Relationship Between CRISPR–Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria
Abstract
1. Introduction
2. Results
2.1. Phenotypic and Genotypic Tetracycline Resistance
2.2. Detection of CRISPR–Cas Loci
2.3. PCR Detection of the Genetic Determinants for HGT
2.3.1. Aggregation Substances
2.3.2. Inducible Pheromones
2.3.3. MGEs
2.4. Bioinformatic Analyses of E. faecalis Genomes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions and DNA Isolation
4.2. Phenotypic Determination of Tetracycline Resistance
4.3. PCR-Based Methods
4.3.1. Detection of Tetracycline Resistance Genes
4.3.2. Detection of CRISPR Loci and CRISPR-Associated (Cas) Genes
4.3.3. Identification of Mobile Genetic Elements (MGEs):
4.4. Genome Sequencing and Bioinformatic Analyses
4.5. Statistical Analysis
5. Conclusions
6. Strengths and Limitations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tannock, G.W.; Cook, G. Enterococci as members of the intestinal microflora of humans. Enterococci:Pathog. Mol.Biol. Antibiot. Resist. 2002, 3, 101–132. [Google Scholar] [CrossRef]
- LPSN - List of Prokaryotic names with Standing in Nomenclature, genus Enterococcus. Available online: https://lpsn.dsmz.de/genus/enterococcus (accessed on 16 December 2024).
- Top, J.; Willems, R.; Bonten, M. Emergence of CC17 Enterococcus faecium: From commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 2008, 52, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance 2008, 13, 19046. [Google Scholar] [CrossRef] [PubMed]
- Hegstad, K.; Mikalsen, T.; Coque, T.; Werner, G.; Sundsfjord, A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010, 16, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Cebeci, T. Species prevalence, virulence genes, and antibiotic resistance of enterococci from food-producing animals at a slaughterhouse in Turkey. Sci. Rep. 2024, 14, 13191. [Google Scholar] [CrossRef] [PubMed]
- Doherty, N.; Trzcinski, K.; Pickerill, P.; Zawadzki, P.; Dowson, C.G. Genetic Diversity of the tet(M) Gene in Tetracycline-Resistant Clonal Lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 2979–2984. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Köse, Ş.; Dao, S.; Ganbarov, K.; Tanomand, A.; Dal, T.; Aghazadeh, M.; Ghotaslou, R.; Rezaee, M.A.; Yousefi, B.; et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect. Drug Resist. 2020, 13, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Cabral, A.S.; Lacerda, F.F.; Leite, V.L.M.; de Miranda, F.M.; da Silva, A.B.; Dos Santos, B.A.; Lima, J.L.D.C.; Teixeira, L.M.; Neves, F.P.G. CRISPR-Cas systems in enterococci. Braz. J. Microbiol. 2024, 55, 3945–3957. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.; Yosef, I.; Edgar, R.; Qimron, U. The bacterial CRISPR/Cas system as analog of the mammalian adaptive immune system. RNA Biol. 2012, 9, 549–554. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-resistant enterococci lack CRISPR-cas. mBio 2010, 1, e00227-10. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Aghazadeh, M.; Ghotaslou, R.; Rezaee, M.A.; Pirzadeh, T.; Cui, L.; Watanabe, S.; Feizi, H.; Kadkhoda, H.; Kafil, H.S. Role of CRISPR-Cas system on antibiotic resistance patterns of Enterococcus faecalis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 49. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Fang, Y.; Xu, Y.; Liang, W. Association of CRISPR-Cas System with the Antibiotic Resistance and Virulence Genes in Nosocomial Isolates of Enterococcus. Infect. Drug Resist. 2022, 15, 6939–6949. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Agerso, Y.; Gerner-Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; D’Haene, K.; Collard, J.M.; Swings, J. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 2004, 70, 1555–1562. [Google Scholar] [CrossRef]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Ayeni, F.A.; Odumosu, B.T.; Oluseyi, A.E.; Ruppitsch, W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 2016, 8, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Costache, C.; Colosi, I.; Toc, D.A.; Daian, K.; Damacus, D.; Botan, A.; Toc, A.; Pana, A.G.; Panaitescu, P.; Neculicioiu, V.; et al. CRISPR-Cas System, Antimicrobial Resistance, and Enterococcus Genus—A Complicated Relationship. Biomedicines 2024, 12, 1625. [Google Scholar] [CrossRef]
- Pandova, M.; Kizheva, Y.; Tsenova, M.; Rusinova, M.; Borisova, T.; Hristova, P. Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings. Pathogens 2024, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Tsvetanova, Z.; Boshnakov, R. Antimicrobial Resistance of Waste Water Microbiome in an Urban Waste Water Treatment Plant. Water 2025, 17, 39. [Google Scholar] [CrossRef]
- Popova, V.P.; Sredkova, M.P.; Hitkova, H.H.; Ivanov, K.T.; Popov, V.G. Multidrug Resistance Among Enterococci at a Tertiary Care Hospital in Northern Bulgaria. J. Biomed. Clin. Res. 2013, 6, 12–17. [Google Scholar] [CrossRef]
