Phenotypic and Genotype Patterns of Antimicrobial Resistance in Non-Human Primates: An Overlooked “One Health” Concern
Abstract
1. Introduction
2. Results
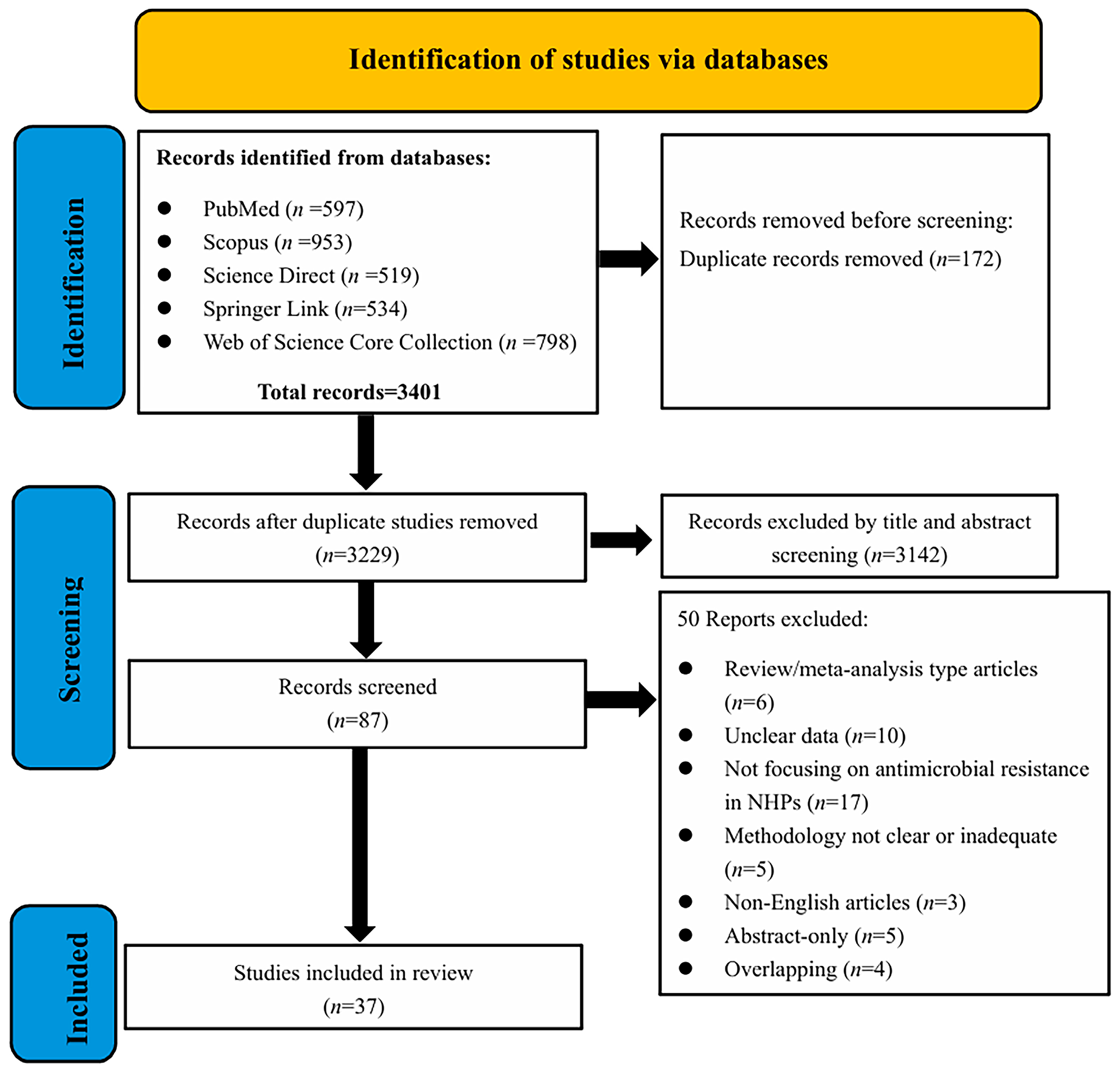
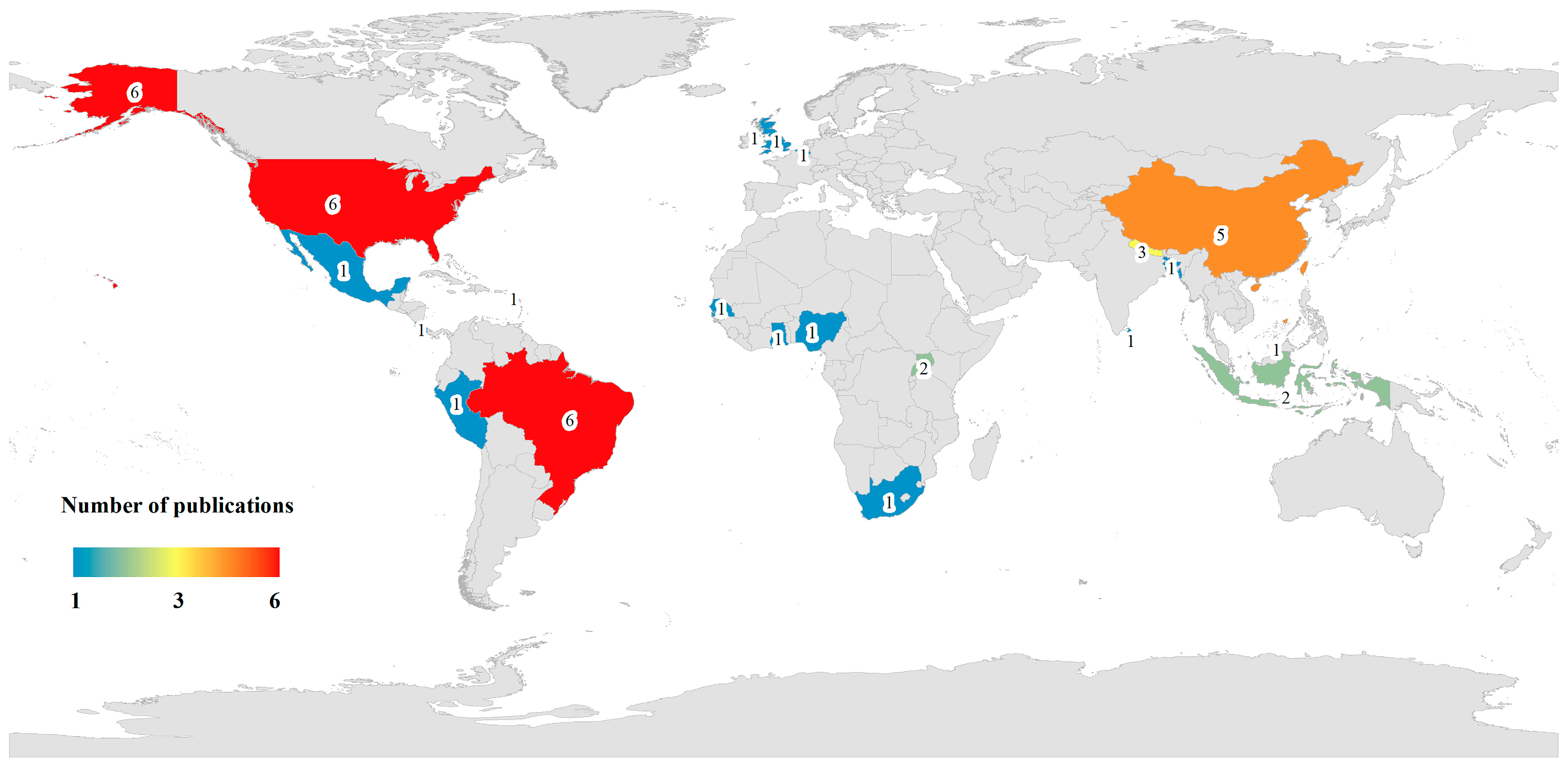
2.1. Descriptive Statistics of Included Studies
2.2. Antibiotic Resistance Studies in NHPs
2.2.1. Basic Characteristics of Selected Studies
