The Potential for Sample Testing at the Pen Level to Inform Prudent Antimicrobial Selection for Bovine Respiratory Disease Treatment: Investigations Using a Feedlot Simulation Tool
Abstract
1. Introduction
2. Materials and Methods
2.1. Model Description
2.2. Purpose
2.3. Key Assumptions
2.3.1. Model Configuration and Diagnostic Paradigms
2.3.2. Responsiveness of BRD Incidence to AMR
2.3.3. Sampling and Testing in Advance of the Need to Treat
2.3.4. Exposure to Antimicrobials
2.3.5. Treatment Change Threshold
2.4. Testing Agent
2.5. Input Data
2.5.1. Testing Agent Parameters
2.5.2. Diagnostic Test Characteristics
| Diagnostic Test Characteristics | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity Estimate (%) | Specificity Estimate (%) | |||||||
| Antimicrobial Class | Diagnostic Test Type | Reference Drug 1 or Gene 2 | Low | Median | High | Low | Median | High |
| Classes with BLCM-derived estimates 3 | ||||||||
| 15-membered ring macrolides | AST | Tulathromycin | 73% | 80% | 86% | 99% | 100% | 100% |
| Metagenomics | msrE-mphE | 56% | 62% | 69% | 96% | 98% | 99% | |
| 16-membered ring macrolides | AST | Tilmicosin | 10% | 23% | 38% | 99% | 100% | 100% |
| Metagenomics | estT | 22% | 43% | 65% | 98% | 99% | 100% | |
| Sulfonamides | AST | Sulfadimethoxine | 83% | 90% | 98% | 98% | 99% | 100% |
| Metagenomics | sul2 | 57% | 63% | 69% | 90% | 92% | 95% | |
| Tetracyclines | AST | Oxytetracycline | 11% | 18% | 25% | 99% | 100% | 100% |
| Metagenomics | tetH | 66% | 82% | 95% | 94% | 97% | 99% | |
| Classes without BLCM-derived estimates 4 | ||||||||
| Diaminopyrimidines | AST | Trimethoprim | 10% | 18% | 25% | 98% | 99% | 100% |
| Metagenomics | dfrA14 | 22% | 43% | 65% | 90% | 92% | 95% | |
| Cephalosporins | AST | Ceftiofur | 10% | 18% | 25% | 98% | 99% | 100% |
| Metagenomics | blaROB-2 | 22% | 43% | 65% | 90% | 92% | 95% | |
| Fluoroquinolones | AST | Enrofloxacin | 10% | 18% | 25% | 98% | 99% | 100% |
| Metagenomics | gyrA mutation | 22% | 43% | 65% | 90% | 92% | 95% | |
| Phenicols | AST | Florfenicol | 10% | 18% | 25% | 98% | 99% | 100% |
| Metagenomics | floR | 22% | 43% | 65% | 90% | 92% | 95% | |
2.6. Key Model Outputs
2.7. Model Verification
2.8. Summary of Monte Carlo Experiments
2.9. Analysis of Model Output
Sensitivity Analyses
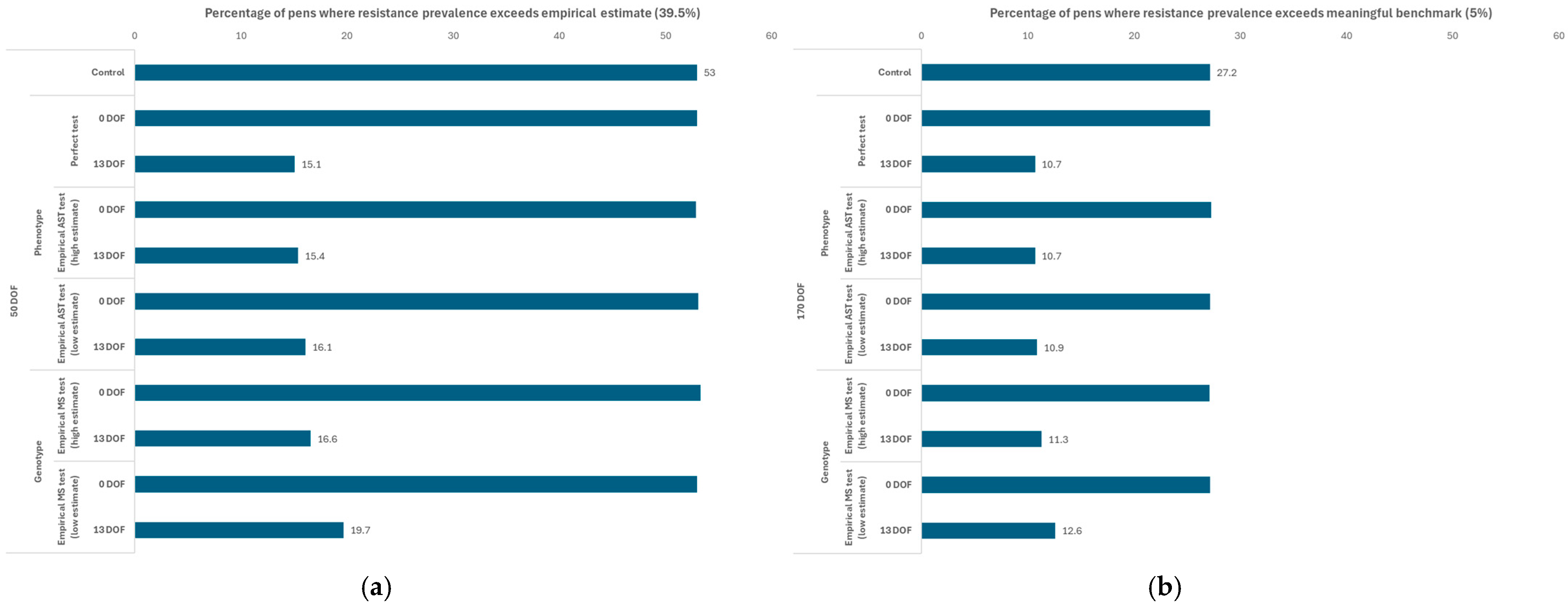
3. Results
3.1. Scenarios Using the Baseline AMU Protocol
3.1.1. Sensitivity of Outputs to Testing Parameters in Baseline Scenario
3.1.2. Impact of Strategy When Incoming Resistance Is High
3.1.3. Sensitivity of Outputs to Testing Parameters When Incoming Resistance Is High
3.2. Scenarios Using the Extreme Macrolide Use Protocol
4. Discussion
Effectiveness of Testing-Informed Treatment in the Modern Feedlot Setting
Theoretical Applications of the Testing-Informed Treatment Strategy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABM | Agent-based Model |
| ADG | Average Daily Growth |
| AMR | Antimicrobial Resistance |
| AMU | Antimicrobial Use |
| ARG | Antimicrobial Resistance Gene |
| AST | Antimicrobial Susceptibility Testing |
| BLCM | Bayesian Latent Class Model |
| BRD | Bovine Respiratory Disease |
| CFAASP | Canadian Feedlot Antimicrobial Use and Antimicrobial Resistance Surveillance Program |
| CI | Confidence Interval |
| CrI | Credible Interval |
| DES | Discrete Event Simulation |
| DOF | Days On Feed |
| IQR | Interquartile Range |
| LCA | Latent Class Analysis |
| MS | Metagenomic Sequencing |
| ODD | Overview, Design Concepts, and Details |
| TI | Testing Informed |
| TO | Testing Only |
| WHO | World Health Organization |
References
- World Health Organization. Antimicrobial Resistance. (November 2023). Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 8 June 2025).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health. Antimicrobial Resistance. Available online: https://www.woah.org/en/what-we-do/global-initiatives/antimicrobial-resistance/ (accessed on 8 June 2025).