- Guan, L.; Beig, M.; Wang, L.; Navidifar, T.; Moradi, S.; Motallebi Tabaei, F.; Teymouri, Z.; Moghadam, M.A.; Sedighi, M. Global status of antimicrobial resistance in clinical Enterococcus faecalis isolates: Systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 80. [Google Scholar] [CrossRef] [PubMed]
- Gołaś-Prądzyńska, M.; Łuszczyńska, M.; Rola, J.G. Dairy Products: A Potential Source of Multidrug-Resistant Enterococcus faecalis and Enterococcus faecium Strains. Foods 2022, 11, 4116. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.; Karkman, A.; Hultman, J.; Lyra, C.; Bengtsson-Palme, J.; Larsson, D.J.; Rautava, S.; Isolauri, E.; Salminen, S.; Kumar, H.; et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018, 9, 3891. [Google Scholar] [CrossRef]
- Alicea-Serrano, A.M.; Contreras, M.; Magris, M.; Hidalgo, G.; Dominguez-Bello, M.G. Tetracycline resistance genes acquired at birth. Arch. Microbiol. 2013, 195, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Laorden, A.; Arraíz-Fernandez, C.; Gonzalez-Fandos, E. Identification and characterisation of antimicrobial resistanceof Enterococcus spp. Isolated from pork and poultry meat. Int. J. Food Sci. Technol. 2023, 58, 4455–4463. [Google Scholar] [CrossRef]
- Macovei, L.; Zurek, L. Ecology of Antibiotic Resistance Genes: Characterization of Enterococci from Houseflies Collected in Food Settings. Appl. Environ. Microbiol. 2006, 72, 4028–4035. [Google Scholar] [CrossRef]
- Soge, O.; Beck, N.; White, T.; No, D.; Roberts, M.C. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J. Antimicrob. Chemother. 2008, 62, 674–680. [Google Scholar] [CrossRef]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Hullahalli, K.; Rodrigues, M.; Nguyen, U.T.; Palmer, K. An Attenuated CRISPR-Cas System in Enterococcus faecalis Permits DNA Acquisition. mBio 2018, 9, e00414-18, Erratum in mBio 2019, 10. [Google Scholar] [CrossRef]
- Pastuszka, A.; Beauruelle, C.; Camiade, E.; Rousseau, G.M.; Moineau, S.; Mereghetti, L.; Horvath, P.; Lanotte, P. Functional Study of the Type II-A CRISPR-Cas System of Streptococcus agalactiae Hypervirulent Strains. CRISPR J. 2021, 4, 233–242. [Google Scholar] [CrossRef]
- Tao, S.; Zhou, D.; Chen, H.; Li, N.; Zheng, L.; Fang, Y.; Xu, Y.; Jiang, Q.; Liang, W. Analysis of genetic structure and function of clustered regularly interspaced short palindromic repeats loci in 110 Enterococcus strains. Front. Microbiol. 2023, 14, 1177841. [Google Scholar] [CrossRef]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010, 26, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Huescas, C.G.Y.; Pereira, R.I.; Prichula, J.; Azevedo, P.A.; Frazzon, J.; Frazzon, A.P.G. Frequency of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) in non-clinical Enterococcus faecalis and Enterococcus faecium strains. Braz. J. Biol. 2019, 79, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Lyons, C.; Raustad, N.; Bustos, M.A.; Shiaris, M. Incidence of Type II CRISPR1-Cas Systems in Enterococcus Is Species-Dependent. PLoS ONE 2015, 10, e0143544. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Peters, J.E.; Makarova, K.S.; Shmakov, S.; Koonin, E.V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl. Acad. Sci. USA 2017, 114, E7358–E7366. [Google Scholar] [CrossRef] [PubMed]
- Pinilla-Redondo, R.; Mayo-Munoz, D.; Russel, J.; Garrett, R.A.; Randau, L.; Sorensen, S.J.; Shah, S.A. Type IV CRISPR–Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 2020, 48, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155 Pt 6, 1749–1757. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2024. [Google Scholar]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Eaton, T.J.; Gasson, M.J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef]
- Billström, H.; Lund, B.; Sullivan, A.; Nord, C.E. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int. J. Antimicrob. Agents 2008, 32, 374–377. [Google Scholar] [CrossRef]
- Heaton, M.P.; Discotto, L.F.; Pucci, M.J.; Handwerger, S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 1996, 171, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.B.; Garcia-Migura, L.; Valenzuela, A.J.; Løhr, M.M.; Hasman, H.; Aarestrup, F.M. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 2010, 80, 25–43. [Google Scholar] [CrossRef]
- Lin, Y.; Yuan, J.; Kolmogorov, M.; Shen, M.W.; Chaisson, M.; Pevzner, P.A. Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA 2016, 113, E8396–E8405. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows—Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Oxford Nanopore Technologies Ltd. Medaka. 2020. Available online: https://github.com/nanoporetech/medaka (accessed on 31 October 2014).
