A Comprehensive Overview of Antimicrobial Peptides: Broad-Spectrum Activity, Computational Approaches, and Applications
Abstract
1. Introduction
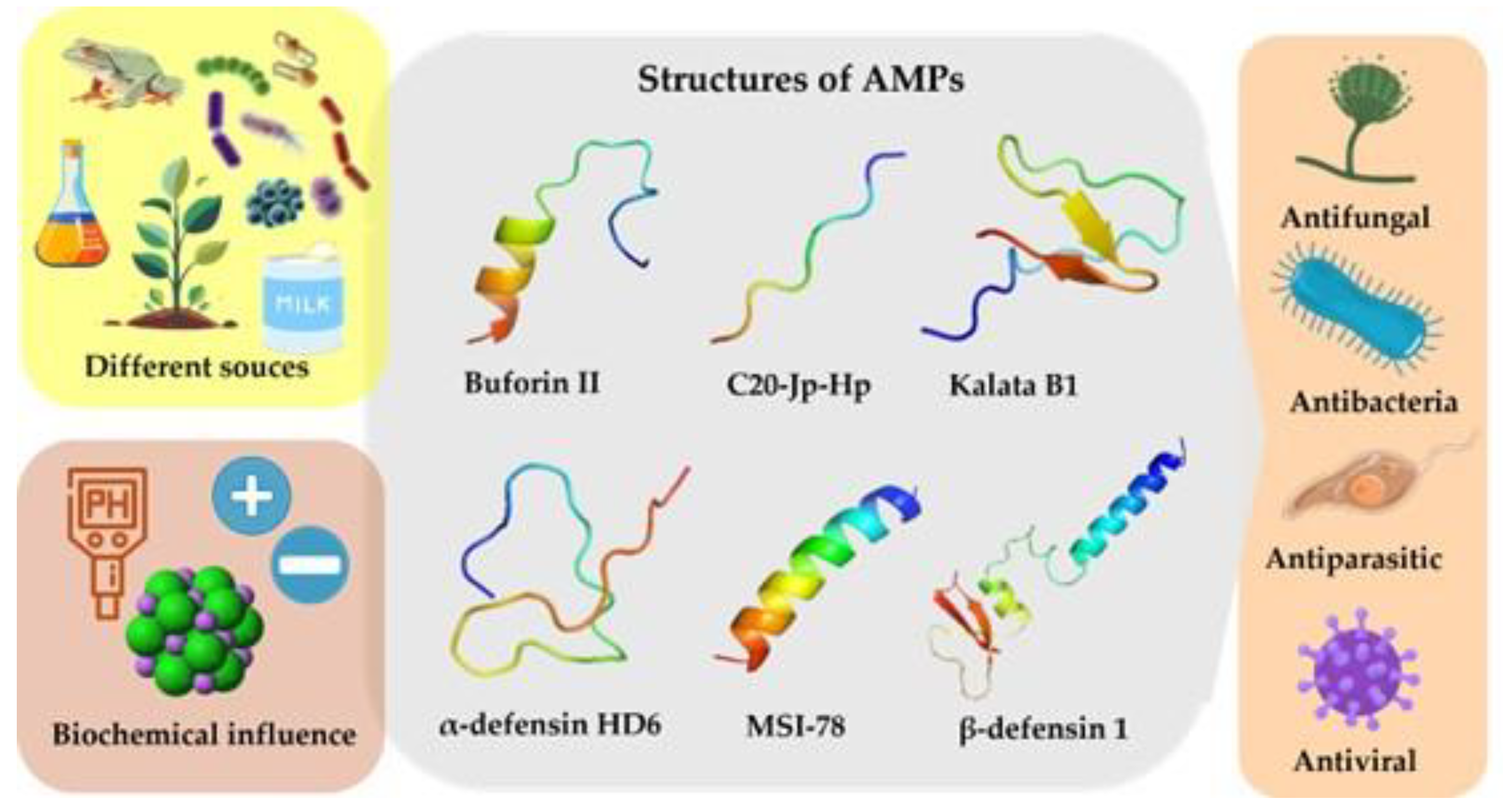
2. Broad-Spectrum Activity of Peptides
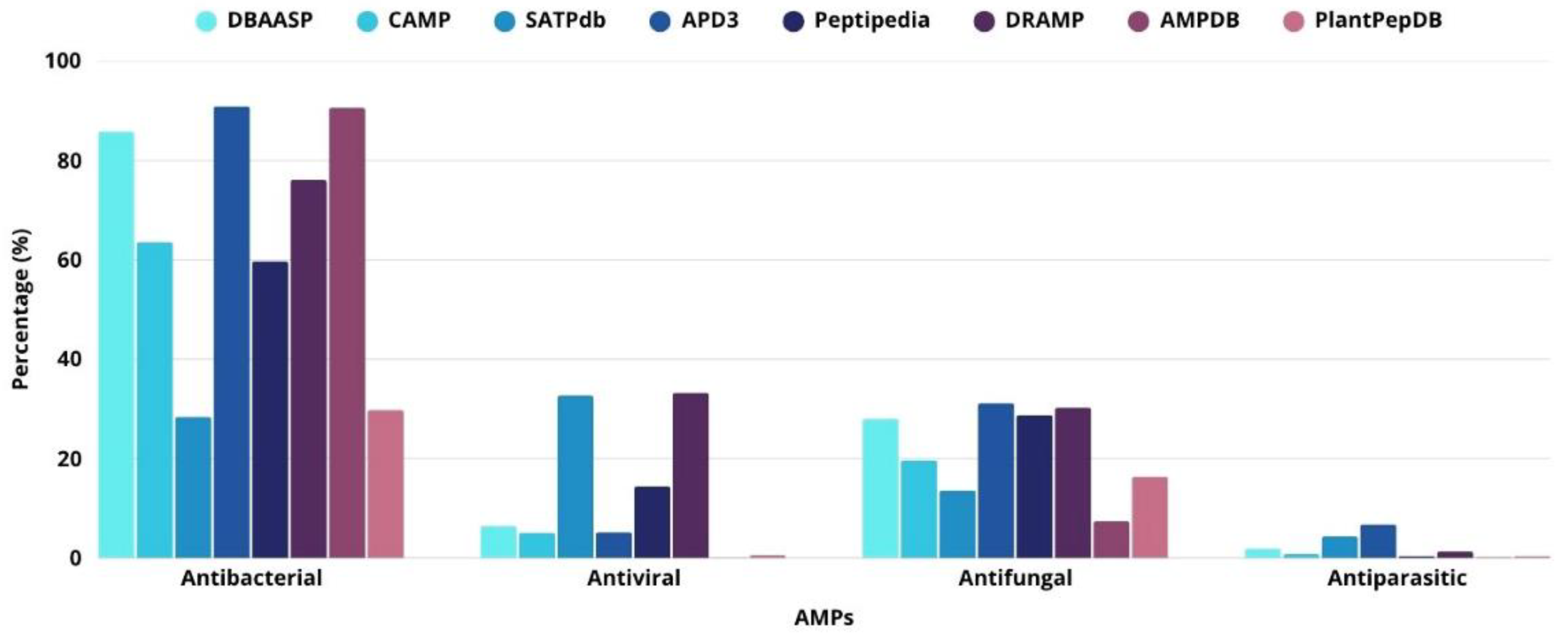
2.1. Antibacterial Activity
2.2. Antiviral Activity
2.3. Antifungal Activity
2.4. Antiparasitic Activity
3. Approaches for Obtaining AMPs
3.1. Peptide Production via Chemical Synthesis
3.2. Peptide Production via Enzymatic Pathways
3.3. Peptide Extraction from Natural Sources
3.4. Peptide Production via Recombinant Technology
4. Biochemical Characteristics of AMPs
4.1. Structure
4.2. Charge, pH, Saline Concentration
4.3. Hydrophobicity
5. Antimicrobial Peptide Action Mechanism
5.1. Extracellular Target AMPs
5.2. Intracellular Target AMPs
5.3. Immunomodulatory AMPs
6. Clinical Trial of AMPs
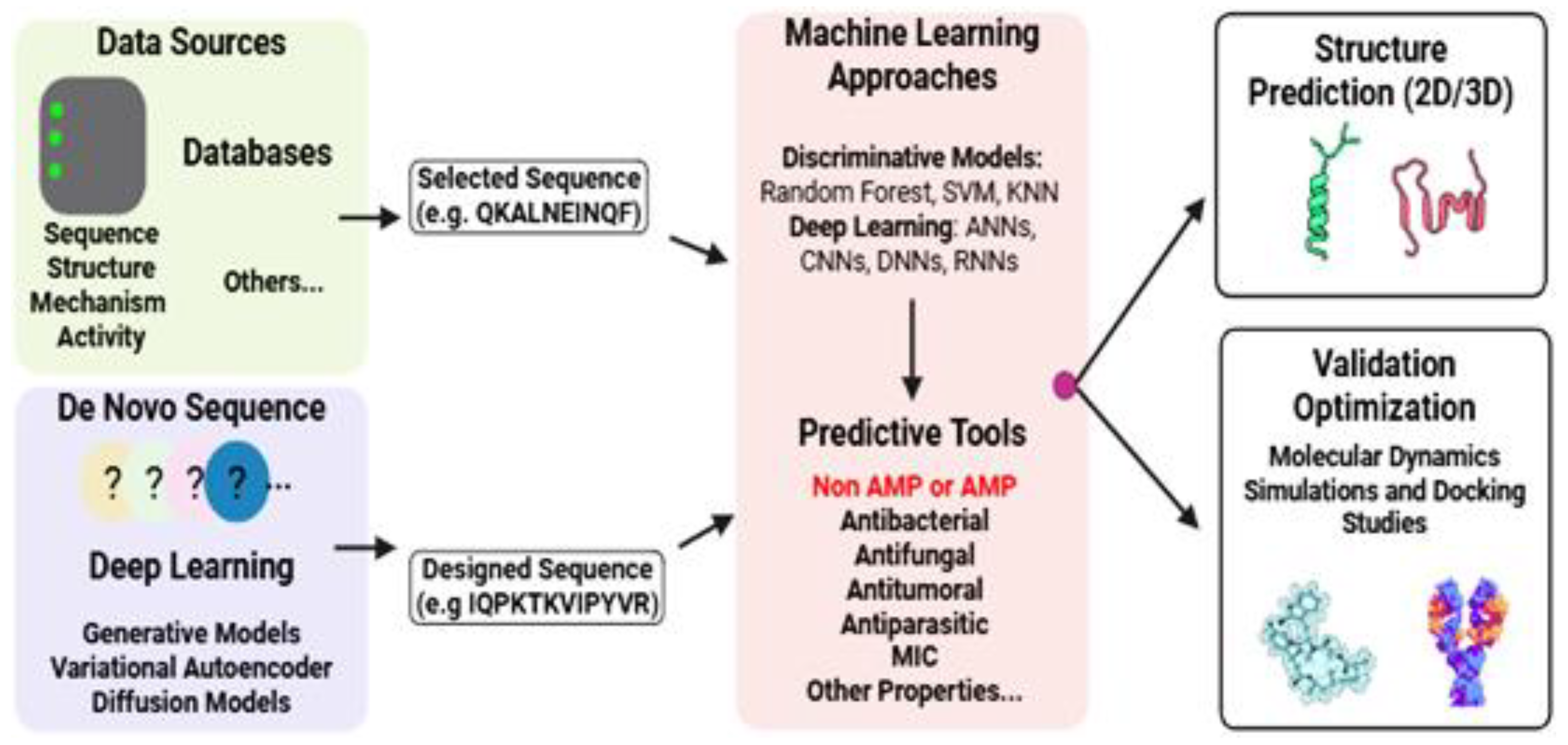
7. Computational Approaches for AMPs Discovery and Design
8. AMPs and Their Use in the Food Industry
9. AMPs and Their Use in Veterinary Medicine
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAP | Bioactive Peptide |
| AMP | Antimicrobial Peptide |
| APD3 | Antimicrobial Peptide Database 3 |
| AMR | Antibiotic Resistance |
| WHO | World Health Organization |
| MERS | Middle East respiratory syndrome |
| SARS | Severe acute respiratory syndrome |
| AVP | Antiviral peptides |
| AFP | Antifungal peptides |
| SPPS | Solid-phase Peptide Synthesis |
| HBTU | O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate |
| HATU | O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate |
| DIC | N,N′-diisopropylcarbodiimide |
| CCK | Cyclic Cystine Knot |
| DMF | Dimethylformamide |
| UAE | Ultrasound-assisted extraction |
| PEF | Pulsed electric fields |
| HHP | High hydrostatic pressure |
| AI | Artificial Intelligence |
| ML | Machine Learning |
| GST | Glutathione S-transferase |
| MBP | Maltose-binding protein |
| GRAS | Generally recognized as safe |
| PG | Phosphatidylglycerol |
| CL | Cardiolipin |
| PS | Phosphatidylserine |
| LPS | Lipopolysaccharides |
| HSPG | Heparan sulfate proteoglycan |
| HCMV | Human cytomegaloviruses |
| MCMV | Murine cytomegaloviruses |
| DHFR | Dihydrofolate reductase |
| TCA | Tricarboxylic acid |
| SQR | Succinate-coenzyme Q reductase |
| ROS | Reactive oxygen species |
| HSV-1 | Simplex virus type 1 |
| HDPs | Host defense peptides |
| LBP | Binding protein |
| FDA | Food and Drug Administration |
| IV | Intravenous injections |
| IM | Intramuscular injections |
| SC | Subcutaneous injections |
| CS/SA | Chitosan/sodium alginate |
| QSAR | Quantitative Structure–Activity Relationship |
| CNNs | Convolutional Neural Networks |
| DNNs | Deep Neural Networks |
| RNNs | Recurrent Neural Networks |
| VAEs | Variational Autoencoders |
| GANs | Generative Adversarial Networks |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| DL | Deep learning |
| SVM | Support vector machines |
| AIR | Ambiguous interaction restraints (AIRs). |
References
- Bizzotto, E.; Zampieri, G.; Treu, L.; Filannino, P.; Di Cagno, R.; Campanaro, S. Classification of bioactive peptides: A systematic benchmark of models and encodings. Comput. Struct. Biotechnol. J. 2024, 23, 2442–2452. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Kanaujia, K.A.; Wagh, S.; Pandey, G.; Phatale, V.; Khairnar, P.; Kolipaka, T.; Rajinikanth, P.; Saraf, S.A.; Srivastava, S.; Kumar, S. Harnessing marine antimicrobial peptides for novel therapeutics: A deep dive into ocean-derived bioactives. Int. J. Biol. Macromol. 2025, 307, 142158. [Google Scholar] [CrossRef]
- Habib, H.M.; Ismail, R.; Agami, M.; El-Yazbi, A.F. Exploring the impact of bioactive peptides from fermented Milk proteins: A review with emphasis on health implications and artificial intelligence integration. Food Chem. 2025, 481, 144047. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Van Epps, H.L. René Dubos: Unearthing antibiotics. J. Exp. Med. 2006, 203, 259. [Google Scholar] [CrossRef]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2019, 27, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Roudi, R.; Syn, N.L.; Roudbary, M. Antimicrobial Peptides as Biologic and Immunotherapeutic Agents against Cancer: A Comprehensive Overview. Front. Immunol. 2017, 8, 1320. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Pirtskhalava, M.; Amstrong, A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Gawde, U.; Chakraborty, S.; Waghu, F.H.; Barai, R.S.; Khanderkar, A.; Indraguru, R.; Shirsat, T.; Idicula-Thomas, S. CAMPR4: A database of natural and synthetic antimicrobial peptides. Nucleic Acids Res. 2023, 51, D377–D383. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016, 44, D1119–D1126. [Google Scholar] [CrossRef]
- Quiroz, C.; Saavedra, Y.B.; Armijo-Galdames, B.; Amado-Hinojosa, J.; Olivera-Nappa, Á.; Sanchez-Daza, A.; Medina-Ortiz, D. Peptipedia: A user-friendly web application and a comprehensive database for peptide research supported by Machine Learning approach. Database 2021, 2021, baab055. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Liu, Y.; Yu, B.; Sun, X.; Yao, H.; Hao, C.; Li, J.; Nawaz, M.; Jiang, X.; Lao, X.; et al. DRAMP 4.0: An open-access data repository dedicated to the clinical translation of antimicrobial peptides. Nucleic Acids Res. 2025, 53, D403–D410. [Google Scholar] [CrossRef] [PubMed]
- Mondal, R.K.; Sen, D.; Arya, A.; Samanta, S.K. Developing anti-microbial peptide database version 1 to provide comprehensive and exhaustive resource of manually curated AMPs. Sci. Rep. 2023, 13, 17843. [Google Scholar] [CrossRef]
- Das, D.; Jaiswal, M.; Khan, F.N.; Ahamad, S.; Kumar, S. PlantPepDB: A manually curated plant peptide database. Sci. Rep. 2020, 10, 2194. [Google Scholar] [CrossRef]
- O’Neill, J.C. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 14 August 2025).
- World Health Organization. Pathogens Prioritization A; WHO: Geneva, Switzerland, 2024.
- Poudel, A.N.; Zhu, S.; Cooper, N.; Little, P.; Tarrant, C.; Hickman, M.; Yao, G. The economic burden of antibiotic resistance: A systematic review and meta-analysis. PLoS ONE 2023, 18, e0285170. [Google Scholar] [CrossRef]
- Al-Madboly, L.A.; Aboulmagd, A.; El-Salam, M.A.; Kushkevych, I.; El-Morsi, R.M. Microbial enzymes as powerful natural anti-biofilm candidates. Microb. Cell Factories 2024, 23, 343. [Google Scholar] [CrossRef]
- Yang, A.; Bai, Y.; Zhang, Y.; Xiao, R.; Zhang, H.; Chen, F.; Zeng, W. Detection and Treatment with Peptide Power: A New Weapon Against Bacterial Biofilms. ACS Biomater. Sci. Eng. 2025, 11, 806–819. [Google Scholar] [CrossRef]
- World Health Organization. Disease Outbreak News; Ebola Virus Disease in the Democratic Republic of the Congo. 2025. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2025-DON580 (accessed on 2 August 2025).
- Qureshi, A. A review on current status of antiviral peptides. Discov. Viruses 2025, 2, 3. [Google Scholar] [CrossRef]
- Puumala, E.; Fallah, S.; Robbins, N.; Cowen, L.E. Advancements and challenges in antifungal therapeutic development. Clin. Microbiol. Rev. 2024, 37, e0014223. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, H.-J. Antimicrobial Activity of Probiotic Bacteria Isolated from Plants: A Review. Foods 2025, 14, 495. [Google Scholar] [CrossRef]
- Rivera-Fernández, N.; Anacleto-Santos, J.; Casarrubias-Tabarez, B.; López-Pérez, T.d.J.; Rojas-Lemus, M.; López-Valdez, N.; Fortoul, T.I. Bioactive Peptides against Human Apicomplexan Parasites. Antibiotics 2022, 11, 1658. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Morán-Marcillo, G.; Monteros-Silva, N.E.d.L.; Bermúdez-Puga, S.; Salazar, M.A.; Blasco-Zúñiga, A.; Cuesta, S.; Molina, C.; Espinosa, F.; Meneses, L.; et al. Bioactivity of synthetic peptides from Ecuadorian frog skin secretions against Leishmania mexicana, Plasmodium falciparum, and Trypanosoma cruzi. Microbiol. Spectr. 2024, 12, e0333923. [Google Scholar] [CrossRef] [PubMed]
- Slezina, M.P.; Odintsova, T.I. Plant Antimicrobial Peptides: Insights into Structure-Function Relationships for Practical Applications. Curr. Issues Mol. Biol. 2023, 45, 3674–3704. [Google Scholar] [CrossRef] [PubMed]
- Shirsat, H.; Datt, M.; Kale, A.; Mishra, M. Plant Defense Peptides: Exploring the Structure–Function Correlation for Potential Applications in Drug Design and Therapeutics. ACS Omega 2025, 10, 7583–7596. [Google Scholar] [CrossRef]
- Meng, T.; Wen, J.; Liu, H.; Guo, Y.; Tong, A.; Chu, Y.; Du, B.; He, X.; Zhao, C. Algal proteins and bioactive peptides: Sustainable nutrition for human health. Int. J. Biol. Macromol. 2025, 303, 140760. [Google Scholar] [CrossRef] [PubMed]
- García-Encinas, J.P.; Ruiz-Cruz, S.; Juárez, J.; Ornelas-Paz, J.d.J.; Del Toro-Sánchez, C.L.; Márquez-Ríos, E. Proteins from Microalgae: Nutritional, Functional and Bioactive Properties. Foods 2025, 14, 921. [Google Scholar] [CrossRef]
- Bosso, A.; Di Nardo, I.; Culurciello, R.; Palumbo, I.; Gaglione, R.; Zannella, C.; Pinto, G.; Siciliano, A.; Carraturo, F.; Amoresano, A.; et al. KNR50: A moonlighting bioactive peptide hidden in the C-terminus of bovine casein αS2 with powerful antimicrobial, antibiofilm, antiviral and immunomodulatory activities. Int. J. Biol. Macromol. 2025, 311, 143718. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, S.K.; Larson, R.G. Binding and insertion of α-helical anti-microbial peptides in POPC bilayers studied by molecular dynamics simulations. Chem. Phys. Lipids 2004, 132, 113–132. [Google Scholar] [CrossRef]