- Food and Agriculture Organization of the United Nations. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. 2016. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/95efae06-4da2-4689-b12a-1b33fffc2244/content (accessed on 8 June 2025).
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [PubMed]
- Lhermie, G.; Grohn, Y.T.; Raboisson, D. Addressing antimicrobial resistance: An overview of priority actions to prevent suboptimal antimicrobial use in food-animal production. Front. Microbiol. 2017, 7, 2114. [Google Scholar] [CrossRef]
- Strong, K.; Emdin, F.; Orubu, S.; Van Katwyk, S.R.; Ganshorn, H.; Grimshaw, J.; Poirier, M.J.P. Government policy interventions to reduce veterinary antimicrobial consumption in production animals: A protocol for a systematic review and evidence map. Syst. Rev. 2025, 14, 122. [Google Scholar] [CrossRef]
- Government of Canada. Pan-Canadian Action Plan on Antimicrobial Resistance. 2023. Available online: https://www.canada.ca/content/dam/phac-aspc/documents/services/publications/drugs-health-products/pan-canadian-action-plan-antimicrobial-resistance/pan-canadian-action-plan-antimicrobial-resistance.pdf (accessed on 8 June 2025).
- Food and Agriculture Organization of the United Nations. The FAO Action Plan on Antimicrobial Resistance 2021–2025. 2021. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/dd6c0ba1-fd85-4a3e-b398-53b610c35318/content (accessed on 8 June 2025).
- Edwards, T.A. Control methods for bovine respiratory disease for feedlot cattle. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 273–284. [Google Scholar] [CrossRef]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for preventive measures? Can. Vet. J. 2010, 51, 1351–1359. [Google Scholar]
- Canadian Feedlot Antimicrobial Use and Antimicrobial Resistance Surveillance Program. World Antimicrobial Awareness Week Presentation. 2024. Available online: https://cahss.ca/CAHSS/Assets/Documents/CIPARS%20Feedlot%20WAAW%20and%20industry%20presentation%20EN%20Final.pdf (accessed on 8 June 2025).
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can. Vet. J. 2010, 51, 1095–1102. [Google Scholar]
- Brault, S.A.; Hannon, S.J.; Gow, S.P.; Warr, B.N.; Withell, J.; Song, J.; Williams, C.M.; Otto, S.; Booker, C.W.; Morley, P.S. Antimicrobial use on 36 beef feedlots in western Canada: 2008–2012. Front. Vet. Sci. 2019, 6, 329. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Booker, C.W. Bovine respiratory disease treatment failure: Definition and impact. Anim. Health Res. Rev. 2020, 21, 172–174. [Google Scholar] [CrossRef]
- Otto, S.J.G.; Pollock, C.M.; Relf-Eckstein, J.; McLeod, L.; Waldner, C.L. Opportunities for laboratory testing to inform antimicrobial use for bovine respiratory disease: Application of Information Quality Value Stream Maps in commercial feedlots. Antibiotics 2024, 13, 903. [Google Scholar] [CrossRef]
- Kamel, M.S.; Davidson, J.L.; Verma, M.S. Strategies for bovine respiratory disease (BRD) diagnosis and prognosis: A comprehensive overview. Animals 2024, 14, 627. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London, UK: The Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 8 June 2025).
- Aidara-Kane, A.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Collignon, P.J.; WHO Guideline Development Group. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control. 2018, 7, 7. [Google Scholar] [CrossRef]
- Confer, A.W. Update on bacterial pathogenesis in BRD. Anim. Health Res. Rev. 2009, 10, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Buczinski, S.; Pardon, B. Bovine respiratory disease diagnosis: What progress has been made in clinical diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 399–423. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Otto, S.J.G.; Gow, S.P.; Woolums, A.R.; Jelinski, M.; Lacoste, S.; Waldner, C.L. Variation in pen-level prevalence of BRD bacterial pathogens and antimicrobial resistance following feedlot arrival in beef calves. Antibiotics 2024, 13, 322. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Gow, S.P.; Woolums, A.R.; Waldner, C.L. Association between respiratory disease pathogens in calves near feedlot arrival with treatment for bovine respiratory disease and subsequent antimicrobial resistance status. Front. Vet. Sci. 2024, 11, 1416436. [Google Scholar] [CrossRef] [PubMed]
- Gröhn, Y.T. Progression to multi-scale models and the application to food system intervention strategies. Prev. Vet. Med. 2015, 118, 238–246. [Google Scholar] [CrossRef]
- Marshall, D.A.; Burgos-Liz, L.; Ijzerman, M.J.; Osgood, N.D.; Padula, W.V.; Higashi, M.K.; Wong, P.K.; Pasupathy, K.S.; Crown, W. Applying dynamic simulation modeling methods in health care delivery research—The SIMULATE checklist: Report of the ISPOR simulation modeling emerging good practices task force. Value Health 2015, 18, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, D.; McDonald, W.; Thompson, M.; Erickson, N.; Gow, S.; Osgood, N.D.; Waldner, C. Contagious acquisition of antimicrobial resistance is critical for explaining emergence in western Canadian feedlots—Insights from an agent-based modelling tool. Front. Vet. Sci. 2025, 11, 1466986. [Google Scholar] [CrossRef]
- Bonabeau, E. Agent-based modeling: Methods and techniques for simulating human systems. Proc. Natl. Acad. Sci. USA 2002, 99, 7280–7287. [Google Scholar] [CrossRef]
- Grimm, V.; Berger, U.; Bastiansen, F.; Eliassen, S.; Ginot, V.; Giske, J.; Goss-Custard, J.; Grand, T.; Heinz, S.K.; Huse, G.; et al. A standard protocol for describing individual-based and agent-based models. Ecol. Model 2006, 198, 115–126. [Google Scholar] [CrossRef]
- Grimm, V.; Railsback, S.F.; Vincenot, C.E.; Berger, U.; Gallagher, C.; DeAngelis, D.L.; Edmonds, B.; Ge, J.; Giske, J.; Groeneveld, J.; et al. The ODD protocol for describing agent-based and other simulation models: A second update to improve clarity, replication and structural realism. J. Artif. Soc. Soc. Simul. 2020, 23, 7. [Google Scholar] [CrossRef]
- Herman, E.K.; Lacoste, S.R.; Freeman, C.N.; Otto, S.J.G.; McCarthy, E.L.; Links, M.G.; Stothard, P.; Waldner, C.L. Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle. Front. Microbiol. 2024, 15, 1386319. [Google Scholar] [CrossRef]
- Abi Younes, J.; Ramsay, D.E.; Lacoste, S.; Deschner, D.; Hill, J.E.; Campbell, J.; Waldner, C.L. Changes in the phenotypic susceptibility of Mannheimia haemolytica isolates to macrolide antimicrobials during the early feeding period following metaphylactic tulathromycin use in western Canadian feedlot calves. Can. Vet. J. 2022, 63, 920–928. [Google Scholar]