- Stothard, P.; Grant, J.R.; Van Domselaar, G. Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 2019, 20, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Gagnon, J.N.; Brouns, S.J.; Fineran, P.C.; Brown, C.M. CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013, 10, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
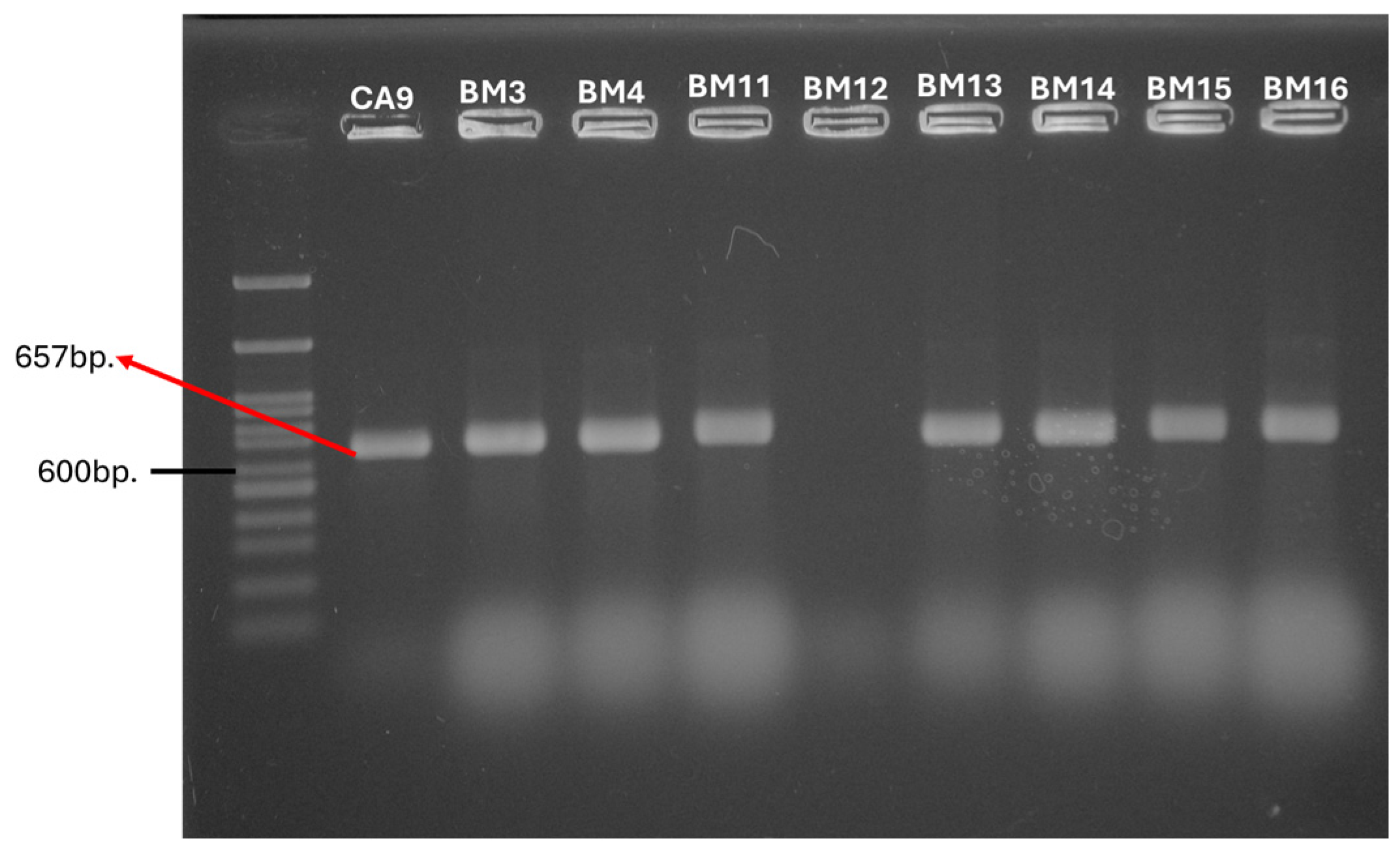
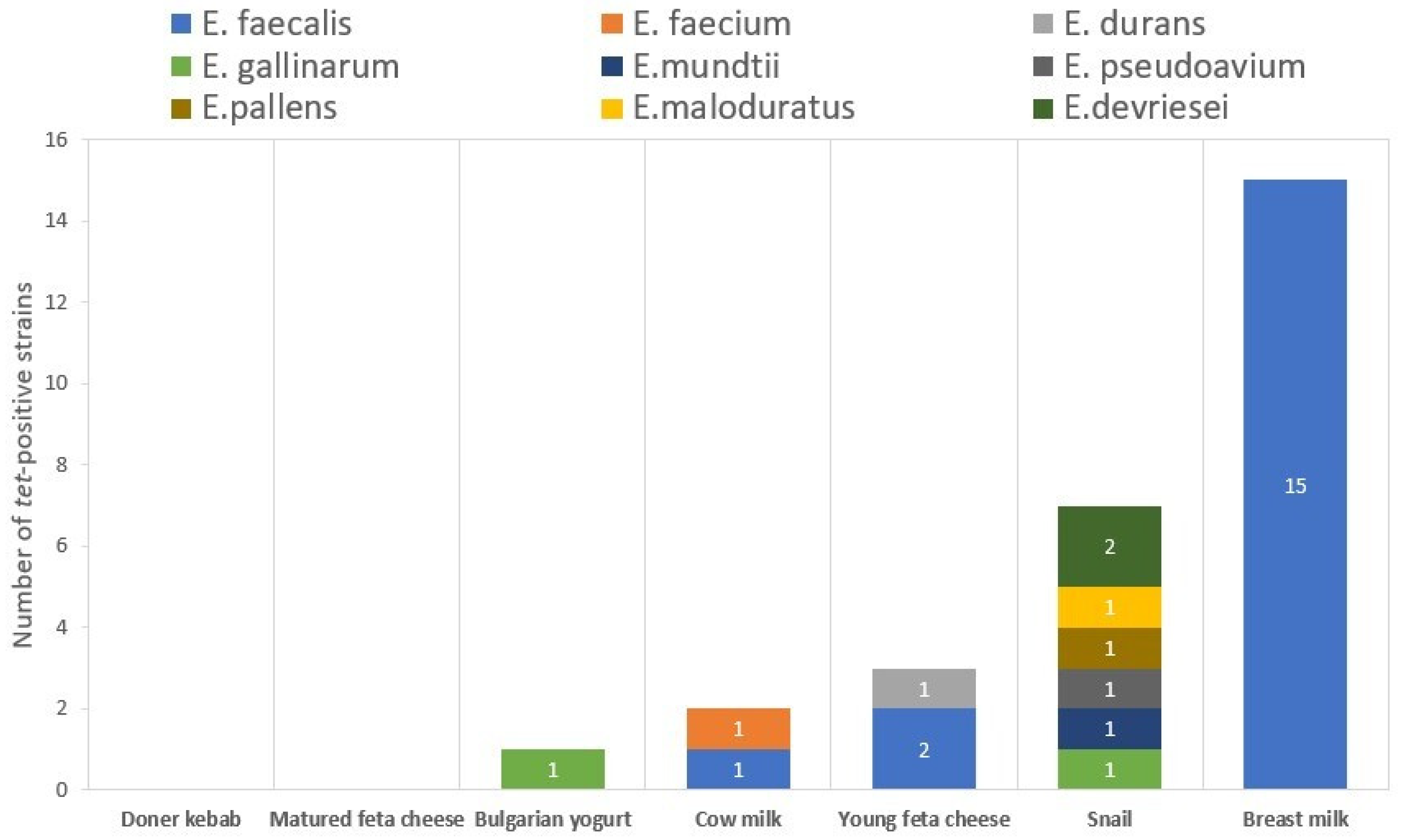

| Presence of tet Genes | Number of Strains (n), % | Strains | Origin | Inhibition Zone (mm) | CLSI Interpretation |
|---|---|---|---|---|---|
| tetM | (n = 22), 31% | E. gallinarum BY17 | Bulgarian yogurt | 14 | R |
| E. faecium CM1 | Cow milk | 19 | S | ||
| E. faecalis CM4 | 15 | I | |||
| E. faecalis YFC1 | Young feta cheese | 14 | R | ||
| E. faecalis YFC3 | 28 | S | |||
| E. pseudoavium CA9 | Cornu aspersum | 14 | R | ||
| E. pallens CA10 | 16 | I | |||
| E. faecalis BM2 | Breast milk | 25 | S | ||
| E. faecalis BM3–BM9 E. faecalis BM14–BM16 | 14 | R | |||
| E. faecalis BM11, BM12 | 13 | R | |||
| E. faecalis BM10, BM13 | 16 | I | |||
| tetS | (n = 6), 8% | E. faecalis YFC3 | Young feta cheese | 28 | S |
| E. mundtii CA8 | Cornu aspersum | 28 | S | ||
| E. pallens CA10 | 16 | I | |||
| E. devriesei CA13 | 49 | S | |||
| E. devriesei CA16 | 17 | I | |||
| E. faecalis BM15 | Breast milk | 14 | R | ||
| tetO | (n = 2), 3% | E. durans YFC5 | Young feta cheese | 32 | S |
| E. malodoratus CA11 | Cornu aspersum | 50 | S | ||
| tetT | (n = 1), 1% | E. gallinarum CA15 | Cornu aspersum | 35 | S |
| tetM+tetS | (n = 3), 4% | E. pallens CA10 | Cornu aspersum | 16 | I |