- García-Beltrán, J.M.; Arizcun, M.; Chaves-Pozo, E. Antimicrobial Peptides from Photosynthetic Marine Organisms with Potential Application in Aquaculture. Mar. Drugs 2023, 21, 290. [Google Scholar] [CrossRef]
- Rodrigues, T.; Guardiola, F.A.; Almeida, D.; Antunes, A. Aquatic Invertebrate Antimicrobial Peptides in the Fight Against Aquaculture Pathogens. Microorganisms 2025, 13, 156. [Google Scholar] [CrossRef]
- Chauhan, K.; Rao, A. Clean-label alternatives for food preservation: An emerging trend. Heliyon 2024, 10, e35815. [Google Scholar] [CrossRef]
- Hao, Y.; Teng, D.; Mao, R.; Yang, N.; Wang, J. Site Mutation Improves the Expression and Antimicrobial Properties of Fungal Defense. Antibiotics 2023, 12, 1283. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Andishmand, H.; Pilevar, Z.; Hashempour-Baltork, F.; Torbati, M.; Dadgarnejad, M.; Rastegar, H.; Mohammadi, S.A.; Azadmard-Damirchi, S. Innovative perspectives on bacteriocins: Advances in classification, synthesis, mode of action, and food industry applications. J. Appl. Microbiol. 2024, 135, lxae274. [Google Scholar] [CrossRef] [PubMed]
- Fabián, J.C.P.; Contreras, A.K.Á.; Bonifacio, I.N.; Robles, M.F.H.; Quiñones, C.R.V.; Ramírez, E.I.Q.; Salinas, C.V. Toward safer and sustainable food preservation: A comprehensive review of bacteriocins in the food industry. Biosci. Rep. 2025, 45, 277–302. [Google Scholar] [CrossRef]
- Esposito, D.; Chatterjee, D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. [Google Scholar] [CrossRef]
- Bernardeau, M.; Vernoux, J.P.; Henri-Dubernet, S.; Guéguen, M. Safety assessment of dairy microorganisms: The Lactobacillus genus. Int. J. Food Microbiol. 2008, 126, 278–285. [Google Scholar] [CrossRef]
- Sorokulova, I. Modern Status and Perspectives of Bacillus Bacteria as Probiotics. J. Probiotics Health 2013, 1, 4. [Google Scholar] [CrossRef]
- Sutcliffe, R.; Doherty, C.P.A.; Morgan, H.P.; Dunne, N.J.; McCarthy, H.O. Strategies for the design of biomimetic cell-penetrating peptides using AI-driven in silico tools for drug delivery. Biomater. Adv. 2025, 169, 214153. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Ludtke, S.J.; He, K.; Heller, W.T.; Harroun, T.A.; Yang, L.; Huang, H.W. Membrane Pores Induced by Magainin. Biochemistry 1996, 35, 13723–13728. [Google Scholar] [CrossRef]
- Sugimoto, A.; Maeda, A.; Itto, K.; Arimoto, H. Deciphering the mode of action of cell wall-inhibiting antibiotics using metabolic labeling of growing peptidoglycan in Streptococcus pyogenes. Sci. Rep. 2017, 7, 1129. [Google Scholar] [CrossRef] [PubMed]
- Cursino, L.; Smajs, D.; Smarda, J.; Nardi, R.; Nicoli, J.; Chartone-Souza, E.; Nascimento, A. Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. J. Appl. Microbiol. 2006, 100, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Mercer, D.K.; Robertson, J.C.; Miller, L.; Stewart, C.S.; O’Neil, D.A. NP213 (Novexatin®): A unique therapy candidate for onychomycosis with a differentiated safety and efficacy profile. Med. Mycol. 2020, 58, 1064–1072. [Google Scholar] [CrossRef]
- Lewies, A.; Wentzel, J.F.; Garmi, J.; Du Plessis, L.H. The Potential Use of Natural and Structural Analogues of Antimicrobial Peptides in the Fight against Neglected Tropical Diseases. Molecules 2015, 20, 15392–15433. [Google Scholar] [CrossRef]
- Grover, A.; Singh, S.; Sindhu, S.; Lath, A.; Kumar, S. Advances in cyclotide research: Bioactivity to cyclotide-based therapeutics. Mol. Divers. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Pei, L.; Hou, Y.; Feng, Y.; Li, F.; Su, H.; Zhang, Y.; Song, Y.; Liu, K.; Cao, G. Equine β-defensin 1 regulates cytokine expression and phagocytosis in S. aureus-infected mouse monocyte macrophages via the Paxillin-FAK-PI3K pathway. Int. Immunopharmacol. 2023, 123, 110793. [Google Scholar] [CrossRef] [PubMed]
- Lundbæk, J.A.; Collingwood, S.A.; Ingólfsson, H.I.; Kapoor, R.; Andersen, O.S. Lipid bilayer regulation of membrane protein function: Gramicidin channels as molecular force probes. J. R. Soc. Interface 2010, 7, 373–395. [Google Scholar] [CrossRef]
- Bahar, A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef]
- Bulger, E.M.; Maier, R.V.; Sperry, J.; Joshi, M.; Henry, S.; Moore, F.A.; Moldawer, L.L.; Demetriades, D.; Talving, P.; Schreiber, M.; et al. A Novel Drug for Treatment of Necrotizing Soft-Tissue Infections. JAMA Surg. 2014, 149, 528. [Google Scholar] [CrossRef]
- Mba, I.E.; Nweze, E.I. Antimicrobial Peptides Therapy: An Emerging Alternative for Treating Drug-Resistant Bacteria. Yale J. Biol. Med. 2022, 95, 445–463. [Google Scholar] [PubMed]
- Jadi, P.K.; Sharma, P.; Bhogapurapu, B.; Roy, S. Alternative Therapeutic Interventions: Antimicrobial Peptides and Small Molecules to Treat Microbial Keratitis. Front. Chem. 2021, 9, 694998. [Google Scholar] [CrossRef] [PubMed]
- Shriwastav, S.; Kaur, N.; Hassan, M.; Ahmed Mohammed, S.; Chauhan, S.; Mittal, D.; Shahbaz, A.; Bibi, A. Antimicrobial peptides: A promising frontier to combat antibiotic resistant pathogens. Ann. Med. Surg. 2025, 87, 2118–2132. [Google Scholar] [CrossRef]
- Kirkpatrick, D.L.; Powis, G. Clinically Evaluated Cancer Drugs Inhibiting Redox Signaling. Antioxid. Redox Signal. 2017, 26, 262–273. [Google Scholar] [CrossRef]
- You, Y.; Liu, H.; Zhu, Y.; Zheng, H. Rational design of stapled antimicrobial peptides. Amino Acids 2023, 55, 421–442. [Google Scholar] [CrossRef] [PubMed]
- North, J.R.; Takenaka, S.; Rozek, A.; Kielczewska, A.; Opal, S.; Morici, L.A.; Finlay, B.; Schaber, C.J.; Straube, R.; Donini, O. A novel approach for emerging and antibiotic resistant infections: Innate defense regulators as an agnostic therapy. J. Biotechnol. 2016, 226, 24–34. [Google Scholar] [CrossRef]
- Chandorkar, G.; Zhan, Q.; Donovan, J.; Rege, S.; Patino, H. Pharmacokinetics of surotomycin from phase 1 single and multiple ascending dose studies in healthy volunteers. BMC Pharmacol. Toxicol. 2017, 18, 24. [Google Scholar] [CrossRef]
- Surur, A.S.; Sun, D. Macrocycle-Antibiotic Hybrids: A Path to Clinical Candidates. Front. Chem. 2021, 9, 659845. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wu, J.; Chen, Y.; Fan, K.; Yu, X.; Li, X.; Zhao, Y.; Li, Y.; Lv, G.; Chen, M.; et al. Efficacy and Safety of PL-5 (Peceleganan) Spray for Wound Infections. Ann. Surg. 2023, 277, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Najeeb, S.; Mali, M.; Moin, S.F.; Raza, S.Q.; Zohaib, S.; Sefat, F.; Zafar, M.S. Histatin peptides: Pharmacological functions and their applications in dentistry. Saudi Pharm. J. 2017, 25, 25–31. [Google Scholar] [CrossRef]
- Wiig, M.E.; Dahlin, L.B.; Fridén, J.; Hagberg, L.; Larsen, S.E.; Wiklund, K.; Mahlapuu, M. PXL01 in Sodium Hyaluronate for Improvement of Hand Recovery after Flexor Tendon Repair Surgery: Randomized Controlled Trial. PLoS ONE 2014, 9, e110735. [Google Scholar] [CrossRef]
- McCullough, P.A.; Bennett-Guerrero, E.; Chawla, L.S.; Beaver, T.; Mehta, R.L.; Molitoris, B.A.; Eldred, A.; Ball, G.; Lee, H.; Houser, M.T.; et al. ABT-719 for the Prevention of Acute Kidney Injury in Patients Undergoing High-Risk Cardiac Surgery: A Randomized Phase 2b Clinical Trial. J. Am. Heart Assoc. 2016, 5, e003549. [Google Scholar] [CrossRef]
- Baker, J.; He, X.; Shi, W. Precision Reengineering of the Oral Microbiome for Caries Management. Adv. Dent. Res. 2019, 30, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Greber, E.K.; Dawgul, M. Antimicrobial Peptides Under Clinical Trials. Curr. Top. Med. Chem. 2016, 17, 620–628. [Google Scholar] [CrossRef]
- Håkansson, J.; Ringstad, L.; Umerska, A.; Johansson, J.; Andersson, T.; Boge, L.; Rozenbaum, R.T.; Sharma, P.K.; Tollbäck, P.; Björn, C.; et al. Characterization of the in vitro, ex vivo, and in vivo Efficacy of the Antimicrobial Peptide DPK-060 Used for Topical Treatment. Front. Cell. Infect. Microbiol. 2019, 9, 174. [Google Scholar] [CrossRef]
- Khatib, M.N.; Shankar, A.H.; Kirubakaran, R.; Gaidhane, A.; Gaidhane, S.; Simkhada, P.; Syed, Z.Q. Ghrelin for the management of cachexia associated with cancer. Cochrane Database Syst. Rev. 2018, 2020, CD012229. [Google Scholar] [CrossRef]
- Parisi, K.; McKenna, J.A.; Lowe, R.; Harris, K.S.; Shafee, T.; Guarino, R.; Lee, E.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. Hyperpolarisation of Mitochondrial Membranes Is a Critical Component of the Antifungal Mechanism of the Plant Defensin, Ppdef1. J. Fungi 2024, 10, 54. [Google Scholar] [CrossRef]
- Donini, O.; A Watkins, B.; Palardy, J.; Opal, S.; Sonis, S.; Abrams, M.J.; North, J.R. Reduced Infection and Mucositis In Chemotherapy-Treated Animals Following Innate Defense Modulation Using a Novel Drug Candidate. Blood 2010, 116, 3781. [Google Scholar] [CrossRef]
- Ming, L.; Huang, J.-A. The Antibacterial Effects of Antimicrobial Peptides OP-145 against Clinically Isolated Multi-Resistant Strains. Jpn. J. Infect. Dis. 2017, 70, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-F.; Han, B.-C.; Lin, W.-Y.; Wang, S.-Y.; Linn, T.Y.; Hsu, H.W.; Wen, C.-C.; Liu, H.-Y.; Chen, Y.-H.; Chang, W.-J. Efficacy of antimicrobial peptide P113 oral health care products on the reduction of oral bacteria number and dental plaque formation in a randomized clinical assessment. J. Dent. Sci. 2024, 19, 2367–2376. [Google Scholar] [CrossRef]