- Noyes, N.R.; Benedict, K.M.; Gow, S.P.; Booker, C.W.; Hannon, S.J.; McAllister, T.A.; Morley, P.S. Mannheimia haemolytica in feedlot cattle: Prevalence of recovery and associations with antimicrobial use, resistance and health outcomes. J. Vet. Intern. Med. 2015, 29, 705–713. [Google Scholar] [CrossRef]
- Erickson, N.E.N.; Ngeleka, M.G.; Lubbers, B.V.; Trokhymchuk, A. Changes in the rates of field isolation and antimicrobial susceptibility of bacterial pathogens collected from fall-placed feedlot steers between arrival at the feedlot and 90 to 120 days on feed. Bov. Pract. 2017, 51, 165–173. [Google Scholar] [CrossRef]
- Timsit, E.; Hallewell, J.; Booker, C.; Tison, N.; Amat, S.; Alexander, T.W. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2017, 208, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Lasheras, S.; Ha, R.; Zaheer, R.; Lee, C.; Booker, C.W.; Dorin, C.; Van Donkersgoed, J.; Deardon, R.; Gow, S.; Hannon, S.J.; et al. Prevalence and risk factors associated with antimicrobial resistance in bacteria related to bovine respiratory disease—A broad cross-sectional study of beef cattle at entry into Canadian feedlots. Front. Vet. Sci. 2021, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Gow, S.; Bergen, R.; Booker, C.; Butters, A.; Dorin, C.; Dimmers, G.; Erickson, N.; Hannon, S.; Hendrick, S.; Ramsay, D.; et al. National Surveillance of Antimicrobial Use and Antimicrobial Resistance in Canadian Feedlots. In Proceedings of the Fifty-Fourth Annual Conference, American Association of Bovine Practitioners, Salt Lake City, UT, USA, 7–9 October 2021; No. 54. pp. 34–41. Available online: https://bovine-ojs-tamu.tdl.org/AABP/article/view/8291 (accessed on 6 February 2024).
- Wennekamp, T.R.; Waldner, C.L.; Windeyer, M.C.; Larson, K.; Trokhymchuk, A.; Campbell, J.R. Antimicrobial resistance in bovine respiratory disease: Auction market- and ranch-raised calves. Can. Vet. J. 2022, 63, 47–54. [Google Scholar] [CrossRef]
- Apley, M.D. Treatment of calves with bovine respiratory disease: Duration of therapy and posttreatment intervals. Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 441–453. [Google Scholar] [CrossRef]
- Smith, K.J.; White, B.J.; Amrine, D.E.; Larson, R.L.; Theurer, M.E.; Szasz, J.I.; Bryant, T.C.; Waggoner, J.W. Evaluation of first treatment timing, fatal disease onset, and days from first treatment to death associated with bovine respiratory disease in feedlot cattle. Vet. Sci. 2023, 10, 204. [Google Scholar] [CrossRef]
- Apley, M.D. Diagnosis and therapy of feedlot lameness. Am. Assoc. Bov. Pract. Conf. Proc. 2020, 53, 54–59. [Google Scholar] [CrossRef]
- Lubbers, B.V.; Turnidge, J. Antimicrobial susceptibility testing for bovine respiratory disease: Getting more from diagnostic results. Vet. J. 2015, 203, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Abi Younes, J.N.; McLeod, L.; Otto, S.J.G.; Chai, Z.; Lacoste, S.; McCarthy, E.L.; Links, M.G.; Herman, E.K.; Stothard, P.; Gow, S.P.; et al. Evaluating the diagnostic performance of long-read metagenomic sequencing compared to culture and antimicrobial susceptibility testing for detection of bovine respiratory bacteria and indicators of antimicrobial resistance. Antibiotics 2025. in submitted. [Google Scholar]
- Ramsay, D.E.; Invik, J.; Checkley, S.L.; Gow, S.P.; Osgood, N.D.; Waldner, C.L. Application of dynamic modelling techniques to the problem of antibacterial use and resistance: A scoping review. Epidemiol. Infect. 2018, 146, 2014–2027. [Google Scholar] [CrossRef]
- Briggs, A.H.; Weinstein, M.C.; Fenwick, E.A.L.; Karnon, J.; Sculpher, M.J.; Paltiel, A.D. Model Parameter Estimation and Uncertainty: A Report of the ISPOR-SMDM Modeling Good Research Practices Task Force-6. Value Health 2012, 15, 835–842. [Google Scholar] [CrossRef]
- Eddy, D.M.; Hollingworth, W.; Caro, J.J.; Tsevat, J.; McDonald, K.M.; Wong, J.B. Model Transparency and Validation: A Report of the ISPOR-SMDM Modeling Good Research Practices Task Force-7. Value Health 2012, 15, 843–850. [Google Scholar] [CrossRef]
- Canadian Feedlot Antimicrobial Use and Antimicrobial Resistance Surveillance Program. Bovine Respiratory Disease (BRD) Pathogen Antimicrobial Resistance (AMR) Update—2022. Available online: https://cfaasp.ca/resources/cfaasp-resources/Bovine-Respiratory-Disease-BRD-Pathogen-Antimicrobial-Resistance-AMR-Update-2022 (accessed on 19 June 2025).
- Rattanapanadda, P.; Ramsay, D.; Butters, A.; Booker, C.W.; Hannon, S.J.; Hendrick, S.; Van Donkersgoed, J.; Warr, B.N.; Gow, S.P.; Morley, P.S. The prevalence and antimicrobial resistance of respiratory pathogens isolated from feedlot cattle in Canada. Front. Microbiol. 2025, 16, 1497402. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Lasheras, S.; Jelinski, M.; Zaheer, R.; McAllister, T.A. Bovine respiratory disease: Conventional to culture-independent approaches to studying antimicrobial resistance in North America. Antibiotics 2022, 11, 487. [Google Scholar] [CrossRef]
- Joseph, L.; Gyorkos, T.W.; Coupal, L. Bayesian estimation of disease prevalence and the parameters of diagnostic tests in the absence of a gold standard. Am. J. Epidemiol. 1995, 141, 263–272. [Google Scholar] [CrossRef]
- Freeman, C.N.; Herman, E.K.; Abi Younes, J.; Ramsay, D.E.; Erickson, N.; Stothard, P.; Links, M.G.; Otto, S.J.G.; Waldner, C. Evaluating the potential of third generation metagenomic sequencing for the detection of BRD pathogens and genetic determinants of antimicrobial resistance in chronically ill feedlot cattle. BMC Vet Res. 2022, 18, 211. [Google Scholar] [CrossRef] [PubMed]
- Kos, D.; Jelinski, M.; Ruzzini, A. Retrospective analysis of antimicrobial resistance associated with bovine respiratory disease. Appl. Environ. Microbiol. 2025, 91, e01909-24. [Google Scholar] [CrossRef] [PubMed]
- Babcock, A.H.; Renter, D.G.; White, B.J.; Dubnicka, S.R.; Scott, H.M. Temporal distributions of respiratory disease events within cohorts of feedlot cattle and associations with cattle health and performance indices. Prev. Vet. Med. 2010, 97, 198–219. [Google Scholar] [CrossRef]
- Canadian Feedlot Antimicrobial Use and Antimicrobial Resistance Surveillance Program. Bovine Respiratory Disease (BRD) Pathogen Antimicrobial Resistance (AMR) Update—CFAASP 2023. Available online: https://cfaasp.ca/resources/cfaasp-resources/Bovine_Respiratory_Disease_BRD_Pathogen_Antimicrobial_Resistance_Update_CFAASP_2023 (accessed on 19 June 2025).