| E. faecalis BM15 | Breast milk | 14 | R | ||
| E. faecalis YFC3 | Young feta cheese | 28 | S |
| Strains | CRISPR | Strains | CRISPR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRISPR1–cas csn1 | CRISPR1–cas loci | CRISPR2 loci | CRISPR3–cas csn1 | CRISPR3–cas loci | CRISPR1–cas csn1 | CRISPR1–cas loci | CRISPR2 loci | CRISPR3–cas csn1 | CRISPR3–cas loci | ||
| E. faecium CM1 | E. faecalis BY25 | ||||||||||
| E. durans CM2 | E. faecalis BY26 | ||||||||||
| E. durans CM3 | E. faecalis BY27 | ||||||||||
| E. faecalis CM4 | E. mundtii CA1 | ||||||||||
| E. faecalis YFC1 | E. casseliflavus CA2 | ||||||||||
| E. durans YFC2 | E. gilvus CA3 | ||||||||||
| E. faecalis YFC3 | E. mundtii CA4 | ||||||||||
| E. durans YFC4 | E. casseliflavus CA5 | ||||||||||
| E. durans YFC5 | E. mundtii CA6 | ||||||||||
| E. faecium MFC1 | E. mundtii CA7 | ||||||||||
| E. faecium MFC2 | E. mundtii CA8 | ||||||||||
| E. faecium DK1 | E. pseudoavium CA9 | ||||||||||
| E. faecium BY1 | E. pallens CA10 | ||||||||||
| E. faecalis BY2 | E. malodoratus CA11 | ||||||||||
| E. faecalis BY3 | E. casseliflavus CA12 | ||||||||||
| E. faecalis BY4 | E. devriesei CA13 | ||||||||||
| E. faecalis BY5 | E. gallinarum CA14 | ||||||||||
| E. faecalis BY6 | E. gallinarum CA15 | ||||||||||
| E. species BY7 | E. devriesei CA16 | ||||||||||
| E. species BY8 | E. mundtii CA17 | ||||||||||
| E. casseliflavus BY9 | E. faecalis BM1 | ||||||||||
| E. faecalis BY10 | E. faecalis BM2 | ||||||||||
| E. faecalis BY11 | E. faecalis BM3 | ||||||||||
| E. faecium BY12 | E. faecalis BM4 | ||||||||||
| E. faecium BY13 | E. faecalis BM5 | ||||||||||
