In Silico Identification and Molecular Characterization of Lentilactobacillus hilgardii Antimicrobial Peptides with Activity Against Carbapenem-Resistant Acinetobacter baumannii
Abstract
1. Introduction
2. Results
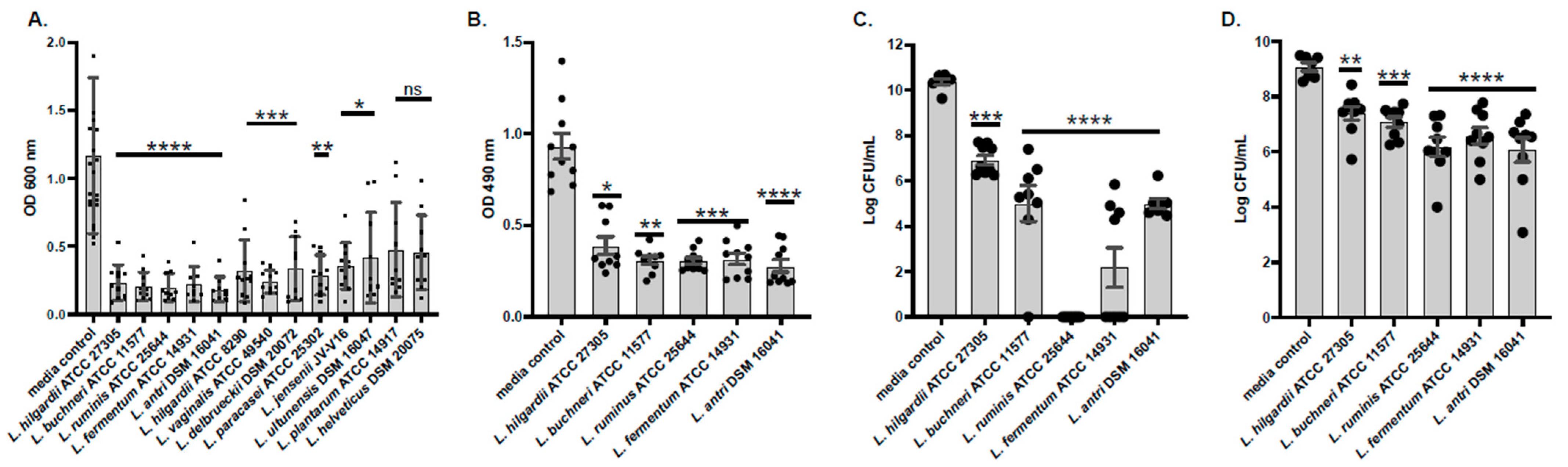
2.1. Conditioned Media Prevents Formation of and Disrupts Pre-Formed A. baumannii Biofilms
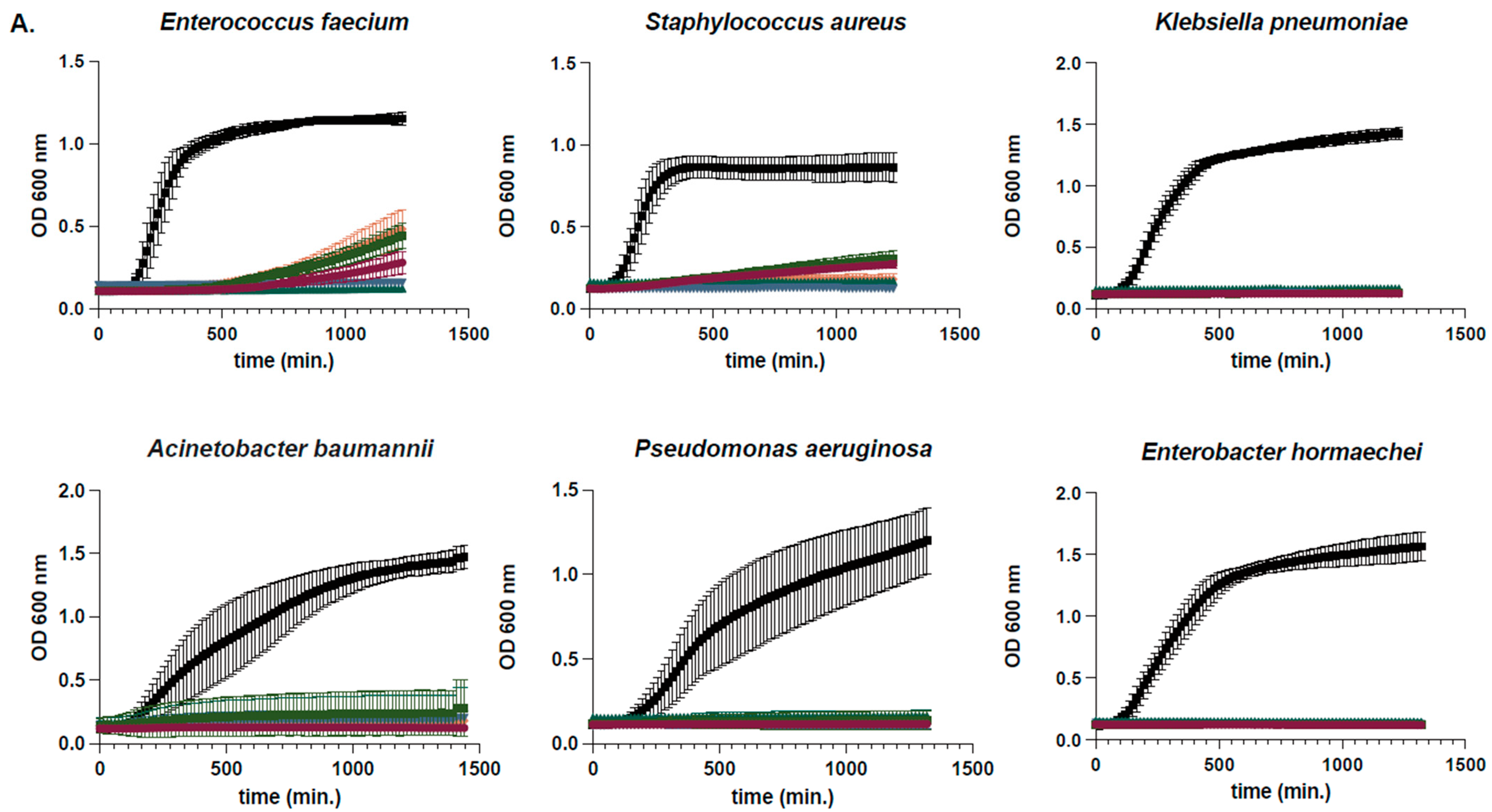
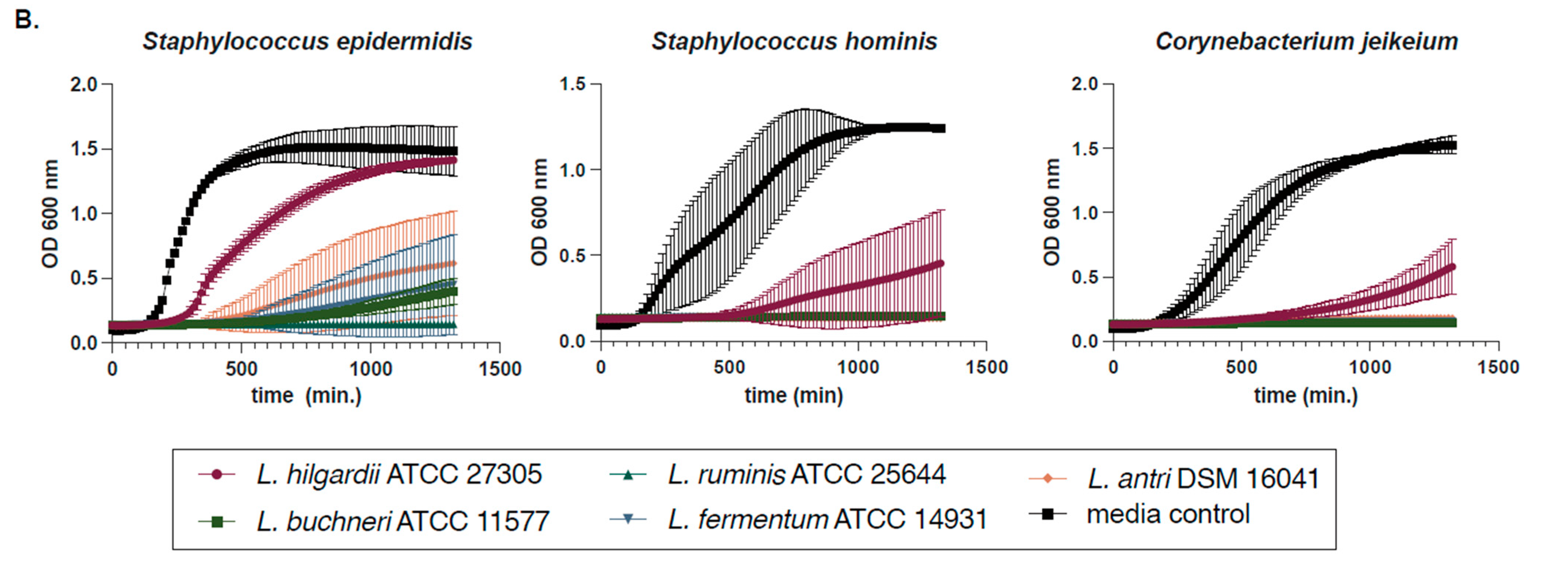
2.2. Determination of Susceptibility Range Within ESKAPEs and Known Skin Commensals
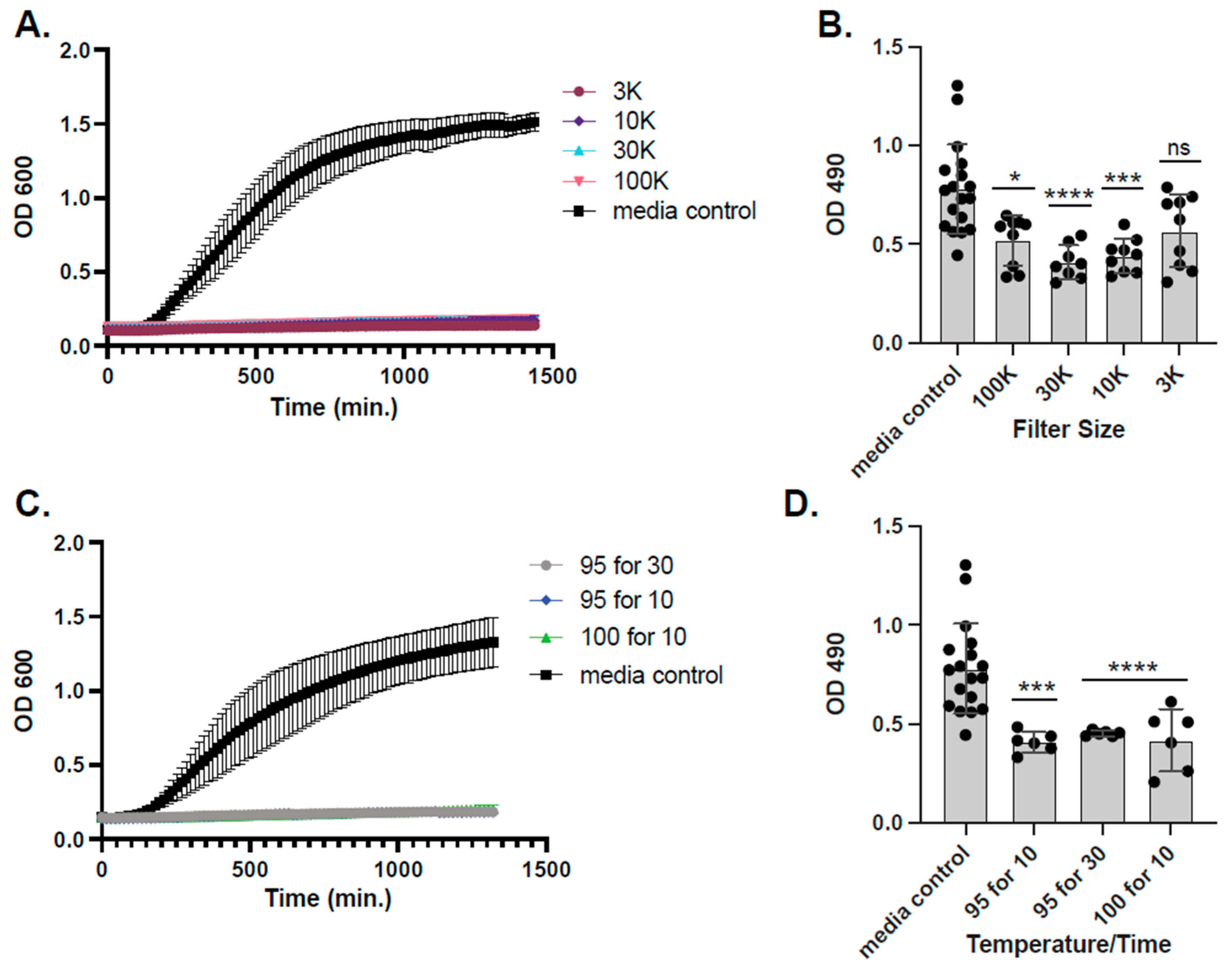
2.3. L. hilgardii Antimicrobial Activity, Size Fractionation, and Thermostability
2.4. Bioinformatic Prediction of L. hilgardii Antimicrobial Peptides
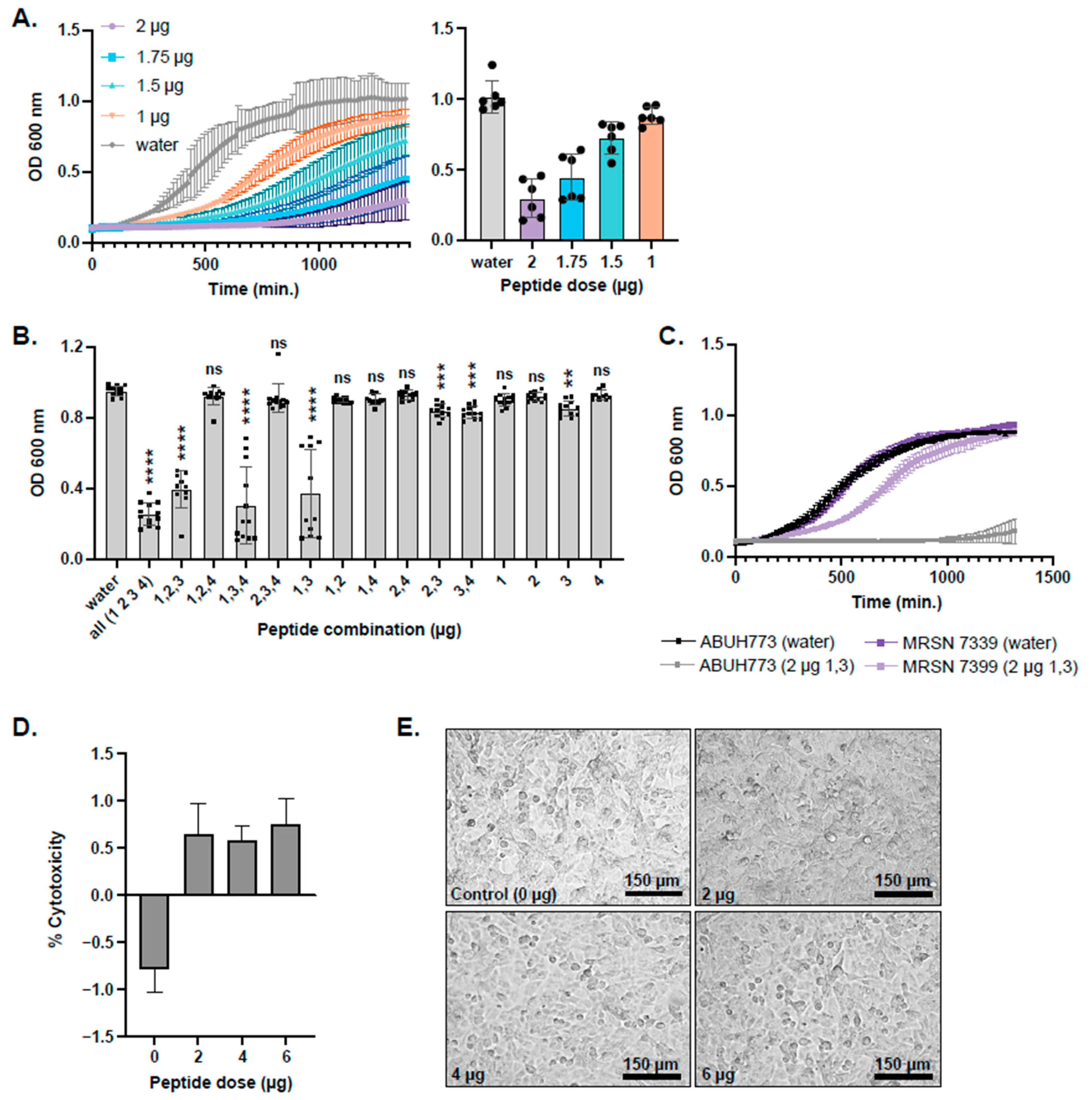
2.5. Synthesis and Testing of Predicted L. hilgardii Peptides on A. baumannii Planktonic Cultures and Epithelial Cells
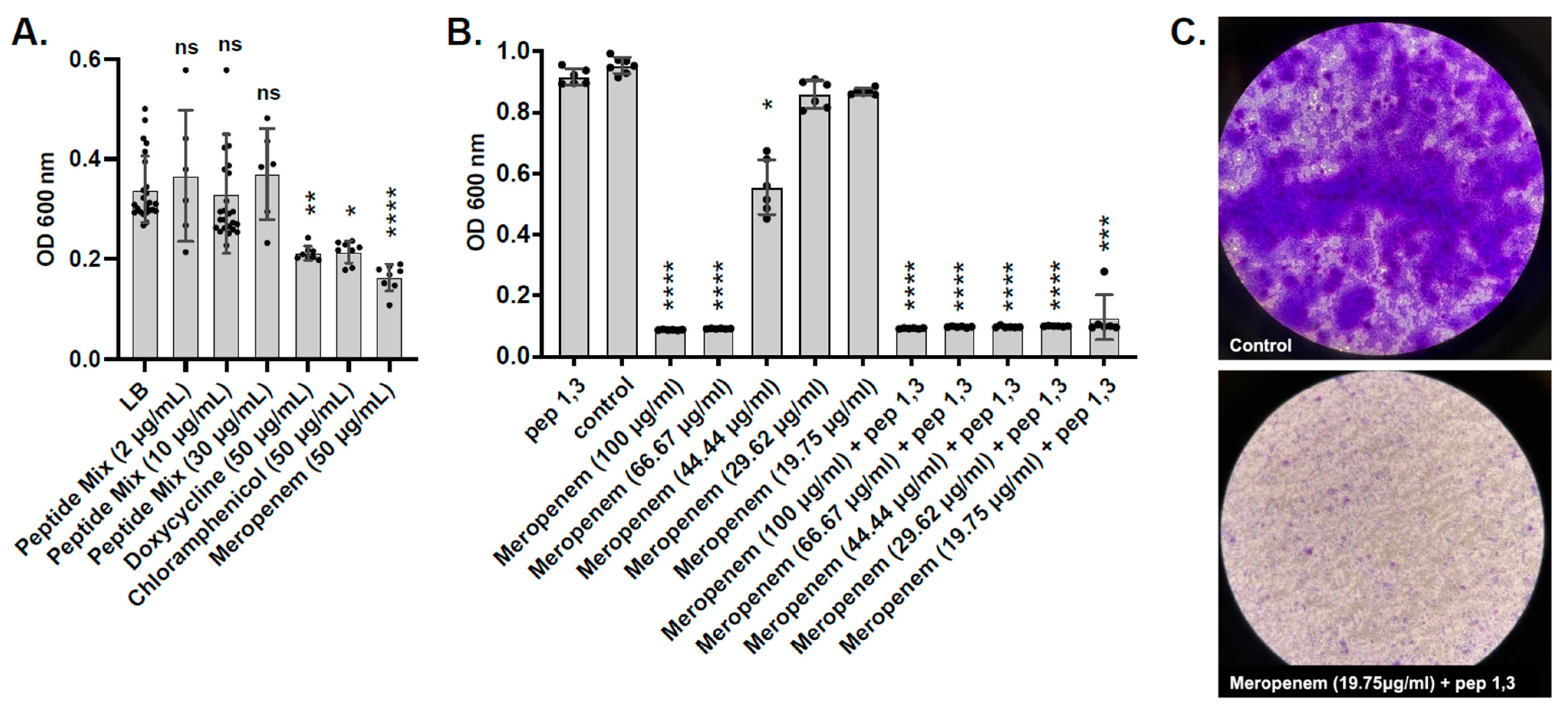
2.6. Testing L. hilgardii Peptides on ESKAPE Pathogens
2.7. Testing of Predicted L. hilgardii Peptides on A. baumannii Pre-Formed Biofilms
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culturing Conditions
4.2. Planktonic Growth Assays
4.3. Biofilm Assays
4.4. Planktonic and Biofilm Viability
4.5. Fractionation Assays
4.6. Bioinformatics
4.7. Inhibition of Planktonic Growth by Chemically Synthesized Peptides
4.8. Synergy Between Peptides and Antibiotics
4.9. Cytotoxicity Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| aa | Amino acid |
| AMPs | Antimicrobial peptides |
| AMR | Antimicrobial resistance |
| ATCC | American Type Culture Collection |
| BEI | Biodefense and Emerging Infections research |
| BHI | Bovine heart infusion |
| BLASTP | Basic Local Alignment Search Tool Protein |
| CAMPdb | Comprehensive antimicrobial peptide database |
| CDC | Centers for Disease Control and Prevention |
| CDSs | Coding sequences |
| CRAb | Carbapenem-resistant A. baumannii |
| DMEM | Dubecco’s Modified Eagle Medium |
| DNA | Deoxyribonucleic acid |
| DOE | Double omission experiment |
| EPS | Extracellular polymeric substance |
| ESKAPE | E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp. |
| GC1 | Global clone 1 |
| GC2 | Global clone 2 |
| HAI | Healthcare-associated infections |
| HMM | Hidden Markov model |
| HMP | Human microbiome project |