- Government of Canada. Categorization of Antimicrobial Drugs Based on Importance in Human Medicine (Version—April) 2009. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/antimicrobial-resistance/categorization-antimicrobial-drugs-based-importance-human-medicine.html (accessed on 18 May 2025).
- European Food Safety Authority. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- Shane, D.D.; McLellan, J.G.; White, B.J.; Larson, R.L.; Armine, D.E.; Sanderson, M.W.; Apley, M.D. Evaluation of animal-to-animal and community contact structures determined by a real-time location system for correlation with and prediction of new bovine respiratory disease diagnoses in beef cattle during the first 28 days after feedlot entry. Am. J. Vet. Res. 2018, 79, 1277–1286. [Google Scholar] [CrossRef]
- Snyder, E.; Credille, B.; Berghaus, R.; Giguère, S. Prevalence of multi drug antimicrobial resistance in Mannheimia haemolytica isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. 2017, 95, 1124–1131. [Google Scholar] [CrossRef]
- Government of Canada. Tackling Antimicrobial Resistance and Antimicrobial Use: A Pan-Canadian Framework for Action. August 2017. Available online: https://www.canada.ca/content/dam/hc-sc/documents/services/publications/drugs-health-products/tackling-antimicrobial-resistance-use-pan-canadian-framework-action/tackling-antimicrobial-resistance-use-pan-canadian-framework-action.pdf (accessed on 22 May 2025).
- Tian, Y.; Osgood, N.D.; Stempien, J.; Onaemo, V.; Danyliw, A.; Fast, G.; Osman, B.A.; Reynolds, J.; Basran, J. The impact of alternate level of care on access block and operational strategies to reduce emergency wait times: A multi-center simulation study. CJEM 2023, 25, 608–616. [Google Scholar] [CrossRef]
- Vázquez-Serrano, J.I.; Peimbert-García, R.E.; Cárdenas-Barrón, L.E. Discrete-event simulation modeling in healthcare: A comprehensive review. Int. J. Environ. Res. Public Health 2021, 18, 12262. [Google Scholar] [CrossRef]
- Sorin-Dupont, B.; Picault, S.; Pardon, B.; Ezanno, P.; Assié, S. Modeling the effects of farming practices on bovine respiratory disease in a multi-batch fattening farm. Prev. Vet. Med. 2023, 219, 106009. [Google Scholar] [CrossRef] [PubMed]
- Picault, S.; Ezanno, P.; Smith, K.; Amrine, D.; White, B.; Assié, S. Modelling the effects of antimicrobial metaphylaxis and pen size on bovine respiratory disease in high and low risk fattening cattle. Vet. Res. 2022, 53, 77. [Google Scholar] [CrossRef]
- Genome Prairie. The ASSETS Project: Fighting Antibiotic Resistance in Livestock. Available online: https://genomeprairie.ca/the-assets-project-fighting-antibiotic-resistance/ (accessed on 2 July 2025).
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial use in Canadian cow-calf herds. Vet. Sci. 2023, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.; Chengappa, M.M.; Kuszak, J.; McVey, D.S. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Cusack, P.M.V.; McMeniman, N.; Lean, I.J. The medicine and epidemiology of bovine respiratory disease in feedlots. Aust. Vet. J. 2003, 81, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, D.; Andrés-Lasheras, S.; Zaheer, R.; McAllister, T.; Homerosky, E.; Anholt, R.M.; Dorin, C. Prevalence, risk factors, and antimicrobial resistance profile of respiratory pathogens isolated from suckling beef calves to reprocessing at the feedlot: A longitudinal study. Front. Vet. Sci. 2021, 8, 764701. [Google Scholar] [CrossRef]
- O’Connor, A.M.; Hu, D.; Totton, S.C.; Scott, N.; Winder, C.B.; Wang, B.; Wang, C.; Glanville, J.; Wood, H.; White, B.; et al. A systematic review and network meta-analysis of injectable antibiotic options for control of bovine respiratory disease in the first 45 days post arrival at the feedlot. Anim. Health Res. Rev. 2019, 20, 163–181. [Google Scholar] [CrossRef]
- Ribble, C.S.; Stitt, T.; Iwasawa, S.; Toews, L.; Stephen, C. A Review of Alternative Practices to Antimicrobial Use for Disease Control in the Commercial Feedlot; National Collaborating Centre for Infectious Diseases: Winnipeg, MB, Canada, 2010; Available online: https://nccid.ca/wp-content/uploads/sites/2/2020/01/CattleFeedlots_Ribble.pdf (accessed on 25 May 2025).
- Van Donkersgoed, J.; Merrill, J.K.; Hendrick, S. Comparative efficacy of tilmicosin, florfenicol, and florfenicol-flunixin meglumine for treatment of undifferentiated fever in backgrounded winter-placed feedlot calves given tilmicosin metaphylactically on arrival. Bov. Pract. 2014, 48, 103–108. [Google Scholar] [CrossRef]
- Canadian Academy of Health Sciences. Antimicrobial Resistance/Antimicrobial Use in Food-Producing Animals in Canada: Strategic Interventions to Strengthen Antimicrobial Stewardship. 2025. Available online: https://cahs-acss.ca/wp-content/uploads/2025/03/CAHS-AMU-Report_EN_Final-1.pdf (accessed on 27 May 2025).