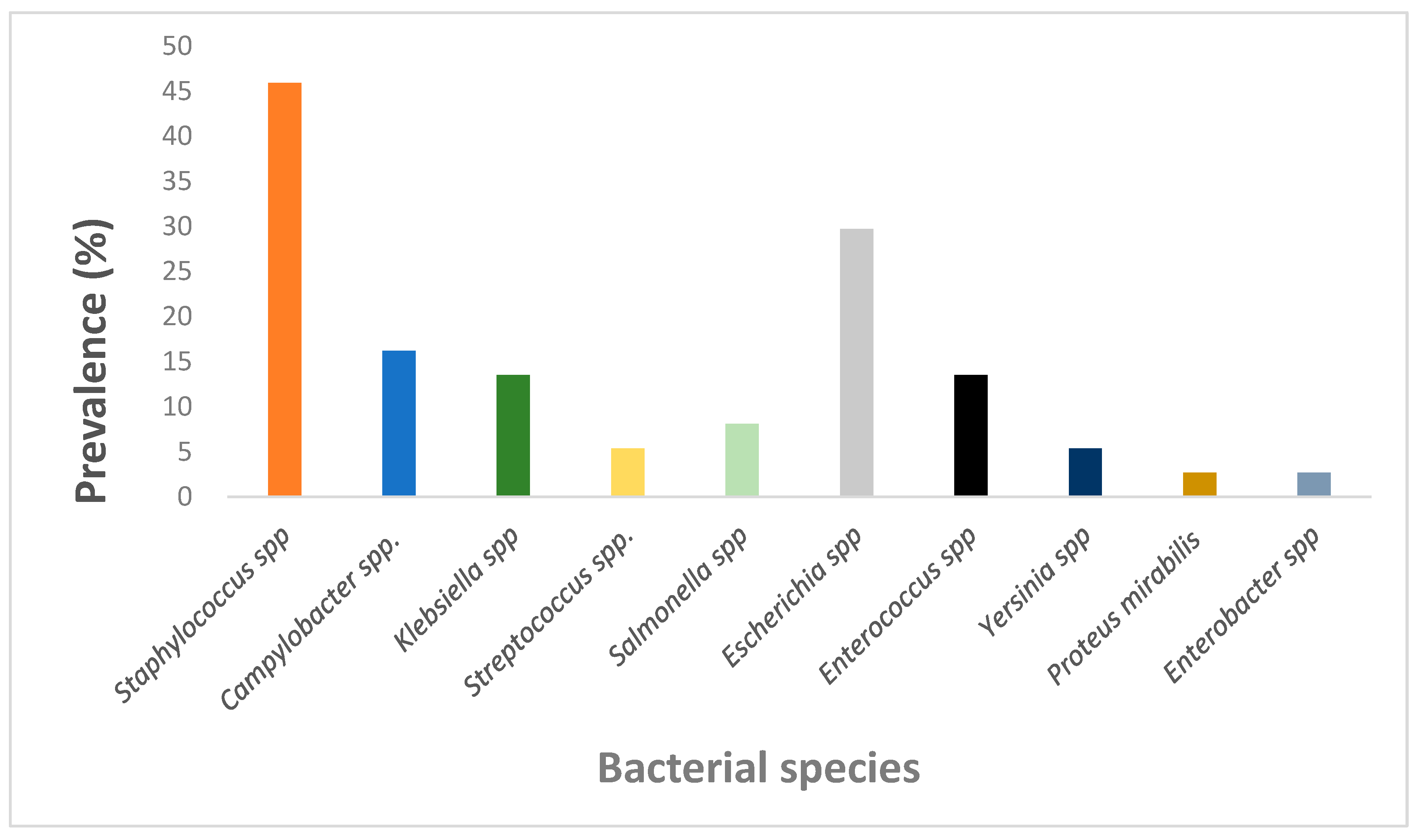
2.2.2. Prevalence of Antimicrobial Bacteria in NHPs
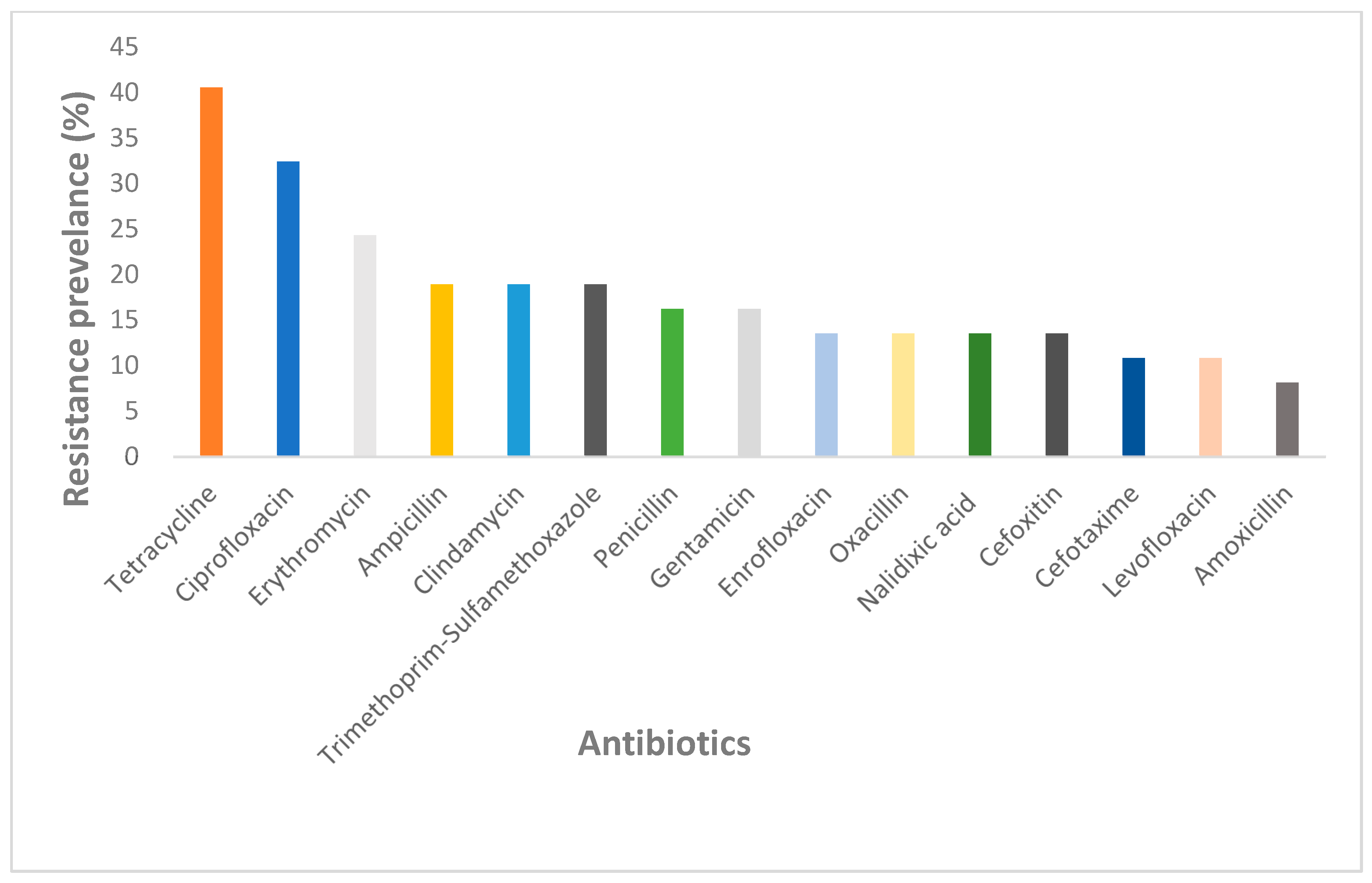
2.2.3. Prevalence of Antibiotic Resistance in NHPs
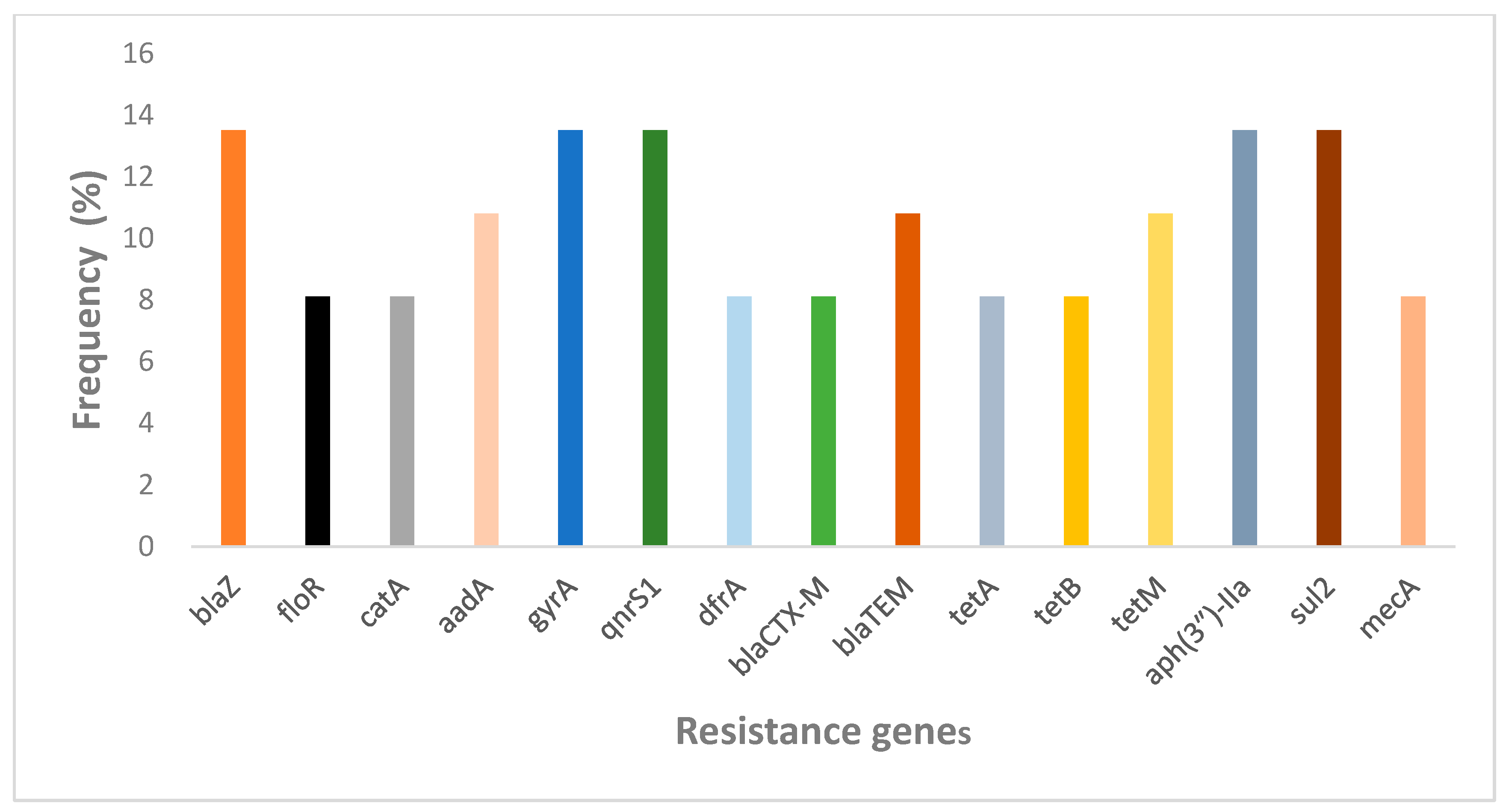
2.2.4. Prevalence of Antibiotic Resistance Genes in NHPs
3. Discussion
3.1. Prevalence of Antimicrobial Resistance Bacteria in NHPs
3.2. Prevalence of Antibiotic Resistance in NHPs
3.3. Prevalence of Antibiotic Resistance Genes in NHPs
4. Limitations
5. Methodology
5.1. Aim and Research Questions
- (A)
- What is the prevalence of AMR in NHPs?
- (B)
- What is the current status of research into phenotypic and genotypic patterns of antimicrobial resistance in NHPs?
- (C)
- What are the technologies being used in detecting AMR in NHPs?
