Emergence of Linezolid Resistance Genes optrA and cfr(D) in an Enterococcus saccharolyticus from Chicken
Abstract
1. Introduction
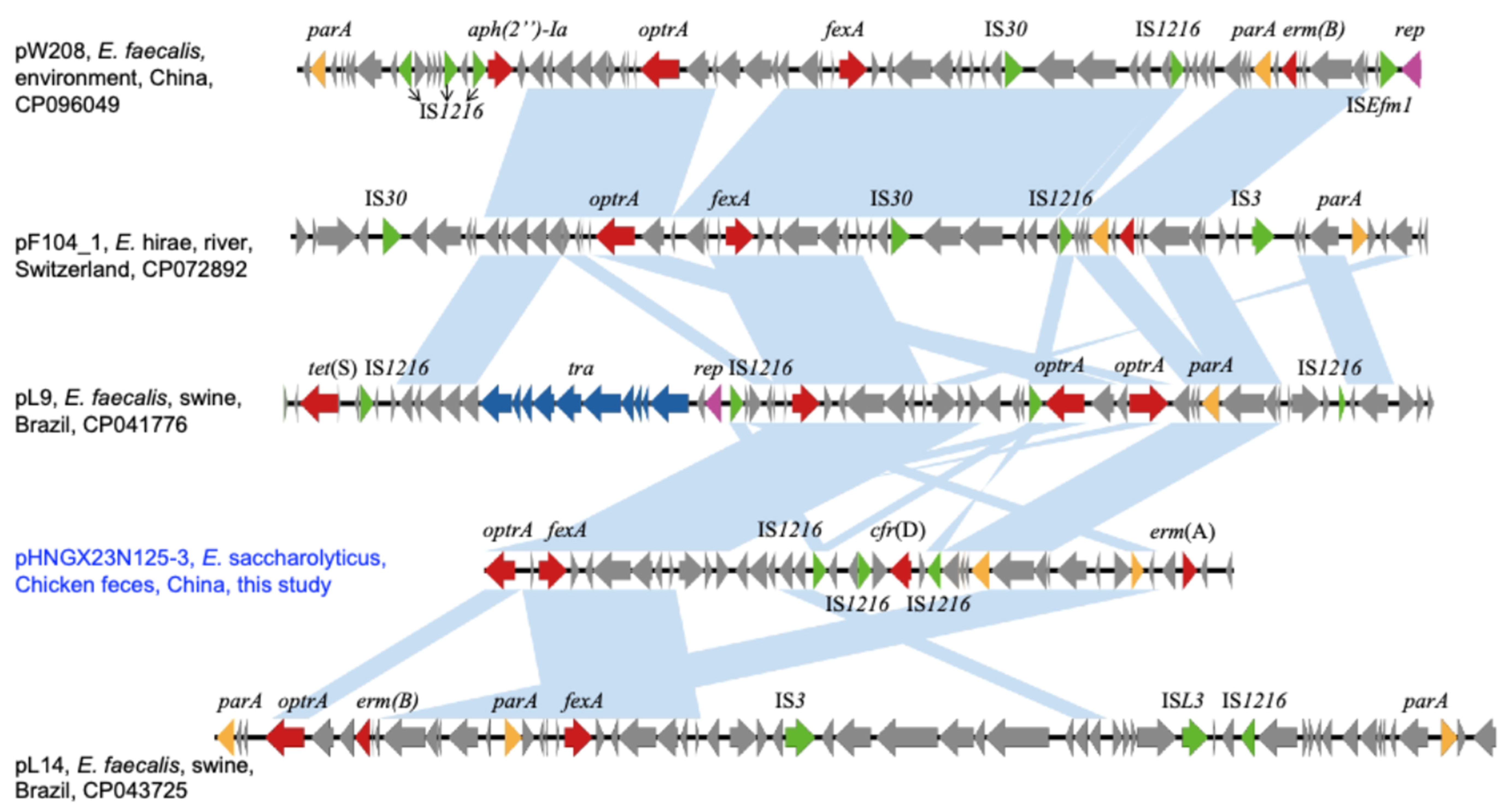
2. Results and Discussions
3. Conclusions
4. Materials and Methods
4.1. Strain Isolation and Detection of Linezolid Resistance Genes
4.2. Antimicrobial Susceptibility Testing
4.3. Whole Genome Sequencing (WGS) and Genome Analysis
4.4. Conjugation Experiments
4.5. Plasmid Stability Testing
4.6. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sadowy, E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [PubMed]
- Schwarz, S.; Zhang, W.; Du, X.D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile oxazolidinone resistance genes in Gram-positive and Gram-negative bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar]
- Nüesch-Inderbinen, M.; Heyvaert, L.; Treier, A.; Zurfluh, K.; Cernela, N.; Biggel, M.; Stephan, R. High occurrence of Enterococcus faecalis, Enterococcus faecium, and Vagococcus lutrae harbouring oxazolidinone resistance genes in raw meat-based diets for companion animals—A public health issue, Switzerland, September 2018 to May 2020. Euro Surveill. 2023, 28, 2200496. [Google Scholar] [CrossRef]
- Brenciani, A.; Morroni, G.; Schwarz, S.; Giovanetti, E. Oxazolidinones: Mechanisms of resistance and mobile genetic elements involved. J. Antimicrob. Chemother. 2022, 77, 2596–2621. [Google Scholar] [PubMed]
- Cinthi, M.; Coccitto, S.N.; Fioriti, S.; Morroni, G.; Simoni, S.; Vignaroli, C.; Magistrali, C.F.; Albini, E.; Brenciani, A.; Giovanetti, E. Occurrence of a plasmid co-carrying cfr(D) and poxtA2 linezolid resistance genes in Enterococcus faecalis and Enterococcus casseliflavus from porcine manure, Italy. J. Antimicrob. Chemother. 2022, 77, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.E.; Deshpande, L.M.; Farrell, D.J.; Spanu, T.; Fadda, G.; Jones, R.N. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J. Antimicrob. Chemother. 2010, 65, 2329–2335. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 2018, 73, 1763–1769. [Google Scholar] [CrossRef]
- Driesen, M.; Timmermans, M.; Cargnel, M.; Simons, X.; Filippitzi, M.E.; Catry, B.; Dal Pozzo, F.; Vanderhaeghen, W.; Callens, B.; Dispas, M.; et al. Risk factor analysis for occurrence of linezolid-resistant bacteria in the digestive and respiratory tract of food-producing Animals in Belgium: A Pilot Study. Antibiotics 2024, 13, 707. [Google Scholar] [CrossRef]
- Gião, J.; Leão, C.; Albuquerque, T.; Clemente, L.; Amaro, A. Antimicrobial Susceptibility of Enterococcus Isolates from Cattle and Pigs in Portugal: Linezolid Resistance Genes optrA and poxtA. Antibiotics 2022, 11, 615. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Q.; Wu, H.; Xia, P.; Tian, R.; Li, R.; Xia, L. Molecular epidemiology, phenotypic and genomic characterization of antibiotic-resistant enterococcal isolates from diverse farm animals in Xinjiang, China. Sci. Total Environ. 2024, 912, 168683. [Google Scholar] [PubMed]