- Holman, D.B.; Timsit, E.; Booker, C.W.; Alexander, T.W. Injectable antimicrobials in commercial feedlot cattle and their effect on the nasopharyngeal microbiota and antimicrobial resistance. Vet. Microbiol. 2018, 214, 140–147. [Google Scholar] [CrossRef]
- Woolums, A.R.; Karisch, B.B.; Frye, J.G.; Epperson, W.; Smith, D.R.; Blanton, J., Jr.; Austin, F.; Kaplan, R.; Hiott, L.; Woodley, T.; et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 2018, 221, 143–152. [Google Scholar] [CrossRef]
- Crosby, W.B.; Karisch, B.B.; Hiott, L.M.; Pinnell, L.J.; Pittman, A.; Frye, J.G.; Jackson, C.R.; Loy, J.D.; Epperson, W.B.; Blanton, J.; et al. Tulathromycin metaphylaxis increases nasopharyngeal isolation of multidrug resistant Mannheimia haemolytica in stocker heifers. Front. Vet. Sci. 2023, 10, 1256997. [Google Scholar] [CrossRef]
- Adewusi, O.O.; Nykiforuk, C.I.J.; Waldner, C.L.; Erickson, N.E.N.; Gow, S.P.; Otto, S.J.G. Laboratory testing to inform antimicrobial use for bovine respiratory disease: Perceptions of Canadian feedlot veterinarians. Vet. Sci. 2025, 12, 409. [Google Scholar] [CrossRef] [PubMed]
- Adewusi, O.O.; Waldner, C.L.; Hanington, P.C.; Hill, J.E.; Freeman, C.N.; Otto, S.J.G. Laboratory tools for the direct detection of bacterial respiratory infections and antimicrobial resistance: A scoping review. J. Vet. Diagn. Investig. 2024, 26, 400–417. [Google Scholar] [CrossRef]
- Alberg, A.J.; Park, J.W.; Hager, B.W.; Brock, M.V.; Diener-West, M. The use of “overall accuracy” to evaluate the validity of screening or diagnostic tests. J. Gen. Intern. Med. 2004, 19, 460–465. [Google Scholar] [CrossRef]
- Klima, C.L.; Holman, D.B.; Cook, S.R.; Conrad, C.C.; Ralston, B.J.; Allan, N.; Anholt, R.M.; Niu, Y.D.; Stanford, K.; Hannon, S.J.; et al. Multidrug resistance in Pasteurellaceae associated with bovine respiratory disease mortalities in North America from 2011 to 2016. Front. Microbiol. 2020, 11, 606438. [Google Scholar] [CrossRef]
- Wickware, C.L.; Ellis, A.C.; Verma, M.; Johnson, T.A. Phenotypic antibiotic resistance prediction using antibiotic resistance genes and machine learning models in Mannheimia haemolytica. Vet. Microbiol. 2025, 302, 110372. [Google Scholar] [CrossRef] [PubMed]
- Dhindwal, P.; Thompson, C.; Kos, D.; Ruzzini, A. A neglected and emerging antimicrobial resistance gene encodes for a serine-dependent macrolide esterase. Proc. Natl. Acad. Sci. USA 2023, 120, e2219827120. [Google Scholar] [CrossRef]
- Owen, J.R.; Noyes, N.; Young, A.E.; Prince, D.J.; Blanchard, P.C.; Lehenbauer, T.W.; Aly, S.S.; Davis, J.H.; O’Rourke, S.M.; Abdo, Z.; et al. Whole-genome sequencing and concordance between antimicrobial susceptibility genotypes and phenotypes of bacterial isolates associated with bovine respiratory disease. G3 Genes Genomes Genet. 2017, 7, 3059–3071. [Google Scholar] [CrossRef] [PubMed]
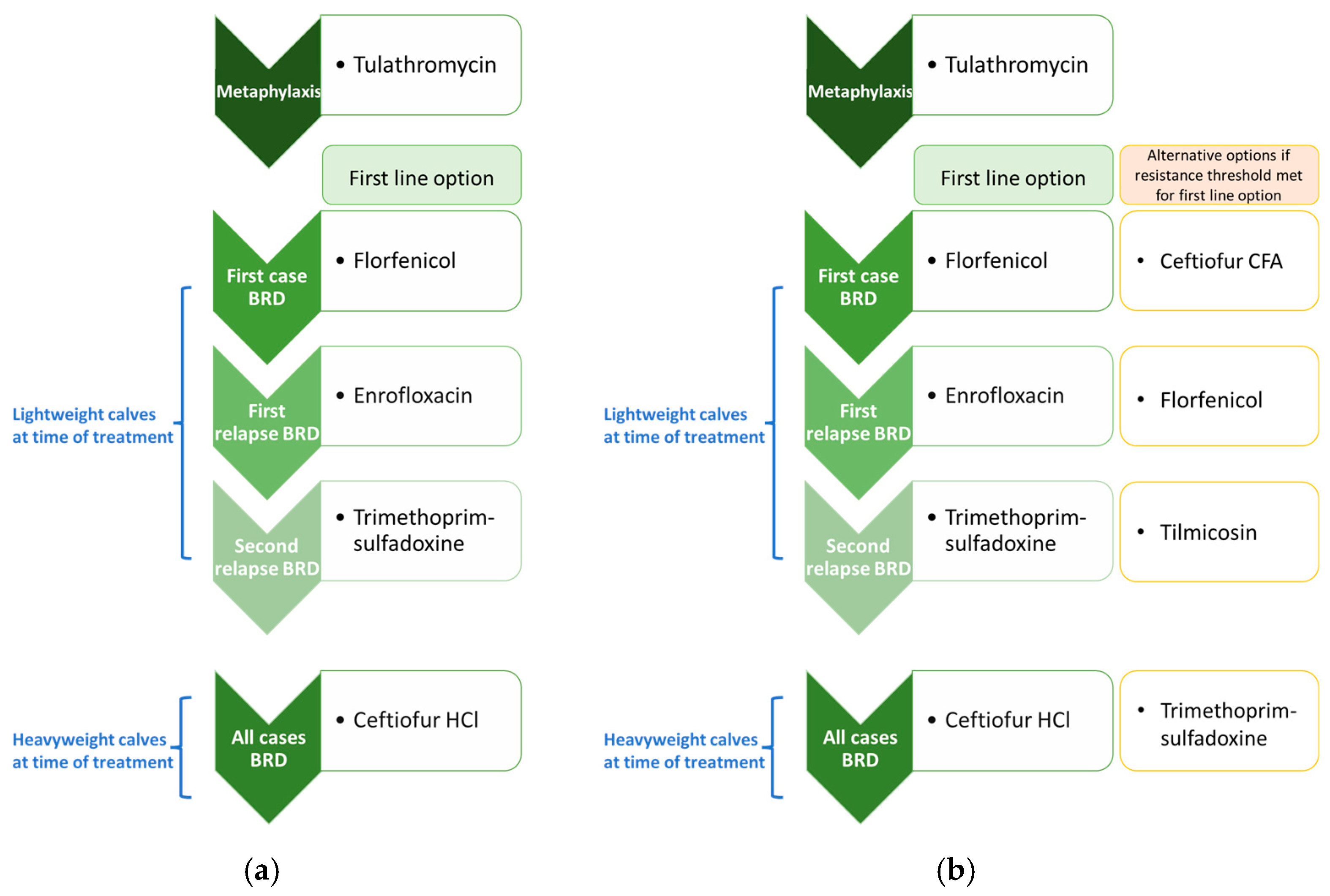

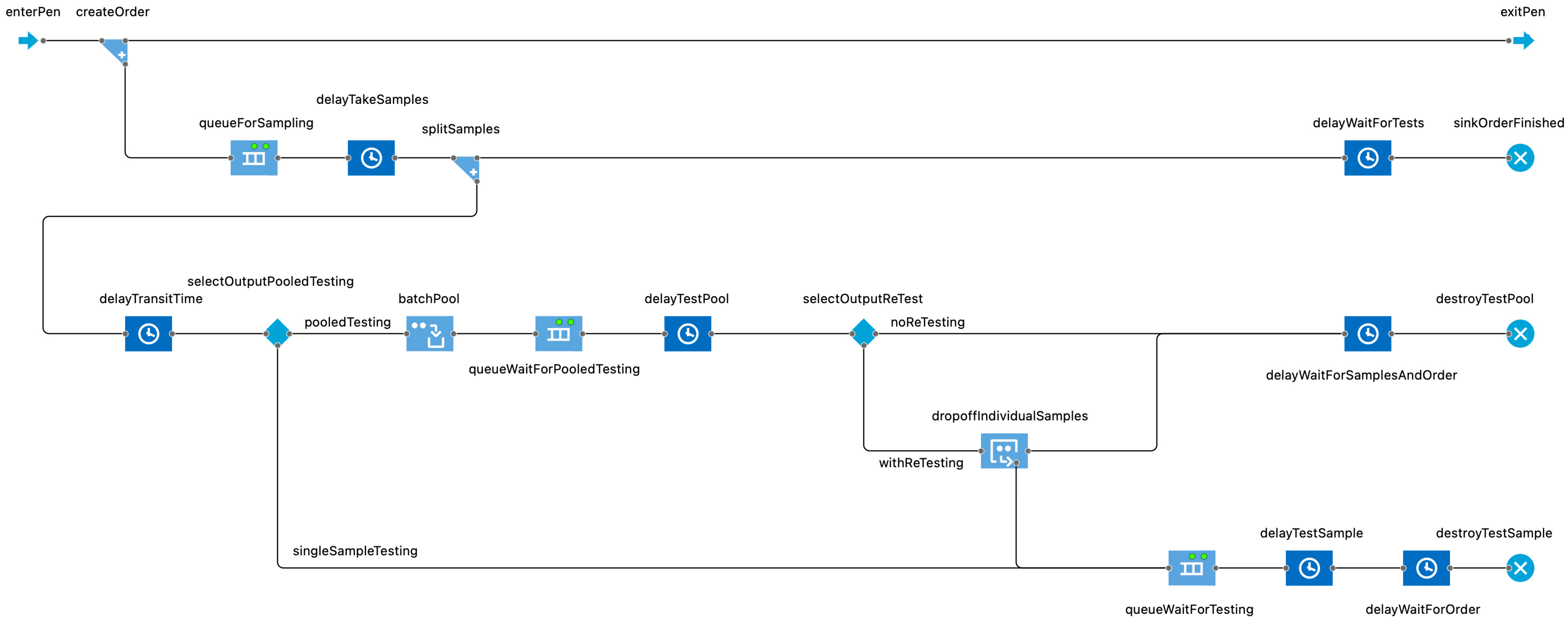



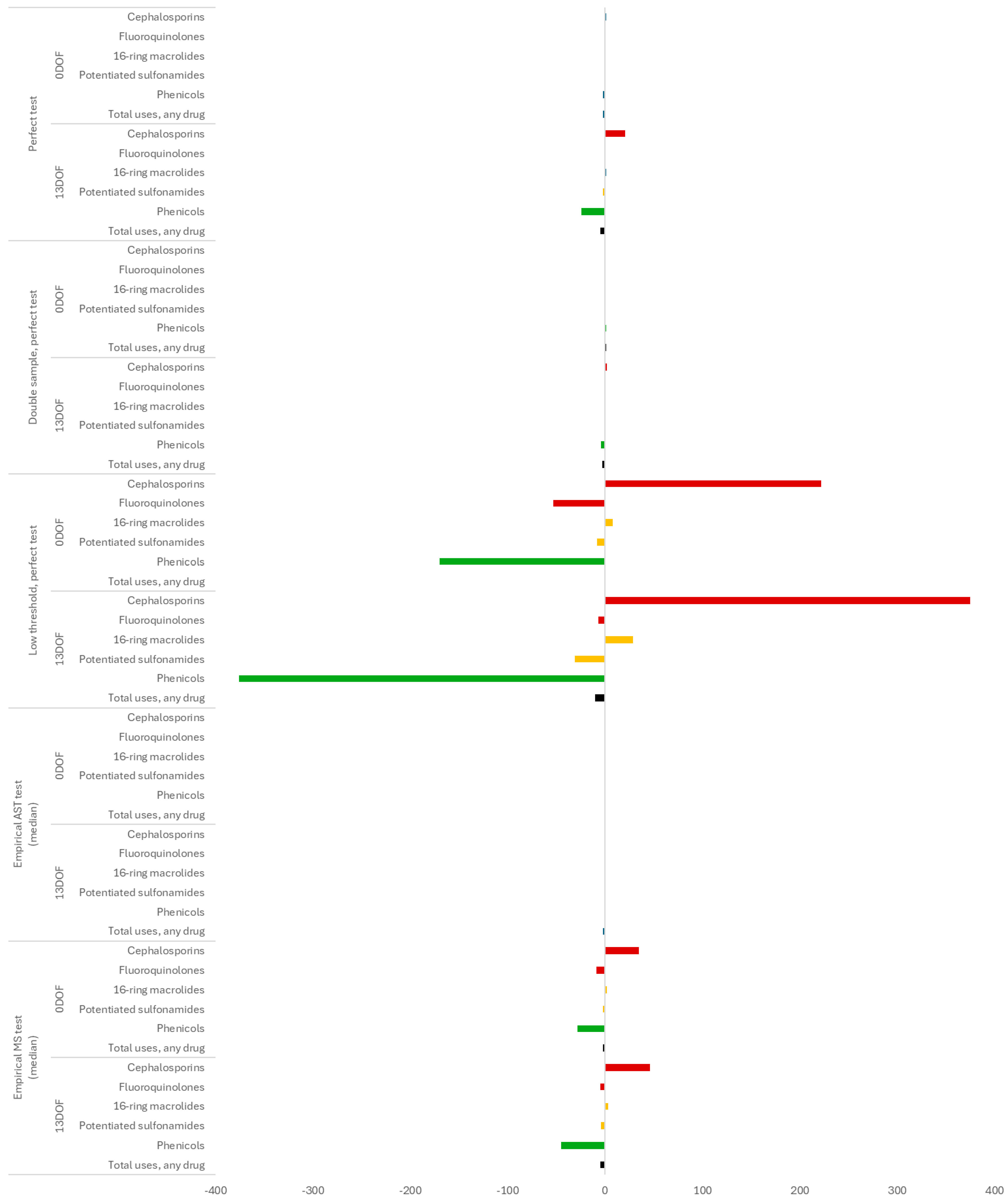
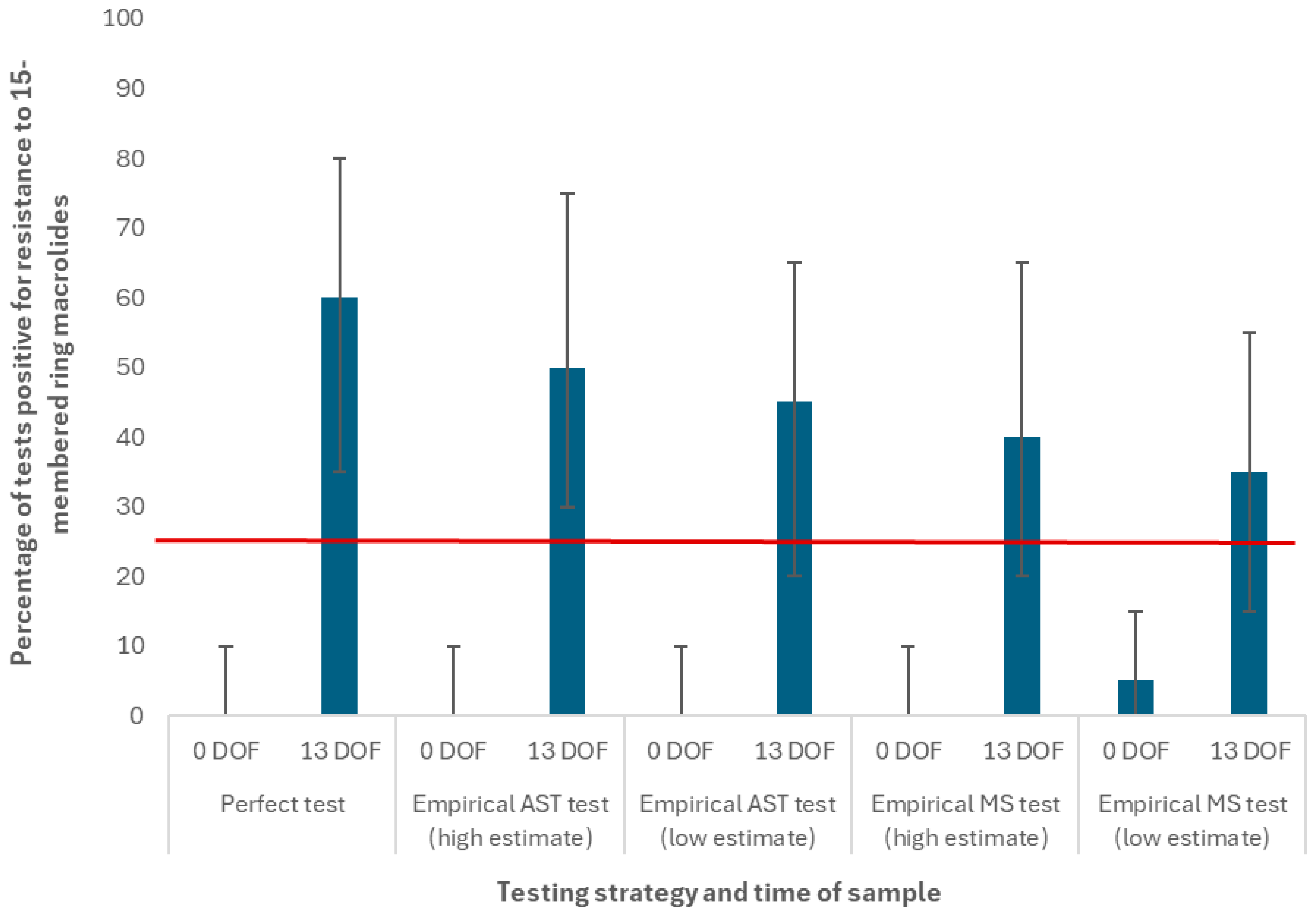
