 | 0.06/4 | 89.0 | 0.06/2 | 90.1 |
| Meropenem | 0.03/0.06 | 98.9 | 0.03/0.12 | 98.3 | 0.03/0.06 | 98.5 |
| Ertapenem b | 0.015/0.25 | 97.3 | 0.06/1 | 86.8 | 0.06/1 | 89.6 |
| Levofloxacin | 0.06/1 | 87.8 | 0.03/0.5 | 93.0 | 0.06/0.5 | 91.6 |
| Gentamicin | 0.5/1 | 93.7 | 0.25/0.5 | 96.1 | 0.25/0.5 | 95.5 |
| Amikacin | 2/2 | 97.7 | 1/2 | 98.8 | 1/2 | 98.5 |
| Tobramycin | 0.5/2 | 92.4 | 0.5/0.5 | 95.0 | 0.5/1 | 94.3 |
| Colistin | 0.25/0.5 | 99.7 c | 0.25/>8 | 79.6 c | 0.25/>8 | 85.1 c |
| Ceph-DS organisms d | (n = 379) d | (n = 1065) d | (n = 1444) d | |||
| Aztreonam–avibactam a | 0.25/0.5 | 100.0 | 0.5/1 | 99.4 | 0.25/1 | 99.6 |
| Ceftazidime–avibactam | 0.5/1 | 99.5 | 0.5/1 | 97.7 | 0.5/1 | 98.2 |
| Meropenem–vaborbactam | 0.03/0.03 | 99.7 | 0.03/0.06 | 98.2 | 0.03/0.06 | 98.5 |
| Ceftolozane–tazobactam | 16/>16 | 17.4 | 8/>16 | 21.9 | 8/>16 | 20.7 |
| Piperacillin–tazobactam | 128/>128 | 9.2 | 64/>128 | 9.1 | 128/>128 | 9.2 |
| Aztreonam | >16/>16 | 2.9 | >16/>16 | 2.5 | >16/>16 | 2.6 |
| Ceftriaxone | >8/>8 | 0.3 | >8/>8 | 0.4 | >8/>8 | 0.3 |
| Ceftazidime | >32/>32 | 1.8 | >32/>32 | 1.1 | >32/>32 | 1.3 |
| Cefepime | 1/8 | 75.2 | 2/32 | 61.3 | 2/32 | 65.0 g |
| Meropenem | 0.06/0.12 | 96.3 | 0.06/0.25 | 94.2 | 0.06/0.25 | 94.7 |
| Ertapenem b | 0.25/0.5 | 91.0 | 0.5/>2 | 60.5 | 0.5/2 | 68.2 |
| Levofloxacin | 0.12/2 | 76.3 | 0.06/2 | 82.7 | 0.06/2 | 81.0 |
| Gentamicin | 0.5/8 | 88.9 | 0.25/4 | 89.5 | 0.25/4 | 89.3 |
| Amikacin | 2/4 | 93.9 | 1/2 | 96.8 | 1/2 | 96.1 |
| Tobramycin | 0.5/8 | 85.9 | 0.5/8 | 85.8 | 0.5/8 | 85.9 |
| Colistin | 0.25/0.5 | 98.9 c | 0.25/>8 | 83.5 c | 0.25/>8 | 87.6 c |
| ESBL producers (excluding CBase) e | (n = 27) | (n = 142) | (n = 169) | |||
| Aztreonam–avibactam a | 0.12/1 | 100.0 | 0.12/0.5 | 100.0 | 0.12/0.5 | 100.0 |
| Ceftazidime–avibactam | 0.5/1 | 100.0 | 0.25/1 | 100.0 | 0.5/1 | 100.0 |
| Meropenem–vaborbactam | 0.03/0.03 | 100.0 | 0.03/0.06 | 100.0 | 0.03/0.06 | 100.0 |
| Ceftolozane–tazobactam | 1/>16 | 63.0 | 1/>16 | 66.9 | 1/>16 | 66.3 |
| Piperacillin–tazobactam | 8/>128 | 55.6 | 16/>128 | 46.8 | 16/>128 | 48.2 |
| Aztreonam | >16/>16 | 7.4 | >16/>16 | 3.5 | >16/>16 | 4.1 |
| Ceftriaxone | >8/>8 | 0.0 | >8/>8 | 1.4 | >8/>8 | 1.2 |
| Ceftazidime | >32/>32 | 11.1 | >32/>32 | 7.0 | >32/>32 | 7.7 |
| Cefepime | 16/>32 | 29.6 | 16/>32 | 23.9 | 16/>32 | 24.9 |
| Meropenem | 0.03/0.12 | 100.0 | 0.03/0.12 | 98.6 | 0.03/0.12 | 98.8 |
| Ertapenem b | 0.03/0.25 | 100.0 | 0.5/1 | 81.1 | 0.12/1 | 85.5 |
| Levofloxacin | 2/16 | 29.6 | 1/16 | 48.6 | 1/16 | 45.6 |
| Gentamicin | 16/>16 | 48.1 | 16/>16 | 43.0 | 16/>16 | 43.8 |
| Amikacin | 2/16 | 74.1 | 1/8 | 87.3 | 2/8 | 85.2 |
| Tobramycin | 8/>16 | 33.3 | 8/>16 | 29.6 | 8/>16 | 30.2 |
| Colistin | 0.25/0.5 | 100.0 c | 0.25/>8 | 87.3 c | 0.25/8 | 89.3 c |