| E. faecium BY14 | E. faecalis BM6 | ||||||||||
| E. faecium BY15 | E. faecalis BM7 | ||||||||||
| E. faecium BY16 | E. faecalis BM8 | ||||||||||
| E. gallinarum BY17 | E. faecalis BM9 | ||||||||||
| E. casseliflavus BY18 | E. faecalis BM10 | ||||||||||
| E. casseliflavus BY19 | E. faecalis BM11 | ||||||||||
| E. casseliflavus BY20 | E. faecalis BM12 | ||||||||||
| E. casseliflavus BY21 | E. faecalis BM13 | ||||||||||
| E. faecalis BY22 | E. faecalis BM14 | ||||||||||
| E. faecalis BY23 | E. faecalis BM15 | ||||||||||
| E. faecalis BY24 | E. faecalis BM16 | ||||||||||
| Strains | Genes Responsible for HGT | Strains | Genes Responsible for HGT | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cpd | cob | ccf | prgW | Int-tn | asa1 | prgB | asa373 | agg | cpd | cob | ccf | prgW | Int-tn | asa1 | prgB | asa373 | agg | ||
| E. faecium CM1 | E. faecalis BY25 | ||||||||||||||||||
| E. durans CM2 | E. faecalis BY26 | ||||||||||||||||||
| E. durans CM3 | E. faecalis BY27 | ||||||||||||||||||
| E. faecalis CM4 | E. mundtii CA1 | ||||||||||||||||||
| E. faecalis YFC1 | E. casseliflavus CA2 | ||||||||||||||||||
| E. durans YFC2 | E. gilvus CA3 | ||||||||||||||||||
| E. faecalis YFC3 | E. mundtii CA4 | ||||||||||||||||||
| E. durans YFC4 | E. casseliflavus CA5 | ||||||||||||||||||
| E. durans YFC5 | E. mundtii CA6 | ||||||||||||||||||
| E. faecium MFC1 | E. mundtii CA7 | ||||||||||||||||||
| E. faecium MFC2 | E. mundtii CA8 | ||||||||||||||||||
| E. faecium DK1 | E. pseudoavium CA9 | ||||||||||||||||||
| E. faecium BY1 | E. pallens CA10 | ||||||||||||||||||