| Antimicrobial Use Type | Indication | Condition, If Applicable | Case Description | Default Antimicrobial Selection(s) | Regimen |
|---|---|---|---|---|---|
| Prophylaxis (in-feed at pen level) | Histophilosis | -- | -- | Chlortetracycline | 2 × 5-day courses of high-dose CTC 1 (18–23 DOF, 25–30 DOF) |
| Liver abscesses | -- | -- | Chlortetracycline Tylosin | Low-dose CTC starts at 42 DOF; switch to TYL 2 28 days before end of feeding period | |
| Foot rot | Detected in 10% of animals in same pen (cumulative) | -- | Chlortetracycline | 7-day course of high-dose CTC | |
| Treatment (injectable at calf level) | Arthritis | Lightweight at time of detection (<1000 lbs) | First case/pull | Oxytetracycline | 3 doses (single dose every 3 days) |
| First relapse (second case/pull) | Trimethoprim–sulfadoxine | 5 doses (single dose per day × 5 days) | |||
| Second relapse (third case/pull) | Ceftiofur CFA 3 | 2 doses (single dose every 4 days) | |||
| Heavyweight at time of detection (>1000 lbs); and selected for treatment (50%) | First or subsequent case/pull | Ceftiofur HCl 3 | Single dose | ||
| Foot rot | Lightweight at time of detection (<1200 lbs) | First or subsequent case/pull | Penicillin G | Single dose | |
| Heavyweight at time of detection (>1200 lbs) | First or subsequent case/pull | Ceftiofur HCl | Single dose |
| Parameter | Condition | Value in Baseline Model | Source or Rationale, If Applicable |
|---|---|---|---|
| Cattle agent parameters | |||
| Average daily gain (ADG) for healthy steer calves | Applies to steers with no BRD or arthritis history that were lighter-weight on arrival | Selected from normal distribution with µ = 3.50 and σ = 0.44 pounds/day | Empirical data from 7685 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with first case of BRD | Applies for remainder of feeding period to animals with single diagnosis (i.e., first case) of BRD | 0.0453 pounds/day (relative to healthy animals) | Empirical data from 1630 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with first relapse of BRD | Applies for remainder of feeding period to animals with first relapse (i.e., second case) of BRD | 0.1759 pounds/day (relative to healthy animals) | Empirical data from 366 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with second relapse of BRD | Applies for remainder of feeding period to animals with second or more relapses (i.e., third or subsequent cases) of BRD | 0.3407 pounds/day (relative to healthy animals) | Empirical data from 162 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with first case of arthritis | Applies for remainder of feeding period to animals with single diagnosis (i.e., first case) of arthritis | 0.2538 pounds/day (relative to healthy animals) | Empirical data from 131 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with first relapse of arthritis | Applies for remainder of feeding period to animals with first relapse (i.e., second case) of arthritis | 0.4625 pounds/day (relative to