| HMP ID | Human microbiome project identifier |
| Kbp | Kilobase pair |
| Kda | Kilodalton |
| LAB | Lactic acid bacteria |
| LDH | Lactate Dehydrogenase |
| LLC | Limited liability corporation |
| MIC | Minimum inhibitory concentration |
| min | Minute |
| mL | Milliliter |
| MRS | DeMan Rogosa Sharpe |
| nm | Nanometer |
| PIA | Polysaccharide intercellular adhesin |
| SOE | Single omission experiment |
| spp. | Species |
| TOE | Triple omission experiment |
| μL | Microliter |
References
- Sikora, A.; Zahra, F. Nosocomial Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Marturano, J.E.; Lowery, T.J. ESKAPE Pathogens in Bloodstream Infections Are Associated with Higher Cost and Mortality but Can Be Predicted Using Diagnoses upon Admission. Open Forum Infect. Dis. 2019, 6, ofz503. [Google Scholar] [CrossRef]
- Seid, M.; Bayou, B.; Aklilu, A.; Tadesse, D.; Manilal, A.; Zakir, A.; Kulyta, K.; Kebede, T.; Alodaini, H.A.; Idhayadhulla, A. Antimicrobial Resistance Patterns of WHO Priority Pathogens at General Hospital in Southern Ethiopia during the COVID-19 Pandemic, with Particular Reference to ESKAPE-Group Isolates of Surgical Site Infections. BMC Microbiol. 2025, 25, 84. [Google Scholar] [CrossRef]
- Bereanu, A.-S.; Bereanu, R.; Mohor, C.; Vintilă, B.I.; Codru, I.R.; Olteanu, C.; Sava, M. Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics 2024, 13, 400. [Google Scholar] [CrossRef]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An Emerging Opportunistic Pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef]
- Bergogne-Bérézin, E.; Towner, K.J. Acinetobacter spp. as Nosocomial Pathogens: Microbiological, Clinical, and Epidemiological Features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Bergogne-Bérézin, E.; Decré, D.; Joly-Guillou, M.L. Opportunistic Nosocomial Multiply Resistant Bacterial Infections--Their Treatment and Prevention. J. Antimicrob. Chemother. 1993, 32 (Suppl. A), 39–47. [Google Scholar] [CrossRef]
- Smolyakov, R.; Borer, A.; Riesenberg, K.; Schlaeffer, F.; Alkan, M.; Porath, A.; Rimar, D.; Almog, Y.; Gilad, J. Nosocomial Multi-Drug Resistant Acinetobacter baumannii Bloodstream Infection: Risk Factors and Outcome with Ampicillin-Sulbactam Treatment. J. Hosp. Infect. 2003, 54, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Chowdhury, G.; Mukhopadhyay, A.K.; Dutta, S.; Basu, S. Convergence of Biofilm Formation and Antibiotic Resistance in Infection. Front. Med. 2022, 9, 793615. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef]