5.2. Data Sources and Search Strategy
5.3. Eligibility Criteria
5.4. Selection of Studies and Data Extraction
6. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic resistance in bacteria associated with food animals: A United States perspective of livestock production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, A.; Klinken, R.D.V.; Jones, D.; Wang, J. Consumers’ perspectives on antibiotic use and antibiotic resistance in food animals: A systematic review. npj Sci. Food 2025, 9, 29. [Google Scholar] [CrossRef]
- Wang, W.J.; Yu, L.M.; Shao, M.Y. Research review on the pollution of antibiotic resistance genes in livestock and poultry farming environments. Chin. J. Appl. Ecol. 2023, 34, 1415–1429. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zhang, Q.; Wang, T.Z.; Xu, N.H.; Lu, T.; Hong, W.J.; Penuelas, J.; Gillings, M.; Wang, M.X.; Gao, W.W.; et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef] [PubMed]
- Varma, J.K.; Oppong-Otoo, J.; Ondoa, P.; Perovic, O.; Park, B.J.; Laxminarayan, R.; Peeling, R.W.; Schultsz, C.; Li, H.; Ihekweazu, C. Africa Centres for Disease Control and Prevention’s framework for antimicrobial resistance control in Africa. Afr. J. Lab. Med. 2018, 7, 4. [Google Scholar] [CrossRef]
- Carroll, D.; Wang, J.; Fanning, S.; Mcmahon, B.J. Antimicrobial resistance in wildlife: Implications for public health. Zoonoses Public Health 2015, 62, 534–542. [Google Scholar] [CrossRef]
- Wang, M.Y.; Lu, X.; Kan, B.; Chen, S.G.; Fan, Y.B. Research progress on bacterial resistance and gene carrying resistance in migratory birds. Chin. J. Prev. Med. 2021, 55, 271–276. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Li, Z.; Song, W.J.; Du, L.N.; Ye, C.; Zhao, B. Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography. Environ. Int. 2019, 127, 371–380. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yu, S.; Li, D.; Gillings, M.R.; Ren, H.; Mao, D.; Guo, J.; Luo, Y. Inter-plasmid transfer of antibiotic resistance genes accelerates antibiotic resistance in bacterial pathogens. ISME J. 2024, 18, wrad032. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jiang, X.; Wu, J.; Wang, Y.; Zhao, L.; Pan, Y.; Xi, Y.; Zhao, G.; Li, Z.; Zhang, L. Synergistic horizontal transfer of antibiotic resistance genes and transposons in the infant gut microbial genome. mSphere 2024, 9, e0060823. [Google Scholar] [CrossRef]
- Lee, S.; Fan, P.; Liu, T.; Yang, A.; Boughton, R.K.; Pepin, K.M.; Miller, R.S.; Jeong, K.C. Transmission of antibiotic resistance at the wildlife-livestock interface. Commun. Biol. 2022, 5, 585. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Benavides, J.A.; Godreuil, S.; Bodenham, R.; Ratiarison, S.; Devos, C.; Petretto, M.-O.; Raymond, M.; Escobar-Páramo, P. No Evidence for Transmission of Antibiotic-Resistant Escherichia coli Strains from Humans to Wild Western Lowland Gorillas in Lopé National Park, Gabon. Appl. Environ. Microbiol. 2012, 78, 4281–4287. [Google Scholar] [CrossRef]
- Medkour, H.; Amona, I.; Akiana, J.; Laidoudi, Y.; Davoust, B.; Bitam, I.; Lafri, I.; Levasseur, A.; Diatta, G.; Sokhna, C.; et al. Bacterial Infections in Humans and Nonhuman Primates from Africa: Expanding the Knowledge. Yale J. Biol. Med. 2021, 94, 227–248. [Google Scholar] [PubMed]
- Huang, H. Captivity and geography influence the antibiotic resistome of non-human primates. Front. Vet. Sci. 2022, 9, 1020276. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K. Clinical veterinarian’s perspective of non-human primate (NHP) use in drug safety studies. J. Immunotoxicol. 2010, 7, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Tanga, C.T.F.; Makouloutou-Nzassi, P.; Mbehang Nguema, P.P.; Düx, A.; Lendzele Sevidzem, S.; Mavoungou, J.F.; Leendertz, F.H.; Mintsa-Nguema, R. Antimicrobial Resistance in African Great Apes. Antibiotics 2024, 13, 1140. [Google Scholar] [CrossRef]
- Zhang, X.L.; Pang, W.; Hu, X.T.; Li, J.L.; Yao, Y.G.; Zheng, Y.T. Experimental primates and non-human primate (NHP) models of human diseases in China: Current status and progress. Dongwuxue Yanjiu 2014, 35, 447–464. [Google Scholar] [CrossRef]
- Estrada, A.; Garber, P.A.; Rylands, A.B.; Roos, C.; Fernandez-Duque, E.; Di Fiore, A.; Nekaris, K.A.; Nijman, V.; Heymann, E.W.; Lambert, J.E.; et al. Impending extinction crisis of the world’s primates: Why primates matter. Sci. Adv. 2017, 3, e1600946. [Google Scholar] [CrossRef]
- Zaniolo, M.M.; Santos, I.C.D.; Barbosa, L.N.; Pachaly, E.M.V.; Gonalves, D.D. Antimicrobial resistance and extended-spectrum beta-lactamase production in enterobacteria isolated from free-living primates. Vector Borne Zoonotic Dis. 2020, 20, 513–516. [Google Scholar] [CrossRef]
- Grassotti, T.T.; de Angelis Zvoboda, D.; da Fontoura Xavier Costa, L.; de Araújo, A.J.G.; Pereira, R.I.; Soares, R.O.; Wagner, P.G.C.; Frazzon, J.; Frazzon, A.P.G. Antimicrobial Resistance Profiles in Enterococcus spp. Isolates from Fecal Samples of Wild and Captive Black Capuchin Monkeys (Sapajus nigritus) in South Brazil. Front. Microbiol. 2018, 9, 2366. [Google Scholar] [CrossRef]
- Zaniolo, M.M.; Santos, I.C.; Bondezan, M.A.D.; Fazoli, K.G.Z.; Tramontin, R.S.; Onaca, F.M.T.; Silva, L.L.; Pachaly, E.M.V.; Pachaly, J.R.; Barbosa, L.N.; et al. Antimicrobial resistance profile of gram-positive isolates belonging to the microbiota of non-human primates. J. Med. Primatol. 2022, 51, 143–148. [Google Scholar] [CrossRef]