| E. faecalis BY2 | E. maloduratus CA11 | ||||||||||||||||||
| E. faecalis BY3 | E. casseliflavus CA12 | ||||||||||||||||||
| E. faecalis BY4 | E. devriesei CA13 | ||||||||||||||||||
| E.faecalis BY5 | E. gallinarum CA14 | ||||||||||||||||||
| E.faecalis BY6 | E. gallinarum CA15 | ||||||||||||||||||
| E. species BY7 | E. devriesei CA16 | ||||||||||||||||||
| E. species BY8 | E. mundtii CA17 | ||||||||||||||||||
| E. casseliflavus BY9 | E. faecalis BM1 | ||||||||||||||||||
| E. faecalis BY10 | E. faecalis BM2 | ||||||||||||||||||
| E. faecalis BY11 | E. faecalis BM3 | ||||||||||||||||||
| E. faecium BY12 | E. faecalis BM4 | ||||||||||||||||||
| E. faecium BY13 | E. faecalis BM5 | ||||||||||||||||||
| E. faecium BY14 | E. faecalis BM6 | ||||||||||||||||||
| E. faecium BY15 | E. faecalis BM7 | ||||||||||||||||||
| E. faecium BY16 | E. faecalis BM8 | ||||||||||||||||||
| E. gallinarum BY17 | E. faecalis BM9 | ||||||||||||||||||
| E. casseliflavus BY18 | E. faecalis BM10 | ||||||||||||||||||
| E. casseliflavus BY19 | E. faecalis BM11 | ||||||||||||||||||
| E. casseliflavus BY20 | E. faecalis BM12 | ||||||||||||||||||
| E. casseliflavus BY21 | E. faecalis BM13 | ||||||||||||||||||
| E. faecalis BY22 | E. faecalis BM14 | ||||||||||||||||||
| E. faecalis BY23 | E. faecalis BM15 | ||||||||||||||||||
| E. faecalis BY24 | E. faecalis BM16 | ||||||||||||||||||
| Isolate | Repeat Sequence | CRISPR Repeat Subtyping | Secondary RNA Structures | * MFE [kcal/mol] | Spacer Sequence | Spacer Target |
|---|---|---|---|---|---|---|
| CM4 | GTTTTAGAGTCATGTTGTTTAGAATGGTACCAAAAC | II-A |  | −2.30 | GGTTATTATGTTACTGGTTACTTTAAAGAC | chromosome |