- Mokoena, M.P.; Omatola, C.A.; Olaniran, A.O. Applications of Lactic Acid Bacteria and Their Bacteriocins against Food Spoilage Microorganisms and Foodborne Pathogens. Molecules 2021, 26, 7055. [Google Scholar] [CrossRef]
- Thuy, T.T.D.; Lu, H.-F.; Bregente, C.J.B.; Huang, F.-C.A.; Tu, P.-C.; Kao, C.-Y. Characterization of the Broad-Spectrum Antibacterial Activity of Bacteriocin-like Inhibitory Substance-Producing Probiotics Isolated from Fermented Foods. BMC Microbiol. 2024, 24, 85. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Nayab, H.; Rehman, T.U.; Williamson, M.P.; Haq, K.U.; Shafi, N.; Shafique, F. Characterisation of Bacteriocins Produced by Spp. Isolated from the Traditional Pakistani Yoghurt and Their Antimicrobial Activity against Common Foodborne Pathogens. Biomed. Res. Int. 2020, 2020, 8281623. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Fu, J.; Xu, B.; Wang, Y.; Jin, M. Interplay between Gut Microbiota and Antimicrobial Peptides. Anim. Nutr. 2020, 6, 389–396. [Google Scholar] [CrossRef]
- Rangel, K.; Lechuga, G.C.; Provance, D.W., Jr.; Morel, C.M.; De Simone, S.G. An Update on the Therapeutic Potential of Antimicrobial Peptides against Infections. Pharmaceuticals 2023, 16, 1281. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, J.; Kim, G.-Y. Synergistic Effects of a Probiotic Culture Extract and Antimicrobial Combinations against Multidrug-Resistant. Medicina 2023, 59, 947. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamiri, M.M.; Wang, J.; Zhang, S.; Li, P.; Odhiambo, W.O.; Chen, Y.; Han, B.; Yang, E.; Xun, M.; Han, L.; et al. Probiotic Species and Their Biosurfactants Eliminate Acinetobacter baumannii Biofilm in Various Manners. Microbiol. Spectr. 2023, 11, e0461422. [Google Scholar] [CrossRef]