- Rojas-Sánchez, E.; Jiménez-Soto, M.; Barquero-Calvo, E.; Duarte-Martnez, F.; Mollenkopf, D.F.; Wittum, T.E.; Muñoz-Vargas, L. Prevalence Estimation, Antimicrobial Susceptibility, and Serotyping of Salmonella enterica Recovered from New World Non-Human Primates (Platyrrhini), Feed, and Environmental Surfaces from Wildlife Centers in Costa Rica. Antibiotics 2023, 12, 844. [Google Scholar] [CrossRef]
- Afiff, U.; Hidayat, R.; Indrawati, A.; Sunartatie, T.; Hardiati, A.; Rotinsulu, D.A.; Arifiantini, R.I.; Naoremisa, D.; Mar’ah, N.; Safika, S. Antibiotic resistance and virulence profile of Klebsiella pneumoniae isolated from wild Sumatran Orangutans (Pongo abelii). J. Adv. Vet. Anim. Res. 2024, 11, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, R.; Kuete Yimagou, E.; Hadjadj, L.; Mediannikov, O.; Ibrahim, A.; Davoust, B.; Barciela, A.; Hernandez-Aguilar, R.A.; Diatta, G.; Sokhna, C.; et al. Population Diversity of Antibiotic Resistant Enterobacterales in Samples from Wildlife Origin in Senegal: Identification of a Multidrug Resistance Transposon Carrying blaCTX–M–15 in Escherichia coli. Front. Microbiol. 2022, 13, 838392. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Wallace, R.M.; Rwego, I.B.; Gillespie, T.R.; Chapman, C.A.; Singer, R.S.; Goldberg, T.L. Antibiotic-Resistant Escherichia coli and Class 1 Integrons in Humans, Domestic Animals, and Wild Primates in Rural Uganda. Appl. Environ. Microbiol. 2018, 84, e01632-18. [Google Scholar] [CrossRef] [PubMed]
- Svenson, E.L.; Coonen, J.; Svenson, J.E.; Simmons, H.A.; Hayes, J.M.; Capuano, S. An Epidemiologic Study of Bacterial Culture and Antibiotic Susceptibility Analyses in Captive Macaques and Marmosets at the Wisconsin National Primate Research Center. J. Am. Assoc. Lab. Anim. Sci. JAALAS 2024, 63, 540–551. [Google Scholar] [CrossRef]
- Bazalar-Gonzales, J.; Silvestre-Espejo, T.; Rodríguez Cueva, C.; Carhuaricra Huamán, D.; Ignación León, Y.; Luna Espinoza, L.; Rosadio Alcántara, R.; Maturrano Hernández, L. Genomic insights into ESBL-producing Escherichia coli isolated from non-human primates in the Peruvian Amazon. Front. Vet. Sci. 2024, 10, 1340428. [Google Scholar] [CrossRef]
- Guerra, M.F.; Teixeira, R.H.; Ribeiro, V.L.; Cunha, M.P.; Oliveira, M.G.; Davies, Y.M.; Silva, K.C.; Silva, A.P.; Lincopan, N.; Moreno, A.M.; et al. Suppurative peritonitis by Klebsiella pneumoniae in captive gold-handed tamarin (Saguinus midas midas). J. Med. Primatol. 2016, 45, 42–46. [Google Scholar] [CrossRef]
- Dos Santos, D.O.; de Campos, B.H.; de Souza, T.G.V.; de Castro, Y.G.; Alves Neto, G.; Vieira, A.D.; Ribeiro, L.N.; de Figueiredo, C.C.C.; Duarte, J.R.; Amaral, V.H.B.; et al. Staphylococcus spp. as part of the microbiota and as opportunistic pathogen in free-ranging black-tuffed marmosets (Callithrix penicillata) from urban areas: Epidemiology, antimicrobial resistance, and pathology. J. Med. Primatol. 2024, 53, e12732. [Google Scholar] [CrossRef] [PubMed]
- Ceccolini, M.E.; Macgregor, S.K.; Spiro, S.; Irving, J.; Hedley, J.; Williams, J.; Guthrie, A. Yersinia pseudotuberculosis infections in primates, artiodactyls, and birds within a zoological facility in the united kingdom. J. Zoo Wildl. Med. 2020, 51, 527–538. [Google Scholar] [CrossRef]
- Sales, I.; Vieira-da-Motta, O.; Tavares, A.; Ruiz-Miranda, C.R.; de Lencastre, H.; Miragaia, M. Impact of human created environments in the pathogenic potential and antimicrobial resistance of staphylococci from wild neotropical primates in Brazil. Comp. Immunol. Microbiol. Infect. Dis. 2024, 104, 102094. [Google Scholar] [CrossRef] [PubMed]
- Berlamont, H.; Smet, A.; De Bruykere, S.; Boyen, F.; Ducatelle, R.; Haesebrouck, F.; De Witte, C. Antimicrobial susceptibility pattern of Helicobacter suis isolates from pigs and macaques. Vet. Microbiol. 2019, 239, 108459. [Google Scholar] [CrossRef]
- Koga, T.; Aoki, W.; Mizuno, T.; Wakazono, K.; Ohno, J.; Nakai, T.; Nomiya, T.; Fujii, M.; Fusegawa, K.; Kinoshita, K.; et al. Antimicrobial resistance in Campylobacter coli and Campylobacter jejuni in cynomolgus monkeys (Macaca fascicularis) and eradication regimens. J. Microbiol. Immunol. Infect. 2017, 50, 75–82. [Google Scholar] [CrossRef]
- Bochart, R.M.; Armantrout, K.; Crank, H.; Tonelli, R.; Shriver-Munsch, C.; Swanson, T.; Fischer, M.; Wu, H.; Axthelm, M.; Sacha, J.; et al. Identification of Vancomycin Resistance in Methicillin-resistant Staphylococcus aureus in two macaque species and decolonization and long-term prevention of recolonization in Cynomolgus Macaques (Macaca fascicularis). Front. Immunol. 2023, 14, 1244637. [Google Scholar] [CrossRef]
- Soge, O.O.; No, D.; Michael, K.E.; Dankoff, J.; Lane, J.; Vogel, K.; Smedley, J.; Roberts, M.C. Transmission of MDR MRSA between primates, their environment and personnel at a United States primate centre. J. Antimicrob Chemother. 2016, 71, 2798–2803. [Google Scholar] [CrossRef]
- Steele, M.; Shazali, S.A.; Cutler, R.R.; Idris, A. High prevalence of multiple drug resistant staphylococci observed in macaque-populated locations in Brunei Darussalam. Trop. Biomed. 2017, 34, 32–36. [Google Scholar]
- Roberts, M.C.; Joshi, P.R.; Monecke, S.; Ehricht, R.; Müller, E.; Gawlik, D.; Diezel, C.; Braun, S.D.; Paudel, S.; Acharya, M.; et al. Staphylococcus aureus and Methicillin Resistant S. aureus in Nepalese Primates: Resistance to Antimicrobials, Virulence, and Genetic Lineages. Antibiotics 2020, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Tegner, C.; Sunil-Chandra, N.P.; Wijesooriya, W.R.P.L.I.; Perera, B.V.; Hansson, I.; Fahlman, Å. Detection, Identification, and Antimicrobial Susceptibility of Campylobacter spp. and Salmonella spp. from Free-ranging Nonhuman Primates in Sri Lanka. J. Wildl. Dis. 2019, 55, 879–884. [Google Scholar] [CrossRef]