- Nüesch-Inderbinen, M.; Biggel, M.; Haussmann, A.; Treier, A.; Heyvaert, L.; Cernela, N.; Stephan, R. Oxazolidinone resistance genes in florfenicol-resistant enterococci from beef cattle and veal calves at slaughter. Front. Microbiol. 2023, 14, 1150070. [Google Scholar]
- Sassi, M.; Guérin, F.; Zouari, A.; Beyrouthy, R.; Auzou, M.; Fines-Guyon, M.; Potrel, S.; Dejoies, L.; Collet, A.; Boukthir, S.; et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006–16. J. Antimicrob. Chemother. 2019, 74, 1469–1472. [Google Scholar]
- Pang, S.; Boan, P.; Lee, T.; Gangatharan, S.; Tan, S.J.; Daley, D.; Lee, Y.T.; Coombs, G.W. Linezolid-resistant ST872 Enteroccocus faecium harbouring optrA and cfr(D) oxazolidinone resistance genes. Int. J. Antimicrob. Agents 2020, 55, 105831. [Google Scholar]
- Shen, W.; Zhang, R.; Cai, J. Co-occurrence of multiple plasmid-borne linezolid resistance genes-optrA, cfr, poxtA2 and cfr(D) in an Enterococcus faecalis isolate from retail meat. J. Antimicrob. Chemother. 2023, 78, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Bamigbade, G.B.; Subhash, A.J.; Al-Ramadi, B.; Kamal-Eldin, A.; Gan, R.Y.; Liu, S.Q.; Ayyash, M. Gut microbiota modulation, prebiotic and bioactive characteristics of date pomace polysaccharides extracted by microwave-assisted deep eutectic solvent. Int. J. Biol. Macromol. 2024, 262, 130167. [Google Scholar]
- Kurban, D.; Roy, J.P.; Kabera, F.; Fréchette, A.; Um, M.M.; Albaaj, A.; Rowe, S.; Godden, S.; Adkins, P.R.F.; Middleton, J.R.; et al. Diagnosing Intra-mammary Infection: Meta-analysis and mapping review on frequency and udder health relevance of microorganism species isolated from Bovine milk samples. Animals 2022, 12, 3288. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, D.; Zhang, J.; Liu, L.; Zhang, Z.; Liu, C.; Hu, S.; Wu, L.; He, Z.; Sun, H. Genomic epidemiology reveals multiple mechanisms of linezolid resistance in clinical enterococci in China. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 41. [Google Scholar]
- Liu, Y.; Wang, Y.; Dai, L.; Wu, C.; Shen, J. First report of multiresistance gene cfr in Enterococcus species casseliflavus and gallinarum of swine origin. Vet. Microbiol. 2014, 170, 352–357. [Google Scholar]
- Tang, B.; Zou, C.; Schwarz, S.; Xu, C.; Hao, W.; Yan, X.M.; Huang, Y.; Ni, J.; Yang, H.; Du, X.D.; et al. Linezolid-resistant Enterococcus faecalis of chicken origin harbored chromosome-borne optrA and plasmid-borne cfr, cfr(D), and poxtA2 genes. Microbiol. Spectr. 2023, 11, e0274122. [Google Scholar]
- Guerin, F.; Sassi, M.; Dejoies, L.; Zouari, A.; Schutz, S.; Potrel, S.; Auzou, M.; Collet, A.; Lecointe, D.; Auger, G.; et al. Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 1699–1703. [Google Scholar]
- Heilbronner, S.; Krismer, B.; Brötz-Oesterhelt, H.; Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 2021, 19, 726–739. [Google Scholar] [PubMed]