- Rodriguez, C.; Ramlaoui, D.; Georgeos, N.; Gasca, B.; Leal, C.; Subils, T.; Tuttobene, M.R.; Sieira, R.; Salzameda, N.T.; Bonomo, R.A.; et al. Phenotypic and Transcriptional Analysis of the Antimicrobial Effect of Lactic Acid Bacteria on Carbapenem-Resistant Acinetobacter baumannii: Lacticaseibacillus rhamnosus CRL 2244 an Alternative Strategy to Overcome Resistance? Res. Sq. 2023; preprint. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Novel Bacteriocins from Lactic Acid Bacteria (LAB): Various Structures and Applications. Microb. Cell Fact. 2014, 13 (Suppl. 1), S3. [Google Scholar] [CrossRef]
- Mondal, R.K.; Sen, D.; Arya, A.; Samanta, S.K. Developing Anti-Microbial Peptide Database Version 1 to Provide Comprehensive and Exhaustive Resource of Manually Curated AMPs. Sci. Rep. 2023, 13, 17843. [Google Scholar] [CrossRef]
- Scarborough, M.J.; Lawson, C.E.; Hamilton, J.J.; Donohue, T.J.; Noguera, D.R. Metatranscriptomic and Thermodynamic Insights into Medium-Chain Fatty Acid Production Using an Anaerobic Microbiome. mSystems 2018, 3, e00221-18. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.; Ruiz-Barba, J.L.; Jiménez-Díaz, R. Purification and Genetic Characterization of Plantaricin NC8, a Novel Coculture-Inducible Two-Peptide Bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 2003, 69, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, Y.; Li, L.; Jiang, X.; Chen, Z.; Zhao, F.; Yi, Y. Biosynthesis and Production of Class II Bacteriocins of Food-Associated Lactic Acid Bacteria. Fermentation 2022, 8, 217. [Google Scholar] [CrossRef]
- Yoon, B.-J. Hidden Markov Models and Their Applications in Biological Sequence Analysis. Curr. Genom. 2009, 10, 402–415. [Google Scholar] [CrossRef]
- Hartzell, J.D.; Kim, A.S.; Kortepeter, M.G.; Moran, K.A. Acinetobacter Pneumonia: A Review. MedGenMed 2007, 9, 4. [Google Scholar] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087-93. [Google Scholar] [CrossRef]
- Gawde, U.; Chakraborty, S.; Waghu, F.H.; Barai, R.S.; Khanderkar, A.; Indraguru, R.; Shirsat, T.; Idicula-Thomas, S. CAMPR4: A Database of Natural and Synthetic Antimicrobial Peptides. Nucleic Acids Res. 2023, 51, D377–D383. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An Automated Web Server for Prediction of Protein Antimicrobial Regions. Bioinformatics 2011, 28, 130–131. [Google Scholar] [CrossRef]