- Cheng, K.-T.; Wu, C.-L.; Yip, B.-S.; Chih, Y.-H.; Peng, K.-L.; Hsu, S.-Y.; Yu, H.-Y.; Cheng, J.-W. The Interactions between the Antimicrobial Peptide P-113 and Living Candida albicans Cells Shed Light on Mechanisms of Antifungal Activity and Resistance. Int. J. Mol. Sci. 2020, 21, 2654. [Google Scholar] [CrossRef]
- Kowalski, R.P.; Romanowski, E.G.; Yates, K.A.; Mah, F.S. An Independent Evaluation of a Novel Peptide Mimetic, Brilacidin (PMX30063), for Ocular Anti-Infective. J. Ocul. Pharmacol. Ther. 2016, 32, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-R.; Yang, L.-M.; Wang, Y.-H.; Pang, W.; Tam, S.-C.; Tien, P.; Zheng, Y.-T. Sifuvirtide, a potent HIV fusion inhibitor peptide. Biochem. Biophys. Res. Commun. 2009, 382, 540–544. [Google Scholar] [CrossRef] [PubMed]
- van der Does, A.M.; Bogaards, S.J.P.; Ravensbergen, B.; Beekhuizen, H.; van Dissel, J.T.; Nibbering, P.H. Antimicrobial Peptide hLF1-11 Directs Granulocyte-Macrophage Colony-Stimulating Factor-Driven Monocyte Differentiation toward Macrophages with Enhanced Recognition and Clearance of Pathogens. Antimicrob. Agents Chemother. 2010, 54, 811–816. [Google Scholar] [CrossRef]
- Ridyard, K.E.; Overhage, J. The Potential of Human Peptide LL-37 as an Antimicrobial and Anti-Biofilm Agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef]
- Isaksson, J.; Brandsdal, B.O.; Engqvist, M.; Flaten, G.E.; Svendsen, J.S.M.; Stensen, W. A Synthetic Antimicrobial Peptidomimetic (LTX 109): Stereochemical Impact on Membrane Disruption. J. Med. Chem. 2011, 54, 5786–5795. [Google Scholar] [CrossRef]
- Chen, X.; Li, S.; Yu, L.; Miller, A.; Du, L. Systematic optimization for production of the anti-MRSA antibiotics WAP-8294A in an engineered strain of Lysobacter enzymogenes. Microb. Biotechnol. 2019, 12, 1430–1440. [Google Scholar] [CrossRef]
- Janec, K.J.; Yuan, H.; Jr, J.E.N.; Kelner, R.H.; Hirt, C.K.; Betensky, R.A.; Guinan, E.C. rBPI 21 (opebacan) promotes rapid trilineage hematopoietic recovery in a murine model of high-dose total body irradiation. Am. J. Hematol. 2018, 93, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Gries, K.; Josten, M.; Wiedemann, I.; Pelzer, S.; Labischinski, H.; Sahl, H.-G. The Lipopeptide Antibiotic Friulimicin B Inhibits Cell Wall Biosynthesis through Complex Formation with Bactoprenol Phosphate. Antimicrob. Agents Chemother. 2009, 53, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.G.; Dullaghan, E.; Mookherjee, N.; Glavas, N.; Waldbrook, M.; Thompson, A.; Wang, A.; Lee, K.; Doria, S.; Hamill, P.; et al. An anti-infective peptide that selectively modulates the innate immune response. Nat. Biotechnol. 2007, 25, 465–472. [Google Scholar] [CrossRef]
- Boakes, S.; Dawson, M.J. Discovery and Development of NVB302, a Semisynthetic Antibiotic for Treatment of Clostridium difficile Infection. In Natural Products; Wiley: Hoboken, NJ, USA, 2014; pp. 455–468. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Hady, W.A.; Deslandes, A.; Rey, A.; Fraisse, L.; Kristensen, H.-H.; Yeaman, M.R.; Bayer, A.S. Efficacy of NZ2114, a Novel Plectasin-Derived Cationic Antimicrobial Peptide Antibiotic, in Experimental Endocarditis Due to Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 5325–5330. [Google Scholar] [CrossRef]
- Iwasaki, M.; Akiba, Y.; Kaunitz, J.D. Recent advances in vasoactive intestinal peptide physiology and pathophysiology: Focus on the gastrointestinal system. F1000Research 2019, 8, 1629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Pimchan, T.; Tian, F.; Thumanu, K.; Rodtong, S.; Yongsawatdigul, J. Isolation, identification, and mode of action of antibacterial peptides derived from egg yolk hydrolysate. Poult. Sci. 2023, 102, 102695. [Google Scholar] [CrossRef]
- Walkenhorst, W.F.; Klein, J.W.; Vo, P.; Wimley, W.C. pH Dependence of Microbe Sterilization by Cationic Antimicrobial Peptides. Antimicrob. Agents Chemother. 2013, 57, 3312–3320. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Chen, N.; Jiang, C. Antimicrobial peptides: Structure, mechanism, and modification. Eur. J. Med. Chem. 2023, 255, 115377. [Google Scholar] [CrossRef]
- Giuliani, A.; Pirri, G.; Nicoletto, S. Antimicrobial peptides: An overview of a promising class of therapeutics. Open Life Sci. 2007, 2, 1–33. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Starke, L.J.; Allolio, C.; Hub, J.S. How pore formation in complex biological membranes is governed by lipid composition, mechanics, and lateral sorting. PNAS Nexus 2025, 4, pgaf033. [Google Scholar] [CrossRef]
- Beck, K.; Nandy, J.; Hoernke, M. Strong Membrane Permeabilization Activity Can Reduce Selectivity of Cyclic Antimicrobial Peptides. J. Phys. Chem. B 2025, 129, 2446–2460. [Google Scholar] [CrossRef]
- Li, J.; Lu, X.; Ma, W.; Chen, Z.; Sun, S.; Wang, Q.; Yuan, B.; Yang, K. Cholesterols Work as a Molecular Regulator of the Antimicrobial Peptide-Membrane Interactions. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef]
- Espeche, J.C.; Martínez, M.; Maturana, P.; Cutró, A.; Semorile, L.; Maffia, P.C.; Hollmann, A. Unravelling the mechanism of action of “de novo” designed peptide P1 with model membranes and gram-positive and gram-negative bacteria. Arch. Biochem. Biophys. 2020, 693, 108549. [Google Scholar] [CrossRef]
- Branco, L.A.C.; Souza, P.F.N.; Neto, N.A.S.; Aguiar, T.K.B.; Silva, A.F.B.; Carneiro, R.F.; Nagano, C.S.; Mesquita, F.P.; Lima, L.B.; Freitas, C.D.T. New Insights into the Mechanism of Antibacterial Action of Synthetic Peptide Mo-CBP3-PepI against Klebsiella pneumoniae. Antibiotics 2022, 11, 1753. [Google Scholar] [CrossRef]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-Stave Model or Toroidal Model? A Case Study on Melittin Pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Et Biophys. Acta (BBA) Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef]
- Gazit, E.; Miller, I.R.; Biggin, P.C.; Sansom, M.S.; Shai, Y. Structure and Orientation of the Mammalian Antibacterial Peptide Cecropin P1 within Phospholipid Membranes. J. Mol. Biol. 1996, 258, 860–870. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 7063. [Google Scholar] [CrossRef]
- Lin, Y.-M.; Wu, S.-J.; Chang, T.-W.; Wang, C.-F.; Suen, C.-S.; Hwang, M.-J.; Chang, M.D.-T.; Chen, Y.-T.; Liao, Y.-D. Outer Membrane Protein I of Pseudomonas aeruginosa Is a Target of Cationic Antimicrobial Peptide/Protein. J. Biol. Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef]
- Chu, H.; Pazgier, M.; Jung, G.; Nuccio, S.-P.; Castillo, P.A.; de Jong, M.F.; Winter, M.G.; Winter, S.E.; Wehkamp, J.; Shen, B.; et al. Human α-Defensin 6 Promotes Mucosal Innate Immunity Through Self-Assembled Peptide Nanonets. Science 2012, 337, 477–481. [Google Scholar] [CrossRef]
- Riciluca, K.C.T.; Oliveira, U.C.; Mendonça, R.Z.; Bozelli Junior, J.C.; Schreier, S.; da Silva Junior, P.I. Rondonin: Antimicrobial properties and mechanism of action. FEBS Open Bio 2021, 11, 2541–2559. [Google Scholar] [CrossRef]
- Lima, P.G.; Souza, P.F.; Freitas, C.D.; Bezerra, L.P.; Neto, N.A.; Silva, A.F.; Oliveira, J.T.; Sousa, D.O. Synthetic peptides against Trichophyton mentagrophytes and T. rubrum: Mechanisms of action and efficiency compared to griseofulvin and itraconazole. Life Sci. 2021, 265, 118803. [Google Scholar] [CrossRef]
- Lopes, F.E.; da Costa, H.P.; Souza, P.F.; Oliveira, J.P.; Ramos, M.V.; Freire, J.E.; Jucá, T.L.; Freitas, C.D. Peptide from thaumatin plant protein exhibits selective anticandidal activity by inducing apoptosis via membrane receptor. Phytochemistry 2019, 159, 46–55. [Google Scholar] [CrossRef]
- Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses 2015, 7, 5428–5442. [Google Scholar] [CrossRef]
- Hoffmann, A.R.; Guha, S.; Wu, E.; Ghimire, J.; Wang, Y.; He, J.; Garry, R.F.; Wimley, W.C. Broad-Spectrum Antiviral Entry Inhibition by Interfacially Active Peptides. J. Virol. 2020, 94, e01682-20. [Google Scholar] [CrossRef]
- Jackson, J.W.; Hancock, T.J.; Dogra, P.; Patel, R.; Arav-Boger, R.; Williams, A.D.; Kennel, S.J.; Wall, J.S.; Sparer, T.E. Anticytomegalovirus Peptides Point to New Insights for CMV Entry Mechanisms and the Limitations of In Vitro Screenings. mSphere 2019, 4, e00586-18. [Google Scholar] [CrossRef]
- Chao, L.; Lu, L.; Yang, H.; Zhu, Y.; Li, Y.; Wang, Q.; Yu, X.; Jiang, S.; Chen, Y.-H. Identification of a Human Protein-Derived HIV-1 Fusion Inhibitor Targeting the gp41 Fusion Core Structure. PLoS ONE 2013, 8, e66156. [Google Scholar] [CrossRef][Green Version]
- Lin, D.; Li, F.; Wu, Q.; Xie, X.; Wu, W.; Wu, J.; Chen, Q.; Liu, S.; He, J. A “building block” approach to the new influenza A virus entry inhibitors with reduced cellular toxicities. Sci. Rep. 2016, 6, 22790. [Google Scholar] [CrossRef]
- Hajigha, M.N.; Hajikhani, B.; Vaezjalali, M.; Kafil, H.S.; Anari, R.K.; Goudarzi, M. Antiviral and antibacterial peptides: Mechanisms of action. Heliyon 2024, 10, e40121. [Google Scholar] [CrossRef]
- Anunthawan, T.; de la Fuente-Núñez, C.; Hancock, R.E.; Klaynongsruang, S. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Et Biophys. Acta (BBA) Biomembr. 2015, 1848, 1352–1358. [Google Scholar] [CrossRef]