| ATAATGATGTACAATTTATTCAAAACCATA | phage | |||||
| GAAAAGCAGTTCGAGCGGAAACTGCGACCA | phage | |||||
| GACTTACAAAAGACTGTGATTTACGTTATA | phage | |||||
| AAACTTTTTTGATTTGGCTTTTTCTCCCT | phage | |||||
| ACAAGGTGACCAAAGGGAACGTTGT | VI-B1 |  | −7.10 | CTCCTCTATGTTAAAACAAACTGCTTAGCCAAAAACATGGAGTAGATGATGAACAGC | phage | |
| ATAGTTGGCGAGCAACAGAAAAAC | III-B | 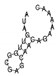 | −3.20 | TCGATATAGAATTGGACGTAGAGCCA | phage | |
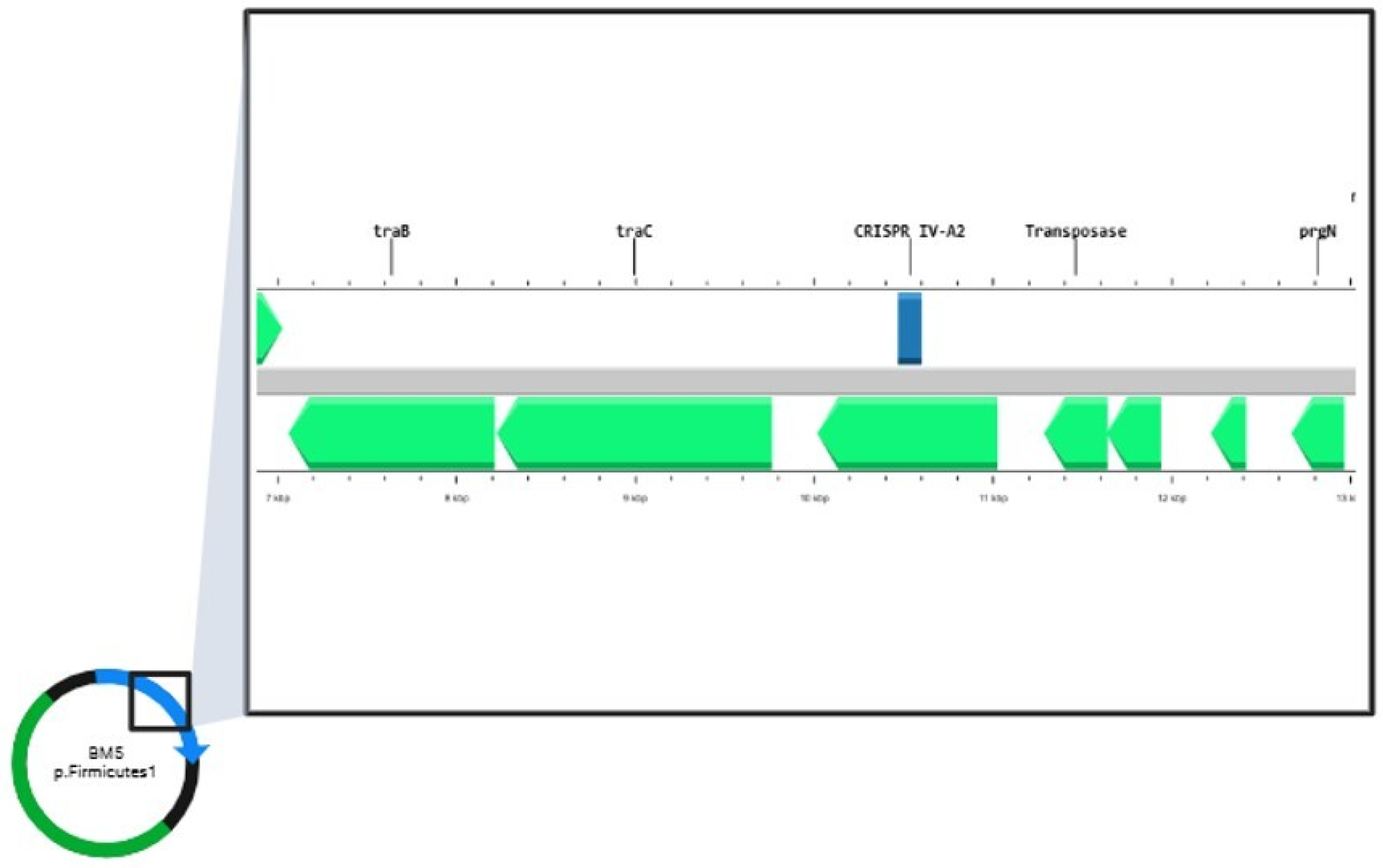
| BM5 | GTTGGTTTTTCCCACTTTCGAACA | IV-A2 |  | −1.40 | AAGTACTGGTATTATTGGATTCTTCTGGAC | plasmid |
| AAACGCCGATTTTATCATGTTTATCCGAAG | plasmid | |||||
| GTTTTGGTACCATTCTAAACAACATGACTCTAAAAC | II-A |  | −3.40 | TCTAATTTTTGAGTAATCGTACCAACTTGG | chromosome | |
| CTACGTCTTAACAAAGATAATTTAAAAGGT | phage | |||||
| GAAGCTACGTTTAAACCCGAAACCCCACTA | chromosome | |||||
| TAGGTAAGTAACTTAACCCTAGGTCAATCG | chromosome | |||||
| ATAGTTGGCGAGCAACAGAAAAAC | III-B |  | −3.20 | TCGATATAGAATTGGACGTAGAGCCA | phage | |
| BM 12 | TAAAACAAACTGCTTAGCCAAAAACATGGAG | II-A | 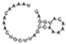 | −1.60 | TAGATGATGAACAGTACAAGGTGACCAAAGGGAACGTTGTCTCCTCTGTGC | phage |
| GTTTTAGAGTCATGTTGTTTAGAATGGTACCAAAAC | II-A |  | −2.30 | AAGTACGGCATTACGCATTCCCCACTTTCT | phage | |
| GTAACAAACGATTAACTTTCGCATAGTCAT | phage | |||||
| AACCGAACTTACACCAACTGCGGATGGTAT | phage | |||||
| TATCGAAAATGATGTATTAATTTTAGGCTA | phage | |||||
| TACCTATGCAGACATTAAGAATTTACCAGA | phage | |||||
| TTATTTGAGAATCTGAAACATTTAGTTCAT | phage | |||||
| ATTTTGATGCATTAGCACCAAAATCAAAAG | phage | |||||