- Lata, S.; Mishra, N.K.; Raghava, G.P.S. AntiBP2: Improved Version of Antibacterial Peptide Prediction. BMC Bioinform. 2010, 11 (Suppl. 1), S19. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Lin, T.-T.; Yang, L.-Y.; Lu, I.-H.; Cheng, W.-C.; Hsu, Z.-R.; Chen, S.-H.; Lin, C.-Y. AI4AMP: An Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning. mSystems 2021, 6, e0029921. [Google Scholar] [CrossRef]
- Xing, W.; Zhang, J.; Li, C.; Huo, Y.; Dong, G. IAMP-Attenpred: A Novel Antimicrobial Peptide Predictor Based on BERT Feature Extraction Method and CNN-BiLSTM-Attention Combination Model. Brief. Bioinform. 2023, 25, bbad443. [Google Scholar] [CrossRef]
- Zhao, F.; Qiu, J.; Xiang, D.; Jiao, P.; Cao, Y.; Xu, Q.; Qiao, D.; Xu, H.; Cao, Y. DeepAMPNet: A Novel Antimicrobial Peptide Predictor Employing AlphaFold2 Predicted Structures and a Bi-Directional Long Short-Term Memory Protein Language Model. PeerJ 2024, 12, e17729. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ren, X.; Luo, X.; Wang, Z.; Li, Z.; Luo, X.; Shen, J.; Li, Y.; Yuan, D.; Nussinov, R.; et al. A Foundation Model Identifies Broad-Spectrum Antimicrobial Peptides against Drug-Resistant Bacterial Infection. Nat. Commun. 2024, 15, 7538. [Google Scholar] [CrossRef] [PubMed]
- Aronica, P.G.A.; Reid, L.M.; Desai, N.; Li, J.; Fox, S.J.; Yadahalli, S.; Essex, J.W.; Verma, C.S. Computational Methods and Tools in Antimicrobial Peptide Research. J. Chem. Inf. Model. 2021, 61, 3172–3196. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60. [Google Scholar] [CrossRef]
- Ekblad, B.; Kyriakou, P.K.; Oppegård, C.; Nissen-Meyer, J.; Kaznessis, Y.N.; Kristiansen, P.E. Structure-Function Analysis of the Two-Peptide Bacteriocin Plantaricin EF. Biochemistry 2016, 55, 5106–5116. [Google Scholar] [CrossRef] [PubMed]
- Teese, M.G.; Langosch, D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry 2015, 54, 5125–5135. [Google Scholar] [CrossRef]
- Otto, M. Bacterial Evasion of Antimicrobial Peptides by Biofilm Formation. Curr. Top. Microbiol. Immunol. 2006, 306, 251–258. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial Peptides and Their Interaction with Biofilms of Medically Relevant Bacteria. Biochim. Biophys. Acta 2015, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Yang, S.-J.; Bayer, A.S.; Mishra, N.N.; Meehl, M.; Ledala, N.; Yeaman, M.R.; Xiong, Y.Q.; Cheung, A.L. The Staphylococcus aureus Two-Component Regulatory System, GraRS, Senses and Confers Resistance to Selected Cationic Antimicrobial Peptides. Infect. Immun. 2012, 80, 74–81. [Google Scholar] [CrossRef]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for Combating Bacterial Biofilms: A Focus on Anti-Biofilm Agents and Their Mechanisms of Action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef]
