Honeydew Honey as a Reservoir of Bacteria with Antibacterial and Probiotic Properties
Abstract
1. Introduction
2. Results
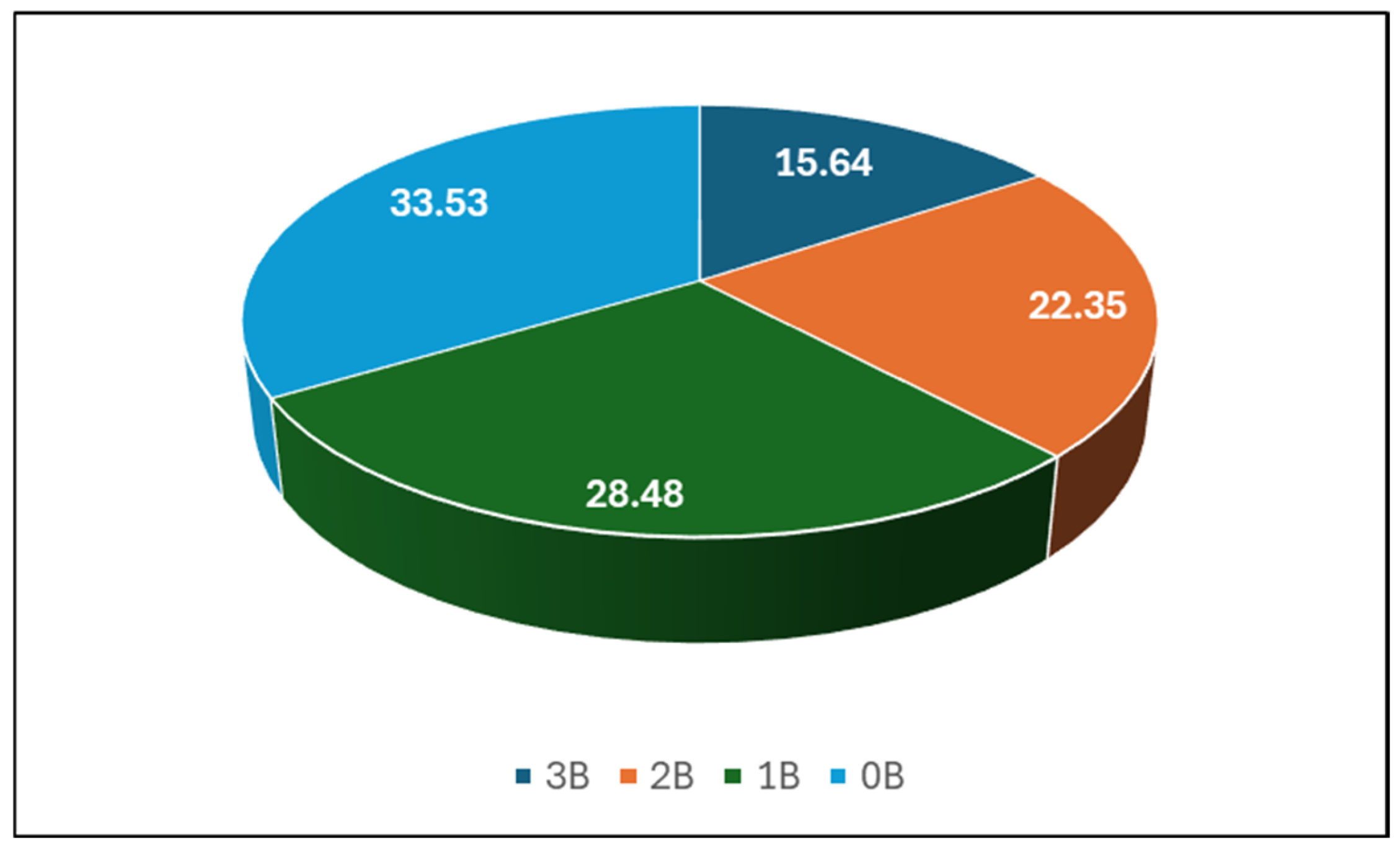
2.1. Total Content of Bacteria
2.2. Antibacterial Potential of Bacteria Isolated from Honey
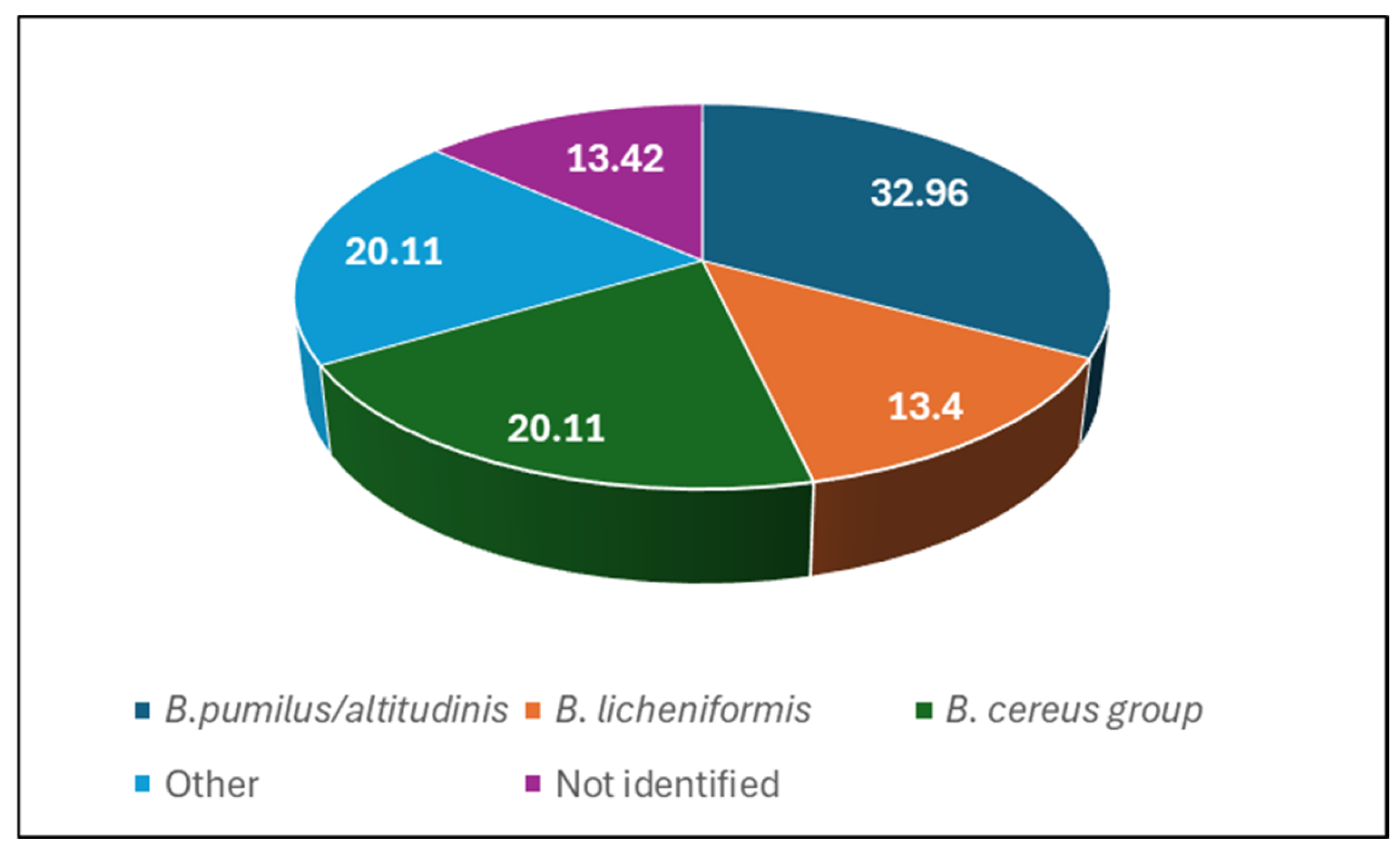
2.3. The Identification of Bacteria Isolated from Honeydew Honey
2.4. Probiotic Properties
2.4.1. Resistance to Bile Salts and Low pH
2.4.2. Antibiotic Sensitivity Test
3. Discussion
4. Materials and Methods
4.1. Microbial Content of Honey
4.2. Antibacterial Potential of Bacteria Isolated from Honey
4.3. Identification of Bacteria by MALDI-TOF MS
4.4. Probiotic Properties
4.4.1. Resistance to Bile Salts and Low pH
4.4.2. Antibiotic Sensitivity Test
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohd Kamal, D.A.; Ibrahim, S.F.; Kamal, H.; Kashim, M.I.A.M.; Mokhtar, M.H. Physicochemical and medicinal properties of Tualang, Gelam and Kelulut honeys: A comprehensive review. Nutrients 2021, 13, 197. [Google Scholar] [