- Woods, S.E.; Lieberman, M.T.; Lebreton, F.; Trowel, E.; de la Fuente-Nu’ñez, C.; Dzink-Fox, J. Characterization of Multi-Drug Resistant Enterococcus faecalis Isolated from Cephalic Recording Chambers in Research Macaques (Macaca spp.). PLoS ONE 2017, 12, e0169293. [Google Scholar] [CrossRef]
- Roberts, M.C.; Joshi, P.R.; Monecke, S.; Ehricht, R.; Müller, E.; Gawlik, D.; Paudel, S.; Acharya, M.; Bhattarai, S.; Pokharel, S.; et al. MRSA Strains in Nepalese Rhesus Macaques (Macaca mulatta) and Their Environment. Front. Microbiol. 2019, 10, 2505. [Google Scholar] [CrossRef] [PubMed]
- Mannion, A.J.; Martin, H.R.; Shen, Z.; Buckley, E.M.; Dzink-Fox, J.L.; Garcia, A.; Marini, R.P.; Patterson, M.M.; Fox, J.G. Plasmid-Mediated Quinolone Resistance in Shigella flexneri Isolated from Macaques. Front. Microbiol. 2018, 9, 311. [Google Scholar] [CrossRef]
- Roberts, M.C.; Joshi, P.R.; Greninger, A.L.; Melendez, D.; Paudel, S.; Acharya, M.; Bimali, N.K.; Koju, N.P.; No, D.; Chalise, M.; et al. The human clone ST22 SCCmec IV methicillin-resistant Staphylococcus aureus isolated from swine herds and wild primates in Nepal: Is man the common source? FEMS Microbiol. Ecol. 2018, 94, fiy052. [Google Scholar] [CrossRef]
- Yang, S.; Liu, Y.; Yang, N.; Lan, Y.; Lan, W.; Feng, J.; Yue, B.; He, M.; Zhang, L.; Zhang, A.; et al. The gut microbiome and antibiotic resistome of chronic diarrhea rhesus macaques (Macaca mulatta) and its similarity to the human gut microbiome. Microbiome 2022, 10, 29. [Google Scholar] [CrossRef]
- Bacon, R.L.; Hodo, C.L.; Wu, J.; Welch, S.; Nickodem, C.; Vinasco, J.; Threadgill, D.; Gray, S.B.; Norman, K.N.; Lawhon, S.D. Diversity of Campylobacter spp. circulating in a rhesus macaque (Macaca mulatta) breeding colony using culture and molecular methods. mSphere 2024, 9, e0056024. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhou, Q.; Qi, H.X.; Lan, W.Q.; Yang, S.J.; Yang, S.Z.; Fan, Z.X.; Zhang, A.Y. Antimicrobial resistance spectrum and virulence characterization of Escherichia coli. Klebsiella pneumoniae and Proteus mirabilis isolated from asymptomatic and diarrheal rhesus monkeys. Microbiol. Res. 2024, 282, 127633. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.K.; Hassan, M.M.; Islam, S.; Rostal, M.K.; Uddin, M.H.; Hagan, E.; Samad, M.A.; Flora, M.S.; Epstein, J.H.; Islam, A. Characterization and epidemiology of antimicrobial resistance patterns of Salmonella spp. and Staphylococcus spp. in free-ranging rhesus macaque (Macaca mulatta) at high-risk interfaces with people and livestock in Bangladesh. Front. Vet. Sci. 2023, 10, 1103922. [Google Scholar] [CrossRef]
- Glover, B.; Wentzel, J.; Jenkins, A.; Van Vuuren, M. The first report of Escherichia fergusonii isolated from non-human primates, in Africa. One health 2017, 3, 70–75. [Google Scholar] [CrossRef]
- Kalule, J.B.; Nakintu, V.Z.; Sendawula, S.P. Nasal carriage of Methicillin-Resistant Staphylococcus aureus among sympatric free-ranging domestic pigs and wild Chlorocebus pygerythrus in a rural African setting. BMC Vet. Res. 2022, 18, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Jiang, S.; Qi, M.; Liu, H.; Zhang, S.; Liu, H.; Zhou, Z.; Wang, L.; Wang, C.; Luo, Y.; et al. Prevalence and characterization of antibiotic resistance genes and integrons in Escherichia coli isolates from captive non-human primates of 13 zoos in China. Sci. Total Environ. 2021, 798, 149268. [Google Scholar] [CrossRef]
- Shidan, Z.; Song, L.; Yumin, Z.; Rong, C.; Siteng, W.; Meirong, L.; Guangjin, L. First report of Streptococcus agalactiae isolated from a healthy captive sichuan golden snub-nosed monkey (Rhinopithecus roxellana) in China. Microb. Pathog. 2024, 195, 106907. [Google Scholar] [CrossRef]
- Okunlade, A.O.; Adekanmbi, A.O.; Olajumoke, J.F.; Osemuohu, O.A. Quinolone resistance markers influoroquinolone-resistant, non-ESBL-producing Escherichia coli isolated from non-human primatesat selected zoological gardens and tourist centres. Int. J. Environ. Stud. 2022, 80, 687–698. [Google Scholar] [CrossRef]
- Hoefer, A.; Boyen, F.; Beierschmitt, A.; Moodley, A.; Roberts, M.C.; Butaye, P. Methicillin-Resistant and Methicillin Susceptible Staphylococcus from Vervet Monkeys (Chlorocebus sabaeus) in Saint Kitts. Antibiotics 2021, 10, 290. [Google Scholar] [CrossRef]
- Adade, E.; Tawiah, P.O.; Roos, C.; Chuma, I.S.; Lubinza, C.C.; Mfinanga, S.G.M.; Knauf, S.; Sylverken, A.A. Antimicrobial susceptibility profile of oral and rectal microbiota of non-human primate species in Ghana: A threat to human health. Vet. Med. Sci. 2023, 10, e1271. [Google Scholar] [CrossRef] [PubMed]
- Foster-Nyarko, E.; Alikhan, N.F.; Ravi, A.; Thilliez, G.; Thomson, N.M.; Baker, D.; Kay, G.; Cramer, J.D.; O’Grady, J.; Antonio, M.; et al. Genomic diversity of Escherichia coli isolates from non-human primates in the Gambia. Microb. Genom. 2020, 6, e000428. [Google Scholar] [CrossRef] [PubMed]
- Vásquez-Aguilar, A.A.; Toledo-Manuel, F.O.; Barbachano-Guerrero, A.; Hernández-Rodríguez, D. Detection of Antimicrobial Resistance Genes in Escherichia coli Isolated from Black Howler Monkeys (Alouatta pigra) and Domestic Animals in Fragmented Rain-Forest Areas in Tabasco, Mexico. J. Wildl. Dis. 2020, 56, 922–927. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Tang, Y.; Qiao, Z.; Wang, Z.; Li, Y.; Ren, J.; Wen, L.; Xu, X.; Yang, J.; Yu, C.; Meng, C. The Prevalence of Staphylococcus aureus and the Occurrence of MRSA CC398 in Monkey Feces in a Zoo Park in Eastern China. Animals 2021, 11, 732. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanism of staphylococcal resistance to clinically relevant antibiotics. Drug Resist. Updat. 2024, 77, 101147. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 1110–1128. [Google Scholar] [CrossRef]