- Sneideris, T.; Erkamp, N.A.; Ausserwöger, H.; Saar, K.L.; Welsh, T.J.; Qian, D.; Katsuya-Gaviria, K.; Johncock, M.L.L.Y.; Krainer, G.; Borodavka, A.; et al. Targeting nucleic acid phase transitions as a mechanism of action for antimicrobial peptides. Nat. Commun. 2023, 14, 7170. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of Action of the Antimicrobial Peptide Buforin II: Buforin II Kills Microorganisms by Penetrating the Cell Membrane and Inhibiting Cellular Functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Yu, X.; Cao, R.; Hong, M.; Xu, Z.; Lu, J.R.; Wang, Y.; Zhu, H. Antimicrobial peptides fight against Pseudomonas aeruginosa at a sub-inhibitory concentration via anti-QS pathway. Bioorganic Chem. 2023, 141, 106922. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, T.; Li, C.; Praveen, P.; Parisi, K.; Beh, C.; Ding, S.; Wade, J.D.; Hong, Y.; Li, S.; et al. Aggregation-prone antimicrobial peptides target gram-negative bacterial nucleic acids and protein synthesis. Acta Biomater. 2025, 192, 446–460. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Suo, H.; Tan, J.; Zhang, Y.; Song, J. Identification and molecular mechanism of action of antibacterial peptides from Flavourzyme-hydrolyzed yak casein against Staphylococcus aureus. J. Dairy Sci. 2023, 106, 3779–3790. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, C.; Zhao, J.; Heng, H.; Peng, M.; Sun, L.; Dai, L.; Chan, E.W.-C.; Chen, S. LTX-315 is a novel broad-spectrum antimicrobial peptide against clinical multidrug-resistant bacteria. J. Adv. Res. 2025, 76, 715–729. [Google Scholar] [CrossRef]
- Choi, H.; Yang, Z.; Weisshaar, J.C. Oxidative stress induced in E. coli by the human antimicrobial peptide LL-37. PLOS Pathog. 2017, 13, e1006481. [Google Scholar] [CrossRef]
- Bermúdez-Puga, S.; Dias, M.; de Oliveira, T.F.; Mendonça, C.M.N.; de Almeida, S.R.Y.; Rozas, E.E.; Nascimento, C.A.O.D.; Mendes, M.A.; Azevedo, P.O.D.S.d.; Almeida, J.R.; et al. Dual antibacterial mechanism of [K4K15]CZS-1 against Salmonella Typhimurium: A membrane active and intracellular-targeting antimicrobial peptide. Front. Microbiol. 2023, 14, 1320154. [Google Scholar] [CrossRef]
- Chadha, S. Combating fungal phytopathogens with human salivary antimicrobial peptide histatin 5 through a multi-target mechanism. World J. Microbiol. Biotechnol. 2023, 39, 215. [Google Scholar] [CrossRef]
- Moghaddam, M.-R.B.; Gross, T.; Becker, A.; Vilcinskas, A.; Rahnamaeian, M. The selective antifungal activity of Drosophila melanogaster metchnikowin reflects the species-dependent inhibition of succinate–coenzyme Q reductase. Sci. Rep. 2017, 7, 8192. [Google Scholar] [CrossRef]
- Maurya, I.K.; Thota, C.K.; Sharma, J.; Tupe, S.G.; Chaudhary, P.; Singh, M.K.; Thakur, I.S.; Deshpande, M.; Prasad, R.; Chauhan, V.S. Mechanism of action of novel synthetic dodecapeptides against Candida albicans. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 5193–5203. [Google Scholar] [CrossRef]
- Di Marino, S.; Scrima, M.; Grimaldi, M.; D’errico, G.; Vitiello, G.; Sanguinetti, M.; De Rosa, M.; Soriente, A.; Novellino, E.; D’ursi, A.M. Antifungal peptides at membrane interaction. Eur. J. Med. Chem. 2012, 51, 154–162. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Chatterjee, D.; Sivashanmugam, K. Immunomodulatory peptides: New therapeutic horizons for emerging and re-emerging infectious diseases. Front. Microbiol. 2024, 15, 1505571. [Google Scholar] [CrossRef]
- Scott, M.G.; Davidson, D.J.; Gold, M.R.; Bowdish, D.; Hancock, R.E.W. The Human Antimicrobial Peptide LL-37 Is a Multifunctional Modulator of Innate Immune Responses. J. Immunol. 2002, 169, 3883–3891. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Dong, N.; Wang, C.; Zhang, T.; Zhang, L.; Xue, C.; Feng, X.; Bi, C.; Shan, A. Bioactivity and Bactericidal Mechanism of Histidine-Rich β-Hairpin Peptide Against Gram-Negative Bacteria. Int. J. Mol. Sci. 2019, 20, 3954. [Google Scholar] [CrossRef]
- de Oliveira, K.B.S.; Leite, M.L.; Melo, N.T.M.; Lima, L.F.; Barbosa, T.C.Q.; Carmo, N.L.; Melo, D.A.B.; Paes, H.C.; Franco, O.L. Antimicrobial Peptide Delivery Systems as Promising Tools Against Resistant Bacterial Infections. Antibiotics 2024, 13, 1042. [Google Scholar] [CrossRef]
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and Recent Advances in Peptide and Protein Drug Delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef]
- Katsila, T.; Siskos, A.P.; Tamvakopoulos, C. Peptide and protein drugs: The study of their metabolism and catabolism by mass spectrometry. Mass Spectrom. Rev. 2012, 31, 110–133. [Google Scholar] [CrossRef]
- Leuthner, K.D.; Yuen, A.; Mao, Y.; Rahbar, A. Dalbavancin (BI-387) for the treatment of complicated skin and skin structure infection. Expert Rev. Anti-Infect. Ther. 2015, 13, 149–159. [Google Scholar] [CrossRef]
- Cortes-Penfield, N.; Oliver, N.T.; Hunter, A.; Rodriguez-Barradas, M. Daptomycin and combination daptomycin-ceftaroline as salvage therapy for persistent methicillin-resistant Staphylococcus aureus bacteremia. Infect. Dis. 2018, 50, 643–647. [Google Scholar] [CrossRef]
- Kapić, E.; Becić, F.; Zvizdić, S. Enfuvirtide, mechanism of action and pharmacological properties. Med. Arh. 2005, 59, 313–316. [Google Scholar]
- Pavithrra, G.; Rajasekaran, R. Identification of Effective Dimeric Gramicidin-D Peptide as Antimicrobial Therapeutics over Drug Resistance: In-Silico Approach. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 575–583. [Google Scholar] [CrossRef]
- Vater, J.; Stein, T.H. Structure, Function, and Biosynthesis of Gramicidin S Synthetase. In Comprehensive Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 1999; pp. 319–352. [Google Scholar] [CrossRef]
- Greig, S.L. Obiltoxaximab: First Global Approval. Drugs 2016, 76, 823–830. [Google Scholar] [CrossRef]
- Kaasch, A.J.; Seifert, H. Oritavancin: A Long-Acting Antibacterial Lipoglycopeptide. Future Microbiol. 2016, 11, 843–855. [Google Scholar] [CrossRef]
- Mazur, N.I.; Löwensteyn, Y.N.; Terstappen, J.; Leusen, J.; Schobben, F.; Cianci, D.; van de Ven, P.M.; Nierkens, S.; Bont, L.J.; Nibbelke, E.E.; et al. Daily intranasal palivizumab to prevent respiratory syncytial virus infection in healthy preterm infants: A phase 1/2b randomized placebo-controlled trial. eClinicalMedicine 2023, 66, 102324. [Google Scholar] [CrossRef]
- Avedissian, S.N.; Liu, J.; Rhodes, N.J.; Lee, A.; Pais, G.M.; Hauser, A.R.; Scheetz, M.H. A Review of the Clinical Pharmacokinetics of Polymyxin B. Antibiotics 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Soon, R.L.; Nation, R.L.; Cockram, S.; Moffatt, J.H.; Harper, M.; Ben Adler, B.; Boyce, J.D.; Larson, I.; Li, J. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J. Antimicrob. Chemother. 2010, 66, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Calic, D.; Schweizer, F.; Zelenitsky, S.; Adam, H.; Lagacé-Wiens, P.R.S.; Rubinstein, E.; Gin, A.S.; Hoban, D.J.; Karlowsky, J.A. New Lipoglycopeptides. Drugs 2010, 70, 859–886. [Google Scholar] [CrossRef]
- Romani, L.; Bistoni, F.; Gaziano, R.; Bozza, S.; Montagnoli, C.; Perruccio, K.; Pitzurra, L.; Bellocchio, S.; Velardi, A.; Rasi, G.; et al. Thymosin α 1 activates dendritic cells for antifungal Th1 resistance through Toll-like receptor signaling. Blood 2004, 103, 4232–4239. [Google Scholar] [CrossRef]
- Vosloo, J.A.; Rautenbach, M. Following tyrothricin peptide production by Brevibacillus parabrevis with electrospray mass spectrometry. Biochimie 2020, 179, 101–112. [Google Scholar] [CrossRef]
- Gramenzi, A.; Cursaro, C.; Andreone, P.; Bernardi, M. Thymalfasin. BioDrugs 1998, 9, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Rybak, M.J. The Pharmacokinetic and Pharmacodynamic Properties of Vancomycin. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S35–S39. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; A Strawbridge, S.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Ooijevaar, R.E.; van Beurden, Y.H.; Terveer, E.M.; Goorhuis, A.; Bauer, M.P.; Keller, J.J.; Mulder, C.J.J.; Kuijper, E.J. Update of treatment algorithms for Clostridium difficile infection. Clin. Microbiol. Infect. 2018, 24, 452–462. [Google Scholar] [CrossRef]
- Rosson, E.; Lux, F.; David, L.; Godfrin, Y.; Tillement, O.; Thomas, E. Focus on therapeutic peptides and their delivery. Int. J. Pharm. 2025, 675, 125555. [Google Scholar] [CrossRef]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020, 10, 13206. [Google Scholar] [CrossRef] [PubMed]
- Prasad, N.K.; Seiple, I.B.; Cirz, R.T.; Rosenberg, O.S. Leaks in the Pipeline: A Failure Analysis of Gram-Negative Antibiotic Development from 2010 to 2020. Antimicrob. Agents Chemother. 2022, 66, e0005422. [Google Scholar] [CrossRef]
- Dale, G.E.; Halabi, A.; Petersen-Sylla, M.; Wach, A.; Zwingelstein, C. Pharmacokinetics, Tolerability, and Safety of Murepavadin, a Novel Antipseudomonal Antibiotic, in Subjects with Mild, Moderate, or Severe Renal Function Impairment. Antimicrob. Agents Chemother. 2018, 62, e00490-18. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Sibuyi, N.R.S.; Madiehe, A.M.; Meyer, M. Nanotechnology-Based Delivery Systems for Antimicrobial Peptides. Pharmaceutics 2021, 13, 1795. [Google Scholar] [CrossRef]
- Haddadzadegan, S.; Dorkoosh, F.; Bernkop-Schnürch, A. Oral delivery of therapeutic peptides and proteins: Technology landscape of lipid-based nanocarriers. Adv. Drug Deliv. Rev. 2022, 182, 114097. [Google Scholar] [CrossRef]
- Tang, T.; Chen, Y.; Zhao, Z.; Bai, Q.; Leisner, J.J.; Liu, T. Nisin-loaded chitosan/sodium alginate microspheres enhance the antimicrobial efficacy of nisin against Staphylococcus aureus. J. Appl. Microbiol. 2024, 135, lxae259. [Google Scholar] [CrossRef] [PubMed]