healthy animals) | Empirical data from 32 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Absolute decrease in ADG for animals with second or third relapse of arthritis | Applies for remainder of feeding period to animals with second or more relapses (i.e., third or subsequent cases) of arthritis | 0.8778 pounds/day (relative to healthy animals) | Empirical data from 3 steer calves with arrival weights ranging from 500–799 pounds (2019–2023) |
| Testing agent parameters | |||
| Number of animals sampled per single pen | Applies to all home pens in the feedlot (not hospital or chronic pens) | 20 * | Simulated data from [23] |
| Time delay for collection of single nasopharyngeal sample | Delay applies to both diagnostic test types (AST and metagenomic sequencing) | 1 min | Observations from sample collection step for multi-year research [32] and CFAASP projects [37,47,48] |
| Time delay for transport of nasopharyngeal sample to diagnostic laboratory | Delay applies to both diagnostic test types (AST and metagenomic sequencing) | 36 h | Observations from streamlined sample transport step for multi-year CFAASP project [37,47,48] |
| Time delay for nasopharyngeal sample processing and diagnostic testing | Delay applies to both diagnostic test types (AST and metagenomic sequencing) | 72 h | Observations from sample processing for multi-year research [31,32] and CFAASP [37,47,48] projects |
| Time delay for reporting of diagnostic result to feedlot veterinarian | Delay applies to both diagnostic test types (AST and metagenomic sequencing) | 0 min (instantaneous) | Model parsimony |
| Threshold at which test result for pen-level AMR triggers a change in BRD treatment | Applies to all antimicrobial drug classes examined in the model | 25% * | Empirical data reported in [27], expert opinion [42] |
| Probability of Resistance at Arrival (CI) | |||
|---|---|---|---|
| Antimicrobial Class | Reference Drug 1 | (a) Baseline Scenarios 2 | (b) High (Worst-Case) Scenarios 3 |
| Cephalosporins | Ceftiofur | 0% (0%, 100%) | 5.1% (0%, 5.1%) |
| Fluoroquinolones | Enrofloxacin | 0.4% (0.2%, 0.9%) | 5.1% (0.2%, 5.1%) |
| 15-membered ring macrolides | Tulathromycin | 2.4% (1.8%, 3.3%) | 9.3% (1.8%, 9.3%) |
| 16-membered ring macrolides | Tilmicosin | 4.3% (3.4%, 5.4%) | 5.1% (3.4%, 5.1%) |
| Potentiated sulfonamides | Trimethoprim | 0.3% (0.1%, 0.6%) | 5.1% (0.1%, 5.1%) |
| Sulfadimethoxine | 4.3% (3.5%, 5.3%) | 74.8% (3.5%, 74.8%) | |
| Phenicols | Florfenicol | 0.1% (0.03%, 0.5%) | 5.1% (0.03%, 5.1%) |
| Tetracyclines | Oxytetracycline | 4.9% (4.1%, 5.7%) | 9.3% (4.1%, 9.3%) |
| Antimicrobial Category 1 | Antimicrobial Class | Reference Drug | Benchmark Percentage at 50 and 70 DOF 2 | Benchmark Percentage at 170 DOF 3 |
|---|---|---|---|---|
| Category I | Cephalosporins | Ceftiofur | 0% | 0% |
| Fluoroquinolones | Enrofloxacin | 0% | 0% | |
| Category II | 15-membered ring macrolides | Tulathromycin | 39.5% | 5% |
| 16-membered ring macrolides | Tilmicosin | 37.5% | 5% | |