- Mueller, M.; Tainter, C.R. Escherichia coli infection. In StatPearls; StatPearls Publishing: Petersburg, FL, USA, 2023. [Google Scholar]
- Kim, J.; Coble, D.J.; Salyards, G.W.; Habing, G.G. Comparative Review of Antimicrobial Resistance in Humans and Nonhuman Primates. Comp. Med. 2018, 68, 124–130. [Google Scholar]
- López, L.; Calderón, D.; Salinas, L.; Graham, J.P.; Blount, Z.D.; Trueba, G. A plasmid with the bla CTX-M gene enhances the fitness of Escherichia coli strains under laboratory conditions. Microbiology 2025, 171, 001525. [Google Scholar] [CrossRef]
- Wasimuddin; Malik, H.; Ratovonamana, Y.R.; Rakotondranary, S.J.; Ganzhorn, J.U.; Sommer, S. Anthropogenic Disturbance Impacts Gut Microbiome Homeostasis in a Malagasy Primate. Front. Microbiol. 2022, 13, 911275. [Google Scholar] [CrossRef]
- Morelli, T.L.; Smith, A.B.; Mancini, A.N.; Balko, E.A.; Borgerson, C.; Dolch, R. The fate of Madagascar’s rainforest habitat. Nat. Clim. Chang. 2019, 10, 89–96. [Google Scholar] [CrossRef]
- Clayton, J.B.; Vangay, P.; Huang, H. Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Enshaie, E.; Nigam, S.; Patel, S.; Rai, V. Livestock Antibiotics Use and Antimicrobial Resistance. Antibiotics 2025, 14, 621. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Beltrán, J.; DelaFuente, J.; León-Sampedro, R.; MacLean, R.C.; San Millán, Á. Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.Q.; Li, H.; Zhang, F.Y.; Mao, H.M.; LÜ, L.B. Occupational Health Risk Management Measures for Personnel Handling Non-Human Primate Laboratory Animals: An Overvie. Lab. Anim. Comp. Med. 2025, 45, 197–205. [Google Scholar]
- Shrum Davis, S.; Salazar-Hamm, P.; Edge, K. Multidrug-resistant Shigella flexneri outbreak affecting humans and non-human primates in New Mexico, USA. Nat. Commun. 2025, 16, 4680. [Google Scholar] [CrossRef]
- Chong, C.W.; Alkatheeri, A.H.S.; Ali, N.; Tay, Z.H.; Lee, Y.L.; Paramasivam, S.J.; Jeevaratnam, K.; Low, W.Y.; Lim, S.H.E. Association of antimicrobial resistance and gut microbiota composition in human and non-human primates at an urban ecotourism site. Gut Pathog. 2020, 12, 14. [Google Scholar] [CrossRef]
- Carne, C.; Semple, S.; MacLarnon, A.; Majolo, B.; Maréchal, L. Implications of tourist-Macaque interactions for disease transmission. EcoHealth 2017, 14, 704–717. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251. [Google Scholar] [CrossRef]
- Trinh, P.; Zaneveld, J.R.; Safranek, S.; Rabinowitz, P.M. One health relationships between human, animal, and environmental microbiomes: A mini-review. Front. Public Health 2018, 6, 235. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.M.; Turku, S.; Lehrfield, L.; Shoman, A. The impact of human activities on zoonotic infection transmissions. Animals 2023, 13, 1646. [Google Scholar] [CrossRef]
- Kweka, E.J.; Kimaro, E.E.; Munga, S. Effect of Deforestation and land use changes on mosquito productivity and development in Western Kenya highlands: Implication for malaria risk. Front. Public Health 2016, 4, 238. [Google Scholar] [CrossRef] [PubMed]
- Duval, P.; Antonellim, P.; Aschan-Leygonie, C.; Valiente Moro, C. Impact of Human Activities on Disease-Spreading Mosquitoes in Urban Areas. J. Urban. Health 2023, 100, 591–611. [Google Scholar] [CrossRef]
- Zainab, S.M.; Junaid, M.; Xu, N.; Malik, R.N. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020, 187, 116455. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, C.; Miranda-Madera, V.; Serrano-Silva, N.; Bernal, J.E.; Rios-Montes, K.; González-Jiménez, F.E.; Ojeda-Juárez, D.; Sarria-Guzmán, Y. Antibiotic-Resistant Bacteria Isolated from Street Foods: A Systematic Review. Antibiotics 2024, 13, 481. [Google Scholar] [CrossRef] [PubMed]
| Species | Locations | Life Context | Type of Sample | Resistant Bacteria | Antibiotic Resistance | Detection Test | Resistance Genes | Ref |
|---|---|---|---|---|---|---|---|---|
| Black-horned capuchin monkey (Sapajus nigritus) | Brazil | Wild | Oral and rectal swabs | Enterobacteriaceae | AMX, AMP, FOX, IPM | ND | ND | [21] |
| Brazil | Captive and wild | Faeces | E. durans, E. faecium, E. faecalis, E. hirae | RFP, TC, E, CI, NFX, CAP, AMP | PCR | msrC, tetM, tetL | [22] | |
| Brazil | Captive | Oral, ocular, nasal swabs | Streptococcus spp. Staphylococcus spp. | GM, ENR, CTX, PG, OX | PCR | mecA | [23] | |
| Central American squirrel monkey (Saimiri oerstedii) | Costa Rica | Captive | Faeces | Salmonella enterica | CI, NIT | ND | ND | [24] |
| Species | Locations | Life Context | Type of Sample | Resistant Bacteria | Antibiotic Resistance | Detection Test | Resistance Genes | Ref |
|---|---|---|---|---|---|---|---|---|
| Sumatran orangutan (Pongo abelii) | Indonesia | Wild | Faeces | K. pneumoniae | AMP, NA, SM | PCR | blaTEM, blaSHV, blaCTX-M, tetA | [25] |
| Chimpanzee (Pan troglodytes) | Senegal | Wild | Faeces | E. coli, Enterobacter spp. | CTX, COL, ETP | WGS | oqxA, oqxB, fosA, blaCMH-3, blaACT-6, blaCMG1,blaCTM-X,qnrS1, sul2, tetA | [26] |
| Mountain gorilla (Gorilla beringei beringei) | Uganda | Wild | Faeces | E. coli | TMP-SMX, CI, GM, EFT | PCR | dfrA, aadA, blaOXA, catB | [27] |
| Species | Locations | Life Context | Type of Sample | Resistant Bacteria | Antibiotic Resistance | Detection Test | Resistance Genes | Ref |