- Hayes, H.C.; Luk, L.Y.P.; Tsai, Y.-H. Approaches for peptide and protein cyclisation. Org. Biomol. Chem. 2021, 19, 3983–4001. [Google Scholar] [CrossRef] [PubMed]
- Bellavita, R.; Braccia, S.; Galdiero, S.; Falanga, A. Glycosylation and Lipidation Strategies: Approaches for Improving Antimicrobial Peptide Efficacy. Pharmaceuticals 2023, 16, 439. [Google Scholar] [CrossRef]
- Erak, M.; Bellmann-Sickert, K.; Els-Heindl, S.; Beck-Sickinger, A.G. Peptide chemistry toolbox—Transforming natural peptides into peptide therapeutics. Bioorganic Med. Chem. 2018, 26, 2759–2765. [Google Scholar] [CrossRef] [PubMed]
- Biondi, S.; Chugunova, E.; Panunzio, M. Chapter 8—From Natural Products to Drugs: Glyco- and Lipoglycopeptides, a New Generation of Potent Cell Wall Biosynthesis Inhibitors. In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 50, pp. 249–297. [Google Scholar] [CrossRef]
- Yan, J.; Cai, J.; Zhang, B.; Wang, Y.; Wong, D.F.; Siu, S.W.I. Recent Progress in the Discovery and Design of Antimicrobial Peptides Using Traditional Machine Learning and Deep Learning. Antibiotics 2022, 11, 1451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, Y.; Wen, H.; Lin, Y.; Hu, Y.; Zhang, Y.; Xia, Q.; Lin, Z. QSAR Modeling and Design of Cationic Antimicrobial Peptides Based on Structural Properties of Amino Acids. Comb. Chem. High Throughput Screen. 2012, 15, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Nedyalkova, M.; Paluch, A.S.; Vecini, D.P.; Lattuada, M. Progress and future of the computational design of antimicrobial peptides (AMPs): Bio-inspired functional molecules. Digit. Discov. 2024, 3, 9–22. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Orozco, R.M.Q.; Rezende, S.B.; Rodrigues, G.; Oshiro, K.G.N.; Cândido, E.D.S.; Franco, O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020, 10, 3097. [Google Scholar] [CrossRef]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A review on antimicrobial peptides databases and the computational tools. Database 2022, 2022, baac011. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging Computational Approaches for Antimicrobial Peptide Discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef]
- Mwangi, J.; Kamau, P.M.; Thuku, R.C.; Lai, R. Design Methods for An.timicrobial Peptides with Improved Performance. Zool. Res. 2023, 44, 1095–1114. [Google Scholar] [CrossRef]
- Brizuela, C.A.; Liu, G.; Stokes, J.M.; de la Fuente-Nunez, C. Methods for Antimicrobial Peptides: Progress and Challenges. Microb. Biotechnol. 2025, 18, e70072. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, G.; Cao, S.; Lv, J. Deep Learning for Antimicrobial Peptides: Computational Models and Databases. J. Chem. Inf. Model. 2025, 65, 1708–1717. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; OvchinBnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Humphreys, I.R.; Pei, J.; Baek, M.; Krishnakumar, A.; Anishchenko, I.; Ovchinnikov, S.; Zhang, J.; Ness, T.J.; Banjade, S.; Bagde, S.R.; et al. Computed structures of core eukaryotic protein complexes. Science 2021, 374, 1340. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Giulini, M.; Reys, V.; Teixeira, J.M.C.; Jiménez-García, B.; Honorato, R.V.; Kravchenko, A.; Xu, X.; Versini, R.; Engel, A.; Verhoeven, S.; et al. HADDOCK3: A modular and versatile platform for integrative modelling of biomolecular complexes. bioRxiv 2025. [Google Scholar] [CrossRef]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Fu, H.; Cao, Z.; Li, M.; Wang, S. ACEP: Improving antimicrobial peptides recognition through automatic feature fusion and amino acid embedding. BMC Genom. 2020, 21, 597. [Google Scholar] [CrossRef]
- Ramos-Martín, F.; Annaval, T.; Buchoux, S.; Sarazin, C.; D’aMelio, N. ADAPTABLE: A comprehensive web platform of antimicrobial peptides tailored to the user’s research. Life Sci. Alliance 2019, 2, e201900512. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, F.; Wei, L.; Jiang, Y.; Chen, J.; Wei, L.; Wei, D.-Q. AFP-MFL: Accurate identification of antifungal peptides using multi-view feature learning. Brief. Bioinform. 2023, 24, bbac606. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Improving Recognition of Antimicrobial Peptides and Target Selectivity through Machine Learning and Genetic Programming. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 300–313. [Google Scholar] [CrossRef]
- Torrent, M.; Nogués, V.M.; Boix, E. A theoretical approach to spot active regions in antimicrobial proteins. BMC Bioinform. 2009, 10, 373. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef]
- Salem, M.; Arshadi, A.K.; Yuan, J.S. AMPDeep: Hemolytic activity prediction of antimicrobial peptides using transfer learning. BMC Bioinform. 2022, 23, 389. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.J.; Carper, D.L.; Spangler, M.K.; A Carrell, A.; A Rush, T.; Minter, S.J.; Weston, D.J.; Labbé, J.L. amPEPpy 1.0: A portable and accurate antimicrobial peptide prediction tool. Bioinformatics 2021, 37, 2058–2060. [Google Scholar] [CrossRef]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Chilimoniuk, J.; Rödiger, S.; Gagat, P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram. Int. J. Mol. Sci. 2020, 21, 4310. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yan, J.; Un, C.; Wang, Y.; Campbell-Valois, F.-X.; Siu, S.W.I. BERT-AmPEP60: A BERT-Based Transfer Learning Approach to Predict the Minimum Inhibitory Concentrations of Antimicrobial Peptides for Escherichia coli and Staphylococcus aureus. J. Chem. Inf. Model. 2025, 65, 3186–3202. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Pires, Á.S.; Franco, O.L. CS-AMPPred: An Updated SVM Model for Antimicrobial Activity Prediction in Cysteine-Stabilized Peptides. PLoS ONE 2012, 7, e51444. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Shelenkov, A.A.; Rogozhin, E.A. Computational Prediction and Structural Analysis of α-Hairpinins, a Ubiquitous Family of Antimicrobial Peptides, Using the Cysmotif Searcher Pipeline. Antibiotics 2024, 13, 1019. [Google Scholar] [CrossRef] [PubMed]
- Azim, S.M.; Sharma, A.; Noshadi, I.; Shatabda, S.; Dehzangi, I. DeepAmp: A Convolutional Neural Network Based Tool for Predicting Protein AMPylation Sites from Binary Profile Representation. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Xu, J.; Li, F.; Li, C.; Guo, X.; Landersdorfer, C.; Shen, H.-H.; Peleg, A.Y.; Li, J.; Imoto, S.; Yao, J.; et al. iAMPCN: A deep-learning approach for identifying antimicrobial peptides and their functional activities. Brief. Bioinform. 2023, 24, bbad240. [Google Scholar] [CrossRef]
- Tang, W.; Dai, R.; Yan, W.; Zhang, W.; Bin, Y.; Xia, E.; Xia, J. Identifying multi-functional bioactive peptide functions using multi-label deep learning. Brief. Bioinform. 2022, 23, bbab414. [Google Scholar] [CrossRef]
- Tang, W.; Dai, R.; Yan, W.; Zhang, W.; Bin, Y.; Xia, E.; Xia, J. Comprehensive assessment of machine learning-based methods for predicting antimicrobial peptides. Brief. Bioinform. 2021, 22, bbab083. [Google Scholar] [CrossRef]
- Bajiya, N.; Choudhury, S.; Dhall, A.; Raghava, G.P.S. AntiBP3: A Method for Predicting Antibacterial Peptides against Gram-Positive/Negative/Variable Bacteria. Antibiotics 2024, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Fensterseifer, I.C.; Ribeiro, S.M.; Franco, O.L. Joker: An algorithm to insert patterns into sequences for designing antimicrobial peptides. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 2043–2052. [Google Scholar] [CrossRef]
- Müller, A.T.; Gabernet, G.; A Hiss, J.; Schneider, G. modlAMP: Python for antimicrobial peptides. Bioinformatics 2017, 33, 2753–2755. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wuyun, Q.; Li, Y.; Liu, Q.; Zhou, X.; Peng, C.; Zhu, Y.; Freddolino, L.; Zhang, Y. Deep-learning-based single-domain and multidomain protein structure prediction with D-I-TASSER. Nat. Biotechnol. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Singh, H.; Singh, S.; Singh Raghava, G.P. Peptide Secondary Structure Prediction using Evolutionary Information. bioRxiv 2019. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Fang, Y.; Ma, Y.; Yu, K.; Dong, J.; Zeng, W. Integrated computational approaches for advancing antimicrobial peptide development. Trends Pharmacol. Sci. 2024, 45, 1046–1060. [Google Scholar] [CrossRef]
- Lee, H.T.; Lee, C.C.; Yang, J.R.; Lai, J.Z.; Chang, K.Y. A Large-Scale Structural Classification of Antimicrobial Peptides. BioMed Res. Int. 2015, 2015, 475062. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; A Wlodarski, M.; Edalatmand, A.; Petkau, A.; A Syed, S.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Novković, M.; Simunić, J.; Bojović, V.; Tossi, A.; Juretić, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Guan, J.; Xie, P.; Chung, C.-R.; Zhao, Z.; Dong, D.; Guo, Y.; Zhang, W.; Deng, J.; Pang, Y.; et al. dbAMP 3.0: Updated resource of antimicrobial activity and structural annotation of peptides in the post-pandemic era. Nucleic Acids Res. 2025, 53, D364–D376. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Singh, S.; Joshi, A.; Anand, P.; Tuknait, A.; Raghava, G.P.S. Hemolytik: A database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res. 2014, 42, D444–D449. [Google Scholar] [CrossRef]