- Adams, M.D.; Wright, M.S.; Karichu, J.K.; Venepally, P.; Fouts, D.E.; Chan, A.P.; Richter, S.S.; Jacobs, M.R.; Bonomo, R.A. Rapid Replacement of Acinetobacter baumannii Strains Accompanied by Changes in Lipooligosaccharide Loci and Resistance Gene Repertoire. mBio 2019, 10, e00356-19. [Google Scholar] [CrossRef]
- Chan, A.P.; Choi, Y.; Clarke, T.H.; Brinkac, L.M.; White, R.C.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D.; Fouts, D.E. AbGRI4, a Novel Antibiotic Resistance Island in Multiply Antibiotic-Resistant Acinetobacter baumannii Clinical Isolates. J. Antimicrob. Chemother. 2020, 75, 2760–2768. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.P.; Sutton, G.; DePew, J.; Krishnakumar, R.; Choi, Y.; Huang, X.-Z.; Beck, E.; Harkins, D.M.; Kim, M.; Lesho, E.P.; et al. A Novel Method of Consensus Pan-Chromosome Assembly and Large-Scale Comparative Analysis Reveal the Highly Flexible Pan-Genome of Acinetobacter baumannii. Genome Biol. 2015, 16, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Jumpstart Reference Strains Consortium; Nelson, K.E.; Weinstock, G.M.; Highlander, S.K.; Worley, K.C.; Creasy, H.H.; Wortman, J.R.; Rusch, D.B.; Mitreva, M.; Sodergren, E.; et al. A Catalog of Reference Genomes from the Human Microbiome. Science 2010, 328, 994–999. [Google Scholar] [CrossRef]
- Endimiani, A.; Carias, L.L.; Hujer, A.M.; Bethel, C.R.; Hujer, K.M.; Perez, F.; Hutton, R.A.; Fox, W.R.; Hall, G.S.; Jacobs, M.R.; et al. Presence of Plasmid-Mediated Quinolone Resistance in Klebsiella pneumoniae Isolates Possessing blaKPC in the United States. Antimicrob. Agents Chemother. 2008, 52, 2680–2682. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Yasmin, M.; Fouts, D.E.; Jacobs, M.R.; Haydar, H.; Marshall, S.H.; White, R.; D’Souza, R.; Lodise, T.P.; Rhoads, D.D.; Hujer, A.M.; et al. Monitoring Ceftazidime-Avibactam and Aztreonam Concentrations in the Treatment of a Bloodstream Infection Caused by a Multidrug-Resistant Enterobacter sp. Carrying Both Klebsiella pneumoniae Carbapenemase-4 and New Delhi Metallo-β-Lactamase-1. Clin. Infect. Dis. 2020, 71, 1095–1098. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Ben Hamida, J.; Fliss, I. BACTIBASE: A New Web-Accessible Database for Bacteriocin Characterization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Zouhir, A.; Hammami, R.; Fliss, I.; Hamida, J.B. A New Structure-Based Classification of Gram-Positive Bacteriocins. Protein J. 2010, 29, 432–439. [Google Scholar] [CrossRef]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An Enhanced Comprehensive Data Repository of Antimicrobial Peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef]
- Jhong, J.-H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.-R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. DbAMP 2.0: Updated Resource for Antimicrobial Peptides with an Enhanced Scanning Method for Genomic and Proteomic Data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Armougom, F.; Wallace, I.M.; Higgins, D.G.; Jongeneel, C.V.; Notredame, C. The M-Coffee Web Server: A Meta-Method for Computing Multiple Sequence Alignments by Combining Alternative Alignment Methods. Nucleic Acids Res. 2007, 35, W645-8. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Llorente, C.; Lang, S.; Brandl, K.; Chu, H.; Jiang, L.; White, R.C.; Clarke, T.H.; Nguyen, K.; Torralba, M.; et al. Bacteriophage Targeting of Gut Bacterium Attenuates Alcoholic Liver Disease. Nature 2019, 575, 505–511. [Google Scholar] [CrossRef] [PubMed]

| Locus_id | Peptide | Amino Acid Sequence of Prepeptides * | Size (aa), (KDa), pI of Prepeptide | Size (aa), (Kda), pI of Mature Peptide | Charge, Hydrophobicity of Mature Peptide | Best BLASTP Match | BLASTP (id, E-Val) | Full Seq HMM (E-Val, Score) | Domain HMM (E-Val, Score) |
|---|---|---|---|---|---|---|---|---|---|
| |-----------leader--------GG|▼|---------mature peptide--------| | |||||||||
| HMPREF0496_RS14880 | 1 | MFNQEKENMSQRYEELSADELSHISGG VTRYRHHEKKSWIDDFMKGFKKTFC | 52, 6.3, 7.4 | 25, 3.2, 10.2 | 6, 8.7 | AMPDB_45803 | 100%, 3 × 10−36 | 1.5 × 10−4, 19.6 | 3 × 10−4, 18.6 |
| HMPREF0496_RS15215 | 2 | MSRNNLTILSTHKLVSVIGG QTFPIPNKPFGDRYPITIQPIIRNAYSF | 48, 5.4, 10.8 | 28, 3.3, 10.1 | 2, 10.1 | No match | N.A. | 3.8 × 10−2, 12 | 4.8 × 10−2, 11.7 |