| Transferable AmpC producers | (n = 15) | (n = 9) | (n = 24) | |||
| Aztreonam–avibactam a | 0.12/0.5 | 100.0 | 0.25/- | 100.0 | 0.25/1 | 100.0 |
| Ceftazidime–avibactam | 0.25/4 | 100.0 | 0.5/- | 100.0 | 0.5/4 | 100.0 |
| Meropenem–vaborbactam | ≤0.015/0.25 | 100.0 | 0.03/- | 100.0 | 0.03/0.12 | 100.0 |
| Ceftolozane–tazobactam | 16/>16 | 40.0 | 0.5/- | 55.6 | 8/>16 | 45.8 |
| Piperacillin–tazobactam | 32/>128 | 26.7 | 16/- | 44.4 | 32/>128 | 33.3 |
| Aztreonam | >16/>16 | 13.3 | 16/- | 44.4 | >16/>16 | 25.0 |
| Ceftriaxone | >8/>8 | 0.0 | 8/- | 0.0 | >8/>8 | 0.0 |
| Ceftazidime | >32/>32 | 6.7 | >32/- | 11.1 | >32/>32 | 8.3 |
| Cefepime | 1/32 | 66.7 | 0.12/- | 66.7 | 1/32 | 66.7 |
| Meropenem | 0.03/0.12 | 100.0 | 0.12/- | 100.0 | 0.06/0.25 | 100.0 |
| Ertapenem b | 0.25/- | 80.0 | 0.5/- | 60.0 | 0.5/1 | 70.0 |
| Levofloxacin | 0.5/32 | 53.3 | 1/- | 44.4 | 0.5/8 | 50.0 |
| Gentamicin | 1/>16 | 73.3 | 0.5/- | 77.8 | 0.5/>16 | 75.0 |
| Amikacin | 2/16 | 86.7 | 2/- | 77.8 | 2/16 | 83.3 |
| Tobramycin | 1/16 | 60.0 | 0.5/- | 66.7 | 1/16 | 62.5 |
| Colistin | 0.25/0.25 | 100.0 c | 0.25/- | 66.7 | 0.25/>8 | 87.5 |
| Carbapenem-resistant f | (n = 12) | (n = 52) | (n = 64) | |||
| Aztreonam–avibactam a | 0.12/0.5 | 100.0 | 0.25/1 | 100.0 | 0.25/1 | 100.0 |
| Ceftazidime–avibactam | 1/4 | 91.7 | 2/>32 | 67.3 | 1/>32 | 71.9 |
| Meropenem–vaborbactam | 0.03/1 | 91.7 | 0.06/32 | 65.4 | 0.06/32 | 70.3 |
| Imipenem–relebactam g | 0.12/0.5 | 91.7 | 0.25/>8 | 67.3 | 0.25/>8 | 71.9 |
| Cefiderocol f | 0.12/1 | 100.0 | 2/16 | 78.8 | 1/16 | 82.8 |
| Levofloxacin | 8/32 | 8.3 | 1/32 | 40.4 | 2/32 | 34.4 |
| Gentamicin | 8/>16 | 25.0 | 0.5/16 | 73.1 | 0.5/16 | 64.1 |
| Amikacin | 8/16 | 41.7 | 2/8 | 82.7 | 2/16 | 75.0 |
| Colistin | 0.25/0.25 | 100.0 c | 0.25/>8 | 86.5 c | 0.25/>8 | 89.1 c |
| MBL producers | (n = 1) | (n = 14) | (n = 15) | |||
| Aztreonam–avibactam a | 100.0 | 0.12/0.25 | 100.0 | 0.12/0.5 | 100.0 | |
| Ceftazidime–avibactam | 0.0 | >32/>32 | 0.0 | >32/>32 | 0.0 | |
| Meropenem–vaborbactam | 0.0 | 16/>32 | 0.0 | 32/>32 | 0.0 | |
| Imipenem–relebactam g | 0.0 | 8/>8 | 0.0 | 8/>8 | 0.0 | |
| Aztreonam | 100.0 | >16/>16 | 7.1 | >16/>16 | 13.3 | |
| Cefiderocol g | 100.0 | 8/32 | 42.9 | 8/32 | 46.7 | |
| Levofloxacin | 0.0 | 1/16 | 35.7 | 2/32 | 33.3 | |
| Gentamicin | 0.0 | 0.5/2 | 92.9 | 0.5/4 | 86.7 | |
| Amikacin | 100.0 | 4/16 | 71.4 | 4/16 | 73.3 | |
| Colistin | 100.0 c | 0.25/0.25 | 92.9 c | 0.25/0.25 | 93.3 c | |
| Ceph-DS isolates with no ESBL, no transferable AmpC, and no carbapenemase | (n = 324) | (n = 857) | (n = 1181) | |||
| Aztreonam–avibactam a | 0.25/0.5 | 100.0 | 0.5/1 | 99.3 | 0.5/1 | 99.5 |
| Ceftazidime–avibactam | 0.5/1 | 100.0 | 0.5/1 | 99.3 | 0.5/1 | 99.5 |
| Meropenem–vaborbactam | 0.03/0.03 | 100.0 | 0.03/0.06 | 100.0 | 0.03/0.03 | 100.0 |