- Gómez, E.A.; Giraldo, P.; Orduz, S. InverPep: A database of invertebrate antimicrobial peptides. J. Glob. Antimicrob. Resist. 2017, 8, 13–17. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; E Bravo, J.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2022, 51, D744–D752. [Google Scholar] [CrossRef]
- D’aLoisio, V.; Dognini, P.; Hutcheon, G.A.; Coxon, C.R. PepTherDia: Database and structural composition analysis of approved peptide therapeutics and diagnostics. Drug Discov. Today 2021, 26, 1409–1419. [Google Scholar] [CrossRef]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef]
- Piotto, S.P.; Sessa, L.; Concilio, S.; Iannelli, P. YADAMP: Yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents 2012, 39, 346–351. [Google Scholar] [CrossRef]
- Fabbri, L.P.; Cavallero, A.; Vidotto, F.; Gabriele, M. Bioactive Peptides from Fermented Foods: Production Approaches, Sources, and Potential Health Benefits. Foods 2024, 13, 3369. [Google Scholar] [CrossRef] [PubMed]
- Bisht, V.; Das, B.; Hussain, A.; Kumar, V.; Navani, N.K. Understanding of probiotic origin antimicrobial peptides: A sustainable approach ensuring food safety. NPJ Sci. Food 2024, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- de la Lastra, J.M.P.; González-Acosta, S.; Otazo-Pérez, A.; Asensio-Calavia, P.; Rodríguez-Borges, V.M. Antimicrobial Peptides for Food Protection: Leveraging Edible Mushrooms and Nano-Innovation. Dietetics 2025, 4, 9. [Google Scholar] [CrossRef]
- Kumar, L.; Tyagi, P.; Lucia, L.; Pal, L. Innovations in Edible Packaging Films, Coatings, and Antimicrobial Agents for Applications in Food Industry. Compr. Rev. Food Sci. Food Saf. 2025, 24, e70217. [Google Scholar] [CrossRef]
- Choi, D.; Bedale, W.; Chetty, S.; Yu, J. Comprehensive review of clean-label antimicrobials used in dairy products. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13263. [Google Scholar] [CrossRef] [PubMed]
- Min, K.H.; Kim, K.H.; Ki, M.-R.; Pack, S.P. Antimicrobial Peptides and Their Biomedical Applications: A Review. Antibiotics 2024, 13, 794. [Google Scholar] [CrossRef]
- Sengkhui, S.; Klubthawee, N.; Aunpad, R. A novel designed membrane-active peptide for the control of foodborne Salmonella enterica serovar Typhimurium. Sci. Rep. 2023, 13, 3507. [Google Scholar] [CrossRef]
- Wang, L.; Dekker, M.; Heising, J.; Zhao, L.; Fogliano, V. Food matrix design can influence the antimicrobial activity in the food systems: A narrative review. Crit. Rev. Food Sci. Nutr. 2024, 64, 8963–8989. [Google Scholar] [CrossRef]
- Chu, Z.; Wang, H.; Dong, B. Research on Food Preservation Based on Antibacterial Technology: Progress and Future Prospects. Molecules 2024, 29, 3318. [Google Scholar] [CrossRef]
- Swann, M. Report/Joint Committee on the use of Antibiotics in Animal Husbandry and Veterinary Medicine; Her Majesty’s Stationery Office: London, UK, 1969. [Google Scholar]
- Vercelli, C.; Gambino, G.; Amadori, M.; Re, G. Implications of Veterinary Medicine in the comprehension and stewardship of antimicrobial resistance phenomenon. From the origin till nowadays. Vet. Anim. Sci. 2022, 16, 100249. [Google Scholar] [CrossRef]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Valdez-Miramontes, C.; De Haro-Acosta, J.; Aréchiga-Flores, C.; Verdiguel-Fernández, L.; Rivas-Santiago, B. Antimicrobial peptides in domestic animals and their applications in veterinary medicine. Peptides 2021, 142, 170576. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.-M.; Huang, H.-N.; Tsai, T.-Y.; You, M.-F.; Wu, H.-Y.; Rajanbabu, V.; Chang, H.-Y.; Pan, C.-Y.; Chen, J.-Y. Dietary supplementation of recombinant antimicrobial peptide Epinephelus lanceolatus piscidin improves growth performance and immune response in Gallus gallus domesticus. PLoS ONE 2020, 15, e0230021. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-S.; Shao, H.; Li, T.-J.; Tang, Z.-R.; Huang, R.-L.; Wang, S.-P.; Kong, X.-F.; Wu, X.; Yin, Y.-L. Dietary Supplementation with Bovine Lactoferrampin–Lactoferricin Produced by Pichia pastoris Fed-batch Fermentation Affects Intestinal Microflora in Weaned Piglets. Appl. Biochem. Biotechnol. 2012, 168, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Ingale, S.L.; Kim, J.S.; Kim, K.H.; Lohakare, J.; Park, Y.K.; Park, J.C.; Kwon, I.K.; Chae, B.J. Effects of dietary supplementation with antimicrobial peptide-P5 on growth performance, apparent total tract digestibility, faecal and intestinal microflora and intestinal morphology of weanling pigs. J. Sci. Food Agric. 2013, 93, 587–592. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, X.; Zhu, C.; Zhao, Y.; Liu, S.; Xia, X.; Liu, X.; Zhang, H.; Xu, Y.; Hang, B.; et al. The antimicrobial peptide MPX kills Actinobacillus pleuropneumoniae and reduces its pathogenicity in mice. Vet. Microbiol. 2020, 243, 108634. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Q.; Dong, M.; Song, H.; Hang, B.; Sun, Y.; Zhang, H.; Hu, J. Evaluation of the efficacy of the antimicrobial peptide HJH-3 in chickens infected with Salmonella Pullorum. Front. Microbiol. 2023, 14, 1102789. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, O.; Grötzinger, J.; Cascorbi, I.; Jung, S. Antimicrobial peptides and proteins of the horse—Insights into a well-armed organism. Vet. Res. 2011, 42, 98. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Li, Y.; Pi, Q.; Tian, J.; Xu, X.; Huang, Z.; Huang, J.; Pian, C.; Mao, S. Unveiling novel antimicrobial peptides from the ruminant gastrointestinal microbiomes: A deep learning-driven approach yields an anti-MRSA candidate. J. Adv. Res. 2025; in press. [Google Scholar] [CrossRef]
| Criterion | Chemical Synthesis | Enzymatic | Natural | Recombinant |
|---|---|---|---|---|
| Cost | High | Moderate | Variable | Low |
| Yield | Medium | Low | Very Low | High |
| Scalability | High | Moderate | Low | Very High |
| Technical Complexity | High | Low | Medium | High |
| Application Versatility | Very High | Moderate | Low | High |
| Level of Specificity | Very High | High | Moderate | High |
| Stability (Half-life) | High | Moderate | Variable | High |
| Peptide Name | Sequence | Structure | Source | Biological Activity | Target Site | Delivery Path | Molecular Weight | pI | Charge | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Reltecimod, p2TA | ASPMLVAYDA |  | Synthetic | Antibacterial | Immune modulation | IV | 1.0 | 3.8 | −1 | [58] |
| Ramoplanin, NTI-851 | NNXXTXXXFXXXXGLAX | 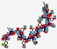 | Actinoplanes | Antibacterial | Disrupt cell wall biosynthesis | Oral/Topical | 1.5 | 5.52 | 0 | [49] |
| XOMA-629 | KLFRXQAKX | 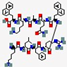 | Synthetic | Antibacterial | Cell membrane | Topical cream | 1.1 | 11.17 | +3 | [59] |
| Mycoprex | Not disclosed | 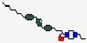 | Insects | Antifungal | Cell membrane | IV | - | - | - | [60] |
| Demegal, D2A21 | FAKKFAKKFKKFAKKFAKFAFAF |  | Synthetic | Antibacterial | Cell membrane | IV | 2.7 | 10.9 | +9 | [61] |
| Glutoxim, NOV-002 | DL-gGlu-DL-Cys-Gly-gGlu-DL-Cys-Gly.2Na+ |  | Synthetic | Antibacterial | Cell membrane | IV | 0.68 | 3.8 | −2 | [62] |
| Omiganan, MBI-226 | ILRWPWWPWRRK | 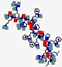 | Synthetic | Antibacterial | Cell membrane | Topical | 1.7 | 12.3 | +4 | [63] |
| Dusquetide SGX942 | RIVPA | 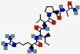 | Synthetic | Antibacterial | Immune modulation | IV | 0.55 | 9.75 | +1 | [64] |
| Surotomycin, MK-426 | Not disclosed |  | Actino bacteria | Antibacterial | Disrupt cytoplasmic membrane | Oral | - | - | - | [65] |
| Cefilavancin, TD-1792 | Not disclosed | 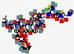 | Synthetic | Antibacterial | Cell wall | IV | - | - | - | [66] |
| Peceleganan, PL-5 | KWKSFLKTF KSA AKTVLHTALKAISS |  | Synthetic | Antibacterial | Cell membrane | Topical spray | 2.8 | 10.7 | +6 | [67] |
| Histatin | DXHEKRHHGYRRKFHEKHHSHREFPFYGDYGSNYLYDN | 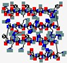 | Human | Anti- candidal | Cell membrane/mitochondrion activity | Mouth wash | 4.8 | 8.32 | +1 | [68] |
| PXL01 | Not disclosed | - | Synthetic | Antibacterial | Inhibits inflammation | IV | - | - | - | [69] |
| Modimelanotide, AP-214 | KKKKKKSYSMEHFRWGKPV | 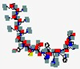 | Synthetic | Antibacterial | Immune modulation | IV | 2.3 | 10.47 | +7 | [70] |
| C16G2 | TFFRLFNRSFTQALGKGGGKNLRIIRK GIHIIKKY | - | Synthetic | Antibacterial | Cell membrane | Mouth rinse | 4 | 12.02 | +9 | [71] |
| Melantropin, CZEN-002 | SYSMEHFRWGKPV |  | Synthetic | Anti-candidal | cAMP induction | Topical | 1.6 | 8.33 | +1 | [72] |
| DPK-060 | Not disclosed | - | Kininogen | Anti -infective | Membrane disruption | Topical | - | - | - | [73] |
| Ghrelin | GSSFLSPEHQRVQQRKESKKPPAKLQPR |  | Synthetic | Anti-infective | Cell membrane | IV | 3.2 | 11.07 | +5 | [74] |
| HXP124, Ppdef1 | Not disclosed | - | Plant | Antifungal | Cell membrane | Topical | - | - | - | [75] |
| Inimex, IMX942 | KSRIVPAIPVSLL | - | Synthetic | Anti-infective | Immune modulation | IV/ | 1.3 | 11 | +2 | [76] |
| Novexatin, NVXT | RRRRRRR | 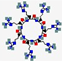 | Synthetic | Antifungal | Cell wall | Topical | 1.1 | 12.78 | +7 | [51] |
| OP-145 | IGKEFKRIVERIKRFLRELVRPLR | - | Synthetic | Antibacterial | Lipid bilayer | Topical | 3 | 11.72 | +6 | [77] |