| Potentiated sulfonamides | Trimethoprim | 1.6% | 5% | |
| Sulfadimethoxine | 51.1% | 5% | ||
| Category III | Phenicols | Florfenicol | 0% | 10% |
| Tetracyclines | Oxytetracycline | 34.3% | 10% |
| Test at | Diagnostic Paradigm | Scenario | Change in Median Number Drug Uses | Median Number BRD 1 | Median Number Cattle at End 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First Relapse | Second Relapse | Chronic Cases | Deaths | Healthy (Target Weight) | Chronic (Target Weight) | Chronic (Reduced Weight) | Euthanize | ||||
| Control (test-only) | -- | 492 | 267 | 141 | 70 | 9236 | 45 | 46 | 47 | ||
| 0 DOF | Phenotype | Perfect test | 0 | 492 | 267 | 141 | 70 | 9236 | 45 | 46 | 47 |
| Empirical AST (high estimate) | 0 | 492 | 267 | 141 | 70 | 9236 | 45 | 46 | 47 | ||
| Empirical AST (low estimate) | 0 | 493 | 267 | 140 | 70 | 9236 | 45 | 46 | 47 | ||
| Genotype | Empirical MS (high estimate) | 0 | 493 | 267 | 141 | 70 | 9235 | 45 | 46 | 48 | |
| Empirical MS (low estimate) | 0 | 493 | 267 | 141 | 70 | 9236 | 44 | 46 | 48 | ||
| 13 DOF | Phenotype | Perfect test | −322 | 304 | 132 | 51 | 72 | 9320 | 17 | 17 | 18 |
| Empirical AST (high estimate) | −322 | 305 | 133 | 51 | 72 | 9320 | 17 | 17 | 18 | ||
| Empirical AST (low estimate) | −319 | 308 | 135 | 53 | 73 | 9318 | 17 | 18 | 18 | ||
| Genotype | Empirical MS (high estimate) | −313 | 312 | 138 | 54 | 72 | 9317 | 18 | 18 | 19 | |
| Empirical MS (low estimate) | −288 | 325 | 148 | 61 | 72 | 9310 | 20 | 21 | 21 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramsay, D.E.; McDonald, W.; Gow, S.P.; McLeod, L.; Otto, S.J.G.; Osgood, N.D.; Waldner, C.L. The Potential for Sample Testing at the Pen Level to Inform Prudent Antimicrobial Selection for Bovine Respiratory Disease Treatment: Investigations Using a Feedlot Simulation Tool. Antibiotics 2025, 14, 1009. https://doi.org/10.3390/antibiotics14101009
Ramsay DE, McDonald W, Gow SP, McLeod L, Otto SJG, Osgood ND, Waldner CL. The Potential for Sample Testing at the Pen Level to Inform Prudent Antimicrobial Selection for Bovine Respiratory Disease Treatment: Investigations Using a Feedlot Simulation Tool. Antibiotics. 2025; 14(10):1009. https://doi.org/10.3390/antibiotics14101009
Chicago/Turabian StyleRamsay, Dana E., Wade McDonald, Sheryl P. Gow, Lianne McLeod, Simon J. G. Otto, Nathaniel D. Osgood, and Cheryl L. Waldner. 2025. "The Potential for Sample Testing at the Pen Level to Inform Prudent Antimicrobial Selection for Bovine Respiratory Disease Treatment: Investigations Using a Feedlot Simulation Tool" Antibiotics 14, no. 10: 1009. https://doi.org/10.3390/antibiotics14101009
APA StyleRamsay, D. E., McDonald, W., Gow, S. P., McLeod, L., Otto, S. J. G., Osgood, N. D., & Waldner, C. L. (2025). The Potential for Sample Testing at the Pen Level to Inform Prudent Antimicrobial Selection for Bovine Respiratory Disease Treatment: Investigations Using a Feedlot Simulation Tool. Antibiotics, 14(10), 1009. https://doi.org/10.3390/antibiotics14101009