- Fioriti, S.; Morroni, G.; Coccitto, S.N.; Brenciani, A.; Antonelli, A.; Di Pilato, V.; Baccani, I.; Pollini, S.; Cucco, L.; Morelli, A.; et al. Detection of oxazolidinone resistance genes and characterization of genetic environments in Enterococci of swine origin, Italy. Microorganisms 2020, 8, 2021. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.W.; Kang, Z.Z.; Wu, S.K.; Chen, Y.P.; Kong, L.H.; Wang, H.N. Detection of the phenicol-oxazolidinone-tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 2019, 74, 2459–2461. [Google Scholar]
- Fukuda, A.; Nakajima, C.; Suzuki, Y.; Usui, M. Transferable linezolid resistance genes (optrA and poxtA) in enterococci derived from livestock compost at Japanese farms. J. Glob. Antimicrob. Resist. 2024, 36, 336–344. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th ed.; CLSI standard M07; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2022. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar]
- McHugh, M.P.; Parcell, B.J.; Pettigrew, K.A.; Toner, G.; Khatamzas, E.; El Sakka, N.; Karcher, A.M.; Walker, J.; Weir, R.; Meunier, D.; et al. Presence of optrA-mediated linezolid resistance in multiple lineages and plasmids of Enterococcus faecalis revealed by long read sequencing. Microbiology (Reading) 2022, 168, 001137. [Google Scholar]
| Strain | Location | Size (bp) | Plasmid Type | Drug Resistance Genes | MIC (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | ERY | TET | FLOR | ENR | VAN | LIN | TED | |||||
| GXN23C125Es | chromosome | 2,639,213 | 16 | >256 | 64 | >128 | 8 | 4 | 8 | 0.5 | ||
| pHNGXN23C125-1 | 41,280 | repUS1-repE(DOp2) | aac(6′)-aph(2″), cat, ant(6)-Ia, lsa(E), lnu(B), aph(3′)-III, erm(B) | |||||||||
| pHNGXN23C125-2 | 38,208 | repUS1-rep(pVEF1) | dfrG, erm(B) | |||||||||
| pHNGXN23C125-3 | 37,500 | / | optrA, fexA, cfr(D), erm(A) | |||||||||
| pHNGXN23C125-4 | 33,200 | / | ||||||||||
| pHNGXN23C125-5 | 9447 | / | ||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Luo, X.; Qian, R.; Gao, G.; Liu, J.; Hong, J.; Yue, C.; Liu, J.-H.; Liu, Y.-Y. Emergence of Linezolid Resistance Genes optrA and cfr(D) in an Enterococcus saccharolyticus from Chicken. Antibiotics 2025, 14, 337. https://doi.org/10.3390/antibiotics14040337
Gao X, Luo X, Qian R, Gao G, Liu J, Hong J, Yue C, Liu J-H, Liu Y-Y. Emergence of Linezolid Resistance Genes optrA and cfr(D) in an Enterococcus saccharolyticus from Chicken. Antibiotics. 2025; 14(4):337. https://doi.org/10.3390/antibiotics14040337
Chicago/Turabian StyleGao, Xun, Xiao Luo, Ruorou Qian, Guolong Gao, Jinghao Liu, Junhao Hong, Chao Yue, Jian-Hua Liu, and Yi-Yun Liu. 2025. "Emergence of Linezolid Resistance Genes optrA and cfr(D) in an Enterococcus saccharolyticus from Chicken" Antibiotics 14, no. 4: 337. https://doi.org/10.3390/antibiotics14040337
APA StyleGao, X., Luo, X., Qian, R., Gao, G., Liu, J., Hong, J., Yue, C., Liu, J.-H., & Liu, Y.-Y. (2025). Emergence of Linezolid Resistance Genes optrA and cfr(D) in an Enterococcus saccharolyticus from Chicken. Antibiotics, 14(4), 337. https://doi.org/10.3390/antibiotics14040337