| P113 | AKRHHGYKRKFH | - | Human | Anti- infective | Cell membrane | Mouth wash | 1.5 | 11.17 | +5 | [78] |
| PAC113 | AKRHHGYKRKFH | - | Human | Antifungal | Target mitochondria | Oral rinse | 1.5 | 11.17 | +5 | [79] |
| Bilacidin, PMX-30063 | Not disclosed |  | Synthetic | Antibacterial | Cell membrane | IV | 1. | - | - | [80] |
| Sifuvirtide | SWETWEREIENYTRQIYRILEESQEQQDRN ERDLLE |  | Synthetic | Anti-HIV | HIV fusion inhibitor | IV | 4.6 | 4.33 | −6 | [81] |
| hLF1-11 | GRRRRSVQWCA | 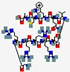 | Humana | Antibacterial Antifungal | Inhibition of growth by iron scavenging | IV/ Topical | 1.3 | 12 | +4 | [82] |
| LL-37 | LLGDFFRKSKEKIGK EFKRIVQRIKDFLR NLVPRTES |  | Human | Antibacterial | Membrane disruption | Topical/inhalation | 4.4 | 10.61 | +6 | [83] |
| LTX-109 | R-Tbt-R-NH-EtPh | 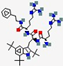 | Synthetic | Antibacterial | Membrane disruption and cell lysis. | Topical (nasal) | 0.78 | - | - | [84] |
| Lotilibcin, WAP-8294A2 | cyclo[D-Asn-D-Trp-D-Orn -N(Me)Val-ObAla(3R-isohexyl) -Ser-D-Asn- Ser-Gly- D- N(Me)Phe-Leu-D-Orn-Glu] | 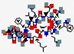 | Lysobacter species | Antibacterial | Membrane disruption | IV | 1.5 | - | - | [85] |
| Opebacan | Not disclosed | - | Recombinant BPI fragment | Antibacterial Antiviral | Permeability increasing protein | IV | - | - | - | [86] |
| Friulimicin B | Not disclosed |  | Actinoplanes friuliensis | Antibacterial | Cell membrane | IV | 1.3 | - | - | [87] |
| IDR-1 | KSRIVPAIPVSLL-NH2 |  | Synthetic | Anti-infective | Reduction of pro-inflammatory cytokines | IV/ Topical | 1.3 | 11 | +2 | [88] |
| NVB302 | Not disclosed | - | Synthetic | Antifungal | Lipid bilayer | IV | - | - | - | [89] |
| Plectasin, NZ2114 | GFGCNGPWDEDDM QCHNHCKSIKGYK GGYCAK GGFVCKCY | - | Pseudoplectania nigrella | Antibacterial | Lipid bilayer | IV/ Topical | 4.4 | 7.77 | +1 | [90] |
| Vasoactive intestinal peptide | HSDAVFTDNYTRLRKQMAVKKYLNSILN |  | Human | Antibacterial | G-protein-coupled receptors | IV/Inhalation | 3.3 | 9.82 | +3 | [91] |
| Peptide Name | Sequence | Structure | Source | Biological Activity | Medical Use | Target Site | Delivery Path | Company | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Bacitracin | Leu-D-Glu Ile-Lys-D-Orn-Ile-D-Phe-His-D-Asp-Asn |  | Bacillus licheniformis | Antibacterial | Prevent pneumonia and empyema in infants, skin and eye infections. | Cell wall | Topical | Various companies | [139] |
| Dalbavacin Xydalba | Not available |  | Semi synthetic | Antibacterial | Acute bacterial skin infections, Osteomyelitis and septic arthritis | Cell wall | IV | DALVANCE® | [140] |
| Daptomycin Cubicin | Decanoyl-WNTGO DaDGsED |  | Streptomyces roseosporus | Antibacterial | Complicated skin infections and bloodstream infections (bacteremia) | Cell membrane | IV | CUBICIN® | [141] |
| Enfuvirtide Fuzeon T20 | YTSLIHSLEESQNQQEKNEQELLELDKWASLWNWF | 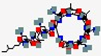 | Synthetic | Anti-HIV | Human Immunodeficiency Virus (HIV) | Fusion protein gp41 | SC | FUZEON® | [142] |
| Gramicidin D | VGALAVVVWLW LWLW-ethanolamine |  | Bacillus brevis | Antibacterial | Skin lesions and eye infections | Cell membrane | Topical | Various companies | [143] |
| Gramicidin S | cyclo[Leu-D -Phe -Pro-Val-Orn-Leu-D-Phe-Pro-Val-Orn] | 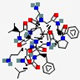 | Bacillus brevis | Antibacterial | Spermicide against bacteria and fungi; genital ulcers | Cell membrane | Topical | Various companies | [144] |
| Obiltoxaximab | Not available | Not available | monoclonal antibody | Antibacterial | Inhalational anthrax | Antitoxin | IV | ANTHIM® | [145] |
| Oritavancin | Not available | Not available | Semi synthetic | Antibacterial | Acute bacterial skin | Cell wall | IV | KIMYRSA™ ORBACTIV® | [146] |
| Palivizumab | Not available | Not available | monoclonal antibody | Antiviral | Prevent serious lung infections caused by respiratory syncytial virus-RSV | Blocking viral replication | IM | SYNAGIS® | [147] |
| Polymyxin B | Not available | Not available | Bacillus polymyxa | Antibacterial | Infections of the urinary tract, meninges, and blood stream | Cell membrane | IV | Various companies | [148] |
| Polymyxin E Colistin | 6-mh-DabTDab [Î3DablLDabDabT] |  | Bacillus polymyxa | Antibacterial | Acute or chronic infections due to Gram-negative bacilli | Cell membrane | Oral | Various companies | [149] |
| Raxibacumab | Not available | Not available | monoclonal antibody | Antibacterial | Inhalational anthrax | Antitoxin | IV | RAXIBACUMAB ® | [11] |
| Telavancin TD-6424 | Not available | Not available - | Semi synthetic | Antibacterial | Osteomyelitis and bacterial infections | Cell membrane | IV | VIBATIV® | [150] |
| Tyrothricin | Not available | Not available - | Brevibacillus parabrevis | Antibacterial Antifungal | Infected skin and infected oropharyngeal mucous membranes | Cell membrane | Topical/ Oral | Various companies | [151,152] |
| Thymalfasin | SDAAVDTSSEITTKDLKEKKEVVEEAE N |  | Synthetic | Antiviral | Hepatitis B and C. | Immuno modulator | IV | ZADAXIN® | [153] |
| Vancomycin | Not available |  | Amycolatopsis orientalis | Antibacterial | Septicemia, infective endocarditis, skin, bone and lower respiratory tract infections. | Cell wall | IV/Oral | Various companies | [154] |
| Database Name | Functionality | Additional Information | URL | Reference |
|---|---|---|---|---|
| Prediction of whether it is an AMP and its potential activity. | ||||
| ACEP | Identification of AMPs. | The classification method is based on deep learning (DL). | https://github.com/Fuhaoyi/ACEP (accessed 30 October 2025) | [187] |
| ADAPTABLE | Designing novel peptides, predicting their activities, and identifying functional motifs. | Webserver and data-miner of antimicrobial peptides. | http://gec.upicardie.fr/adaptable/ (accessed 30 October 2025) | [188] |
| AFP-MFL | A novel deep learning model that can predict antifungal peptides. | It only needs the peptide sequence to run. | https://inner.wei-group.net/AFPMFL/#/ (accessed 30 October 2025) | [189] |
| AMP- Scanner | A web server tool for predicting if it is an AMP based on amino acid sequence. | Only includes bacteria targets. | https://www.dveltri.com/ascan/ (accessed 30 October 2025) | [190] |
| AMPA | A web server tool for identifying active regions in antimicrobial proteins. | The algorithm uses an antimicrobial propensity scale to generate an antimicrobial profile. | https://tcoffee.crg.eu/apps/ampa/do (accessed 30 October 2025) | [191,192] |
| AMPDeep | Deep learning approach to predict hemolytic activity of AMPs. | It was built on Python. | https://github.com/milad73s/AMPDeep (accessed 30 October 2025) | [193] |
| amPEPpy | A python application for predicting antimicrobial peptide sequences. | The classification method is based on random forest. | https://github.com/tlawrence3/amPEPpy (accessed 30 October 2025) | [194] |
| AmpGram | A web server tool for identification of AMPs. | The classification method is based on random forest and n-gram analysis. | http://biongram.biotech.uni.wroc.pl/AmpGram/ (accessed 30 October 2025) | [195] |
| AxPEP | It is a collection of sequence-based machine learning methods for AMPs prediction. | The classification method is based on random forest. | https://app.cbbio.online/ampep/home (accessed 30 October 2025) | [196] |
| CS-AMPPred | An SVM-based (support vector machines) tool to predict antimicrobial activity in cysteine-knotted proteins. | Were based on 310 AMPs and 310 non-antimicrobial peptide sequences. | https://sourceforge.net/projects/csamppred/ (accessed 30 October 2025) | [197] |
| Cysmotif Searcher | A Perl package for revealing peptide sequences possessing cysteine motifs. | Cysteine motifs are common to various families of AMPs and other cysteine-rich peptides. | https://github.com/fallandar/cysmotifsearcher (accessed 30 October 2025) | [198] |
| deepAMP | A tool for predicting protein AMPylation sites from binary profile representation. | The classification method is based on a convolutional neural network. | https://github.com/MehediAzim/DeepAmp (accessed 30 October 2025) | [199] |
| iAMPCN | DL approach to identifying antimicrobial peptides and their functional activities. | It was built on Python. | https://github.com/joy50706/iAMPCN (accessed 30 October 2025) | [200] |
| MLBP | Multi-label DL approach to identifying multi-functional bioactive peptide functions. | It was built on Python. | https://github.com/tangwending/MLBP (accessed 30 October 2025) | [201] |
| sAMPpred-GAT | A web tool for identification of AMPs. | The program uses graphs constructed based on predicted peptide structures. | http://bliulab.net/sAMPpred-GAT/server (accessed 30 October 2025) | [202] |
| Design of Peptides | ||||
| AntiBP 3.0 | A web-tool for predicting, scanning and designing AMPs. | Based on machine learning techniques. | https://webs.iiitd.edu.in/raghava/antibp3/ (accessed 30 October 2025) | [203] |
| Joker | An algorithm to design antimicrobial peptides. | It was developed based on amino acid motifs. | https://github.com/williamfp7/Joker (accessed 30 October 2025) | [204] |
| ModlAMP | A Python package for working with any sequence of natural amino acids. | It includes the following modules: sequence generation, sequence library analysis and description calculation. | https://modlamp.org/ (accessed 30 October 2025) | [205] |