|---|---|---|---|---|---|---|---|---|
| Common marmoset (Callithrix jacchus) | United States | Captive | Oral, skin, swab, faeces | S. aureus, Yersinia spp. Campylobacter spp. | PRM, NIT, OX | ND | ND | [28] |
| Mustached tamarin (Saguinus mystax) | Peru | Semi-captive | Faeces | E. coli | CAP, GM | WGS | blaCTM-X, floR, catA, cmlA1, aac(3″)-IId, aadA, aph(6″)-Id, aph(3″)-IIa, qnrB, qnrS1, sul1, sul2, sul3, tetA, tetB, tetM | [29] |
| Red-handed tamarin (Saguinus midas) | Brazil | Captive | Exudate | Klebsiella pneumoniae | AMX, PG, NV, FLR, SUD, SMX | ND | ND | [30] |
| Black-tufted marmoset (Callithrix pencillata) | Brazil | Wild | Faeces | Staphylococcus epidermidis | PG, FOX, CI, CLI, E | ND | ND | [31] |
| Silvery marmoset (Mico argentatus) | England | Captive | Tissues | Yersinia pseudotuberculosis | CLI | ND | ND | [32] |
| Golden lion tamarin (Leontopithecus rosalia) | Brazil | Wild | Oral and rectal swabs | Staphylococcus spp. | PG, E, OX, FD | ND | ND | [33] |
| Species | Locations | Life Context | Type of Sample | Resistant Bacteria | Antibiotic Resistance | Detection Test | Resistance Genes | Ref |
|---|---|---|---|---|---|---|---|---|
| Long-tailed macaque (Macaca fascicularis) | Belgium | Captive | Gastric mucosa | Helicobacter suis | ENR, LVX, MFX, SH, MY, TC | WGS | gyrA, acrB | [34] |
| China, Indonesia | Captive | Faeces | Campylobacter coli, Campylobacter jejuni | TC, E, CI, AMX | ND | ND | [35] | |
| United States | Captive | Nasal swab | MRSA, VRSA, VISA | FOX, TMP-SMX, CLI, E, PG, VAN | ND | ND | [36] | |
| United States | Captive | Nasal swab | MSSA, MRSA | ND | WGS | blaZ, tet38, aph(3″)-IIa, gyrA | [37] | |
| Brunei Darussalam | Wild | Faeces | Staphylococcus spp. | TMP, SMZ, FD | ND | ND | [38] | |
| Assamese macaque (Macaca assamensis) | Nepal | Wild | Oral | MRSA | PEN, OX, FOX, GM, E | DNA Microarray | blaZ, aacA-aphD, aph(3″)-IIa, erm(C), mph(C), dfrA, msrA | [39] |
| Toque macaque (Macaca sinica) | Sri Lanka | Wild | Faeces | Campylobacter spp. Salmonella spp. | TC, CI, NA | ND | ND | [40] |
| Rhesus macaque (Macaca mulatta) | United States | Captive | Cephalic chambers | E. faecalis | CI, ENR, TMP-SMX, TC, CAP, B, E | WGS | bcrA, bcrB, bcrR, catA, catB, gyrA, aph(3′)-II a, tetM, tetS, tetL, dfrG | [41] |
| Nepal | Wild | Saliva | MRSA | ND | DNA microarray | aacA-aphD, dfrA, ermC, aph(3″)-IIa, blaZ, mecA, msrA | [42] | |
| United States | Captive | Faeces | Shigella flexneri | AMP, AMC, GM, TC, CI, ENR, LVX, NA | WGS | aadA, aac(3″)-IId, blaOXA, oqxA, oqxB, catA, tetB, blaTEM, qnrS1, blaCTX-M | [43] | |
| Nepal | Wild | Saliva | MRSA | CI, GM, E | WGS | gyrA, ermC, aacA-aphD, blaZ | [44] | |
| China | Captive | Faeces, tissue fluid | E. coli, K. pneumoniae, P. mirabilis | FLR, TC, KM, AMP, IPM, FOX, SM | ND | ND | [45] | |
| United States | Captive | Rectal swab | Campylobacter jejuni, Campylobacter coli | CI, AZM, CLI, TC | WGS | aph(3″)-IIa, gyrA, tetO, floR, sul2 | [46] | |
| China | Captive | Faeces | E. coli, P. mirabilis, K. pneumoniae | LVX, ENR, CTX | WGS | qnrS1, blaSHV, blaTEM, blaCTX-M, sul2, floR | [47] | |
| Bangladesh | Wild | Faeces | Salmonella spp. Staphylococcus | TC, AZM, TMP-SMX, NA, AMP, MET, CLI, RFP | ND | ND | [48] | |
| Vervet monkeys (Chlorocebus pygerythrus) | South Africa | Wild | Faeces | Escherichia fergusonii | POL, COL, AMK | ND | ND | [49] |
| Uganda | Wild | Nasal swab | MRSA | TC, SMZ-TMP, PG | ND | ND | [50] | |
| Golden snub-nosed monkeys (Rhinopithecus roxellana) | China | Captive | Faeces | E. coli | DOX, TC | PCR | tetA | [51] |
| China | Captive | Faeces | Streptococcus agalactiae | E, TC, CLI | WGS | mreA, tetM, tet(L), tet(O) | [52] | |
| African green monkeys (Chlorocebus sabaeus) | Nigeria | Captive | Faeces | E. coli | PIP, LVX, TMP-SMX | PCR | qnrD, qnrA, qnrB, qnrS1 | [53] |
| Saint Kitts and Nevis | Captive and wild | Nasal swab | MSSA, MRSA | ND | WGS | mecA, blaZ, mphC, dfrG, ermC, tetK | [54] | |
| Olive Baboons (Papio anubis) | Ghana | Wild | Oral and rectal swabs | E. coli, Staphylococcus spp. | PEN, OX | ND | ND | [55] |
| Guinea baboons (Papio papio) | Gambia | Wild | Faeces | E. coli | AMK, TMP-SMX, CI, CTX, TC | WGS | blaEC, aadA, tetA | [56] |
| Species | Locations | Life Context | Type of Sample | Resistant Bacteria | Antibiotic Resistance | Detection Test | Resistance Genes | Ref |
|---|---|---|---|---|---|---|---|---|
| Black howler monkeys (Alouatta pigra) | Mexico | Wild | Faeces | E. coli | ND | PCR | sul1, sul2, tetB, blaTEM | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Okyere, S.K.; Shi, Y.; Qu, Y.; Chen, C. Phenotypic and Genotype Patterns of Antimicrobial Resistance in Non-Human Primates: An Overlooked “One Health” Concern. Antibiotics 2025, 14, 985. https://doi.org/10.3390/antibiotics14100985
Wen J, Okyere SK, Shi Y, Qu Y, Chen C. Phenotypic and Genotype Patterns of Antimicrobial Resistance in Non-Human Primates: An Overlooked “One Health” Concern. Antibiotics. 2025; 14(10):985. https://doi.org/10.3390/antibiotics14100985
Chicago/Turabian StyleWen, Juan, Samuel Kumi Okyere, Yujie Shi, Yu Qu, and Chaoxi Chen. 2025. "Phenotypic and Genotype Patterns of Antimicrobial Resistance in Non-Human Primates: An Overlooked “One Health” Concern" Antibiotics 14, no. 10: 985. https://doi.org/10.3390/antibiotics14100985
APA StyleWen, J., Okyere, S. K., Shi, Y., Qu, Y., & Chen, C. (2025). Phenotypic and Genotype Patterns of Antimicrobial Resistance in Non-Human Primates: An Overlooked “One Health” Concern. Antibiotics, 14(10), 985. https://doi.org/10.3390/antibiotics14100985