| Ceftolozane–tazobactam | 16/>16 | 14.5 | 8/>16 | 15.6 | 8/>16 | 15.3 |
| Piperacillin–tazobactam | 128/>128 | 5.6 | 64/>128 | 3.3 | 128/>128 | 3.9 |
| Aztreonam | >16/>16 | 1.9 | >16/>16 | 2.1 | >16/>16 | 2.0 |
| Ceftriaxone | >8/>8 | 0.3 | >8/>8 | 0.2 | >8/>8 | 0.3 |
| Ceftazidime | >32/>32 | 1.2 | >32/>32 | 0.2 | >32/>32 | 0.5 |
| Cefepime | 1/4 | 82.7 | 2/4 | 71.1 | 1/4 | 74.3 |
| Meropenem | 0.06/0.06 | 99.7 | 0.06/0.25 | 98.5 | 0.06/0.12 | 98.8 |
| Ertapenem b | 0.25/0.5 | 93.7 | 0.5/2 | 61.4 | 0.5/2 | 69.6 |
| Levofloxacin | 0.12/1 | 84.3 | 0.06/0.5 | 91.9 | 0.06/1 | 89.8 |
| Gentamicin | 0.5/1 | 96.0 | 0.25/0.5 | 98.7 | 0.25/0.5 | 98.0 |
| Amikacin | 2/2 | 98.5 | 1/2 | 99.5 | 1/2 | 99.2 |
| Tobramycin | 0.5/1 | 94.4 | 0.5/0.5 | 98.6 | 0.5/1 | 97.5 |
| Colistin | 0.25/0.5 | 98.8 c | 0.25/>8 | 83.0 c | 0.25/>8 | 87.4 c |
| Organism Subset (No.) | MDR Rate | XDR Rate |
|---|---|---|
| All CFC and ECLC (5106) | 31.5% | 0.5% |
| Ceph-DS isolates (1444) | 96.8% | 2.6% |
| ESBL producers (206) | 97.1% | 12.6% |
| Transferable AmpC producers (24) | 91.7% | 0.0% |
| Carbapenemase producers (74) | 100.0% | 37.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sader, H.S.; Doyle, T.B.; Kimbrough, J.H.; Mendes, R.E.; Castanheira, M. Characterization of Enterobacter cloacae and Citrobacter freundii Species Complex Isolates with Decreased Susceptibility to Cephalosporins from United States Hospitals and Activity of Aztreonam–Avibactam and Comparator Agents (2019–2023). Antibiotics 2025, 14, 382. https://doi.org/10.3390/antibiotics14040382
Sader HS, Doyle TB, Kimbrough JH, Mendes RE, Castanheira M. Characterization of Enterobacter cloacae and Citrobacter freundii Species Complex Isolates with Decreased Susceptibility to Cephalosporins from United States Hospitals and Activity of Aztreonam–Avibactam and Comparator Agents (2019–2023). Antibiotics. 2025; 14(4):382. https://doi.org/10.3390/antibiotics14040382
Chicago/Turabian StyleSader, Helio S., Timothy B. Doyle, John H. Kimbrough, Rodrigo E. Mendes, and Mariana Castanheira. 2025. "Characterization of Enterobacter cloacae and Citrobacter freundii Species Complex Isolates with Decreased Susceptibility to Cephalosporins from United States Hospitals and Activity of Aztreonam–Avibactam and Comparator Agents (2019–2023)" Antibiotics 14, no. 4: 382. https://doi.org/10.3390/antibiotics14040382
APA StyleSader, H. S., Doyle, T. B., Kimbrough, J. H., Mendes, R. E., & Castanheira, M. (2025). Characterization of Enterobacter cloacae and Citrobacter freundii Species Complex Isolates with Decreased Susceptibility to Cephalosporins from United States Hospitals and Activity of Aztreonam–Avibactam and Comparator Agents (2019–2023). Antibiotics, 14(4), 382. https://doi.org/10.3390/antibiotics14040382