| Prediction of Structure | ||||
| AlphaFold | To predict a protein/peptide’s 3D structure based on amino acid sequence. | AI system developed by Google DeepMind. | https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb (accessed 30 October 2025) | [206,207] |
| iTASSER | A web tool for predicting structure of proteins. | It also provides a structure-based function annotation. | https://zhanggroup.org/I-TASSER/ (accessed 30 October 2025) | [208] |
| PEP2D | A web tool for predicting secondary structure of peptides. | The model was trained and tested based on 3100 peptide structures. | https://webs.iiitd.edu.in/raghava/pep2d/ (accessed 30 October 2025) | [209] |
| PEP-FOLD 4 | A de novo approach to predict peptide structure from amino acid sequences. | The peptides should have 5 to 50 amino acids. | https://mobyle2.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::PEP-FOLD4 (accessed 30 October 2025) | [209] |
| RoseTTAFold | A Python-based AI tool for protein/peptide structure prediction. | The structure prediction method is based on DL. | https://github.com/RosettaCommons/RoseTTAFold?tab=readme-ov-file (accessed 30 October 2025) | [179] |
| SWISS-MODEL | A web tool for protein structure prediction and modeling. | It supports interactive modeling for both simple and complex needs. | wissmodel.expasy.org (accessed 30 October 2025) | [210] |
| Molecular Docking and Molecular Modeling | ||||
| AutoDock | A tool to predict how small molecules bind to a receptor of know 3D structure. | It has two options: AutoDock-GPU and AutoDock Vina. | https://autodock.scripps.edu/ (accessed 30 October 2025) | [181] |
| CABS-dock | A web server tool for flexible protein-peptide docking. | It requires the sequence of a protein receptor and a peptide sequence. | https://biocomp.chem.uw.edu.pl/CABSdock (accessed 30 October 2025) | [183] |
| CHARMM36 | Broad-scope molecular simulation software for complex environments. | There is no cost to academic students. | https://academiccharmm.org/ (accessed 30 October 2025) | [185] |
| GROMACS | Powerful open-source suite for molecular dynamics simulation and analysis. | Broad spectrum of calculation types, preparation and analysis tools. | https://manual.gromacs.org/archive/4.6.7/online/speptide.html (accessed 30 October 2025) | [184] |
| HADDOCK | A web platform for biomolecular docking simulations. | A data-driven docking approach guided by ambiguous interaction restraints (AIRs). | https://rascar.science.uu.nl/haddock2.4 (accessed 30 October 2025) | [97] |
| MARTINI | Coarse-grained force field for biomolecular simulations, parameterized using experimental data and atomistic simulations. | Based on a four-to-one mapping scheme. | https://cgmartini.nl/ (accessed 30 October 2025) | [186] |
| NAMD | A highly scalable software for parallel molecular dynamics simulations of large biomolecules. | It uses the program VHD for simulation setup and trajectory analysis. | https://www.ks.uiuc.edu/Research/namd/ (accessed 30 October 2025) | [211] |
| VMD | A program to display, animate and analyze large biomolecules systems. | It has 3-D graphics. | https://www.ks.uiuc.edu/Research/vmd/ (accessed 30 October 2025) | [5] |
| Database Name | Type of data | Additional Information | URL | Reference |
|---|---|---|---|---|
| ADAM | Comprehensive AMPs database. | It contains tools for searching and predicting whether peptides are antimicrobial. | http://bioinformatics.cs.ntou.edu.tw/adam/index.html (accessed 30 October 2025) | [213] |
| AMPDb | Extensively curated AMPs. | It assimilates information from various resources: NCBI, EMBL, UniProt, RCSB-PDB and PubMed. | https://bblserver.org.in/ampdb/ (accessed 30 October 2025) | [17] |
| APD | Manually curated AMPs. | It contains 5099 peptides | https://aps.unmc.edu/ (accessed 30 October 2025) | [8] |
| CAMP | Conserved sequence signatures represented by 45 families. | It includes sequence, protein definition, accession numbers, activity, source organism, target organisms and protein family descriptions | https://classamp.bicnirrh.res.in/ (accessed 30 October 2025) | [13] |
| CARD | Antibiotics, resistance genes, their products and associated phenotypes. | It includes tools as BLAST and can predict resistome based on homology and SNP models. | https://card.mcmaster.ca/ (accessed 30 October 2025) | [214] |
| DADP | Manually curated database of AMPs and defense peptides. | Described minimum-inhibiting concentration (MIC) against at least one microorganism. | https://webs.iiitd.edu.in/raghava/satpdb/catalogs/dadp/ (accessed 30 October 2025) | [215] |
| dbAMP | Manually curated AMPs. | It includes tools that can discover novel AMPs or relate functional activities by predictive models. | http://awi.cuhk.edu.cn/dbAMP (accessed 30 October 2025) | [216] |
| DBAASP | Structure–activity of peptides. | It includes a tool that predicts whether the amino acid sequence has antimicrobial potential. | https://dbaasp.org/home (accessed 30 October 2025) | [12] |
| DRAMP | Manually curated AMPs. | It includes the annotations: sequences, structures, activities, physicochemical, patent, and clinical | http://dramp.cpu-bioinfor.org/ (accessed 30 October 2025) | [16] |
| Hemolytik | Manually curated peptides, including AMPs. | It contains experimentally validated hemolytic and non-hemolytic peptides. | http://crdd.osdd.net/raghava/hemolytik/ (accessed 30 October 2025) | [217] |
| InverPep | Specialized database of AMPs from invertebrates. | It includes extensive annotations: name, sequence, length, secondary structure, molar mass, charge, isoelectric point, hydrophobicity, etc. | https://ciencias.medellin.unal.edu.co/gruposdeinvestigacion/prospeccionydisenobiomoleculas/InverPep/public/home_en (accessed 30 October 2025) | [218] |
| MEGARes | Hand-curated resistance genes for antimicrobial drugs, biocides and metals. | Sources include Antibacterial Biocide and Metal Resistance Genes (BacMet), CARD, ResFinder, PointFinder, AMRFinderPlus. | https://www.meglab.org/megares/ (accessed 30 October 2025) | [219] |
| PlantPepDB | Information about plant peptides | Users can get all information about different functional, physicochemical and structural properties of a single peptide, in a single entry. | http://14.139.61.8/PlantPepDB/pages/simplesearch.php (accessed 30 October 2025) | [18] |
| Peptipedia | Tool for peptide sequence | Uses 76 databases and reports on activity and sequence | https://app.peptipedia.cl/ (accessed 30 October 2025) | [15] |
| PepTherDia | Manually curated database of approved peptide drugs and diagnostic agents. | Annotations: structural, physicochemical and pharmacokinetic properties, indications, routes of administration, production methodologies, marketing authorization and origin of their design. | https://peptherdia.herokuapp.com/ (accessed 30 October 2025) | [220] |
| SATPdb | Structurally annotated therapeutic peptides. | It is possible to search for all the peptides containing a common segment (motif) or well-known motifs. | https://webs.iiitd.edu.in/raghava/satpdb/sub_search.php (accessed 30 October 2025) | [14] |
| ThPDB | Approved and approved/investigational therapeutic peptides. | It includes sequence, indication, mechanism of action, pharmacodynamics, toxicity, metabolism, absorption, half-life, patent information, interaction with other drugs, targets, others. | http://crdd.osdd.net/raghava/thpdb/ (accessed 30 October 2025) | [221] |
| YADAMP | Structurally annotated peptides classified into 10 categories. | Include anticancer, antiviral, antiparasitic, antibacterial, drug delivery and toxic peptides. | https://webs.iiitd.edu.in/raghava/satpdb/index.html (accessed 30 October 2025) | [222] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marciano, C.L.; Félix de Lima, J.V.; Couto Rosa, M.S.d.; do Nascimento, R.A.; Ferraz, A.d.O.; Silva, I.C.d.; Chrysostomo-Massaro, T.N.; Rosa-Garzon, N.G.d.; Cabral, H. A Comprehensive Overview of Antimicrobial Peptides: Broad-Spectrum Activity, Computational Approaches, and Applications. Antibiotics 2025, 14, 1115. https://doi.org/10.3390/antibiotics14111115
Marciano CL, Félix de Lima JV, Couto Rosa MSd, do Nascimento RA, Ferraz AdO, Silva ICd, Chrysostomo-Massaro TN, Rosa-Garzon NGd, Cabral H. A Comprehensive Overview of Antimicrobial Peptides: Broad-Spectrum Activity, Computational Approaches, and Applications. Antibiotics. 2025; 14(11):1115. https://doi.org/10.3390/antibiotics14111115
Chicago/Turabian StyleMarciano, Camila Langer, João Vítor Félix de Lima, Murilo Sousa do Couto Rosa, Rafaelly Avelar do Nascimento, Antonio de Oliveira Ferraz, Iago Castro da Silva, Taís Nader Chrysostomo-Massaro, Nathália Gonsales da Rosa-Garzon, and Hamilton Cabral. 2025. "A Comprehensive Overview of Antimicrobial Peptides: Broad-Spectrum Activity, Computational Approaches, and Applications" Antibiotics 14, no. 11: 1115. https://doi.org/10.3390/antibiotics14111115
APA StyleMarciano, C. L., Félix de Lima, J. V., Couto Rosa, M. S. d., do Nascimento, R. A., Ferraz, A. d. O., Silva, I. C. d., Chrysostomo-Massaro, T. N., Rosa-Garzon, N. G. d., & Cabral, H. (2025). A Comprehensive Overview of Antimicrobial Peptides: Broad-Spectrum Activity, Computational Approaches, and Applications. Antibiotics, 14(11), 1115. https://doi.org/10.3390/antibiotics14111115