| HMPREF0496_RS02405 | 3 | MNKLSKFSKVTDKDLSRINGG GVWWTVITTIGKVGYSAYKDRNDIKSGFNKGFKKP | 56, 6.3, 10.6 | 35, 4.0, 10.4 | 5, 8.4 | BAC103 (Plantaricin NC8β) | 49%, 2 × 10−10 | 2.5 × 10−4, 18.9 | 4.5 × 10−4, 18.1 |
| HMPREF0496_RS02410 | 4 | MKNIKVVKDLDLKAVTGG DWASPFWNSWGYTQGKKATWNLKHPFVRF | 47, 5.5, 10.5 | 29, 3.6, 10.5 | 4, 10.6 | BAC089 (Plantaricin NC8α) | 47%, 1 × 10−13 | 1.6 × 10−3, 16.4 | 2.1 × 10−3, 16.0 |
| HMPREF0496_RS15205 | 5 | MKDNFKNLNSYKKLDVNSLNLIEGG NSVASQVSDIFSRFKRAFSGSFVYKVSGRNQF | 57, 6.5, 9.4 | 32, 3.6, 11.1 | 4, 7.8 | No match | N.A. | 6.3 × 10−4, 17.6 | 1.2 × 10−3, 16.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Appel, A.; Velazco, L.; Alreja, A.B.; LeClair, K.; Duggal, A.P.; Vashee, I.; Taal, A.M.; Gonzalez-Juarbe, N.; Fouts, D.E. In Silico Identification and Molecular Characterization of Lentilactobacillus hilgardii Antimicrobial Peptides with Activity Against Carbapenem-Resistant Acinetobacter baumannii. Antibiotics 2025, 14, 1004. https://doi.org/10.3390/antibiotics14101004
Appel A, Velazco L, Alreja AB, LeClair K, Duggal AP, Vashee I, Taal AM, Gonzalez-Juarbe N, Fouts DE. In Silico Identification and Molecular Characterization of Lentilactobacillus hilgardii Antimicrobial Peptides with Activity Against Carbapenem-Resistant Acinetobacter baumannii. Antibiotics. 2025; 14(10):1004. https://doi.org/10.3390/antibiotics14101004
Chicago/Turabian StyleAppel, Amanda, Lily Velazco, Adit B. Alreja, Kara LeClair, Aryaan P. Duggal, Isha Vashee, Aji Mary Taal, Norberto Gonzalez-Juarbe, and Derrick E. Fouts. 2025. "In Silico Identification and Molecular Characterization of Lentilactobacillus hilgardii Antimicrobial Peptides with Activity Against Carbapenem-Resistant Acinetobacter baumannii" Antibiotics 14, no. 10: 1004. https://doi.org/10.3390/antibiotics14101004
APA StyleAppel, A., Velazco, L., Alreja, A. B., LeClair, K., Duggal, A. P., Vashee, I., Taal, A. M., Gonzalez-Juarbe, N., & Fouts, D. E. (2025). In Silico Identification and Molecular Characterization of Lentilactobacillus hilgardii Antimicrobial Peptides with Activity Against Carbapenem-Resistant Acinetobacter baumannii. Antibiotics, 14(10), 1004. https://doi.org/10.3390/antibiotics14101004