| ATTACTTGTTAAGGCTTCAATTATCAATTC | phage | |||||
| AAACTTTTTTGATTTGGCTTTTTCTCCCCT | chromosome | |||||
| BM 15 | GTTTTAGAGTCATGTTGTTTAGAATGGTACCAAAAC | II-A |  | −2.30 | TAAAGCAGCTTCTAAAACAGAAGGTGAAAT | phage |
| GATTGGTAAGATTACATGATCTTTAGTACG | chromosome | |||||
| AAAGAAATGGACACATTACACAACGCTTTC | phage | |||||
| TAAAAACAAGACGAAATGAGGAAATTAACA | chromosome | |||||
| CAATGTAAATGCTCATTATGATTTACATAT | chromosome | |||||
| GTTTTGGTACCATTCTAAACAACATGACTCTAAAAC | II-A |  | −3.40 | AAATTTTTTGAACTTAATGCAATTTCTTGA | chromosome | |
| TTTGATAATCCAGAATCAACATCTTCACCA | phage | |||||
| TGCATAATAATCTTTTCTCTTAATGTTTTT | phage | |||||
| AACCCTCTTACTATGAGTTCCATTTATTTT | phage |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandova, M.; Kizheva, Y.; Hristova, P. Relationship Between CRISPR–Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria. Antibiotics 2025, 14, 145. https://doi.org/10.3390/antibiotics14020145
Pandova M, Kizheva Y, Hristova P. Relationship Between CRISPR–Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria. Antibiotics. 2025; 14(2):145. https://doi.org/10.3390/antibiotics14020145
Chicago/Turabian StylePandova, Maria, Yoana Kizheva, and Petya Hristova. 2025. "Relationship Between CRISPR–Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria" Antibiotics 14, no. 2: 145. https://doi.org/10.3390/antibiotics14020145
APA StylePandova, M., Kizheva, Y., & Hristova, P. (2025). Relationship Between CRISPR–Cas Systems and Acquisition of Tetracycline Resistance in Non-Clinical Enterococcus Populations in Bulgaria. Antibiotics, 14(2), 145. https://doi.org/10.3390/antibiotics14020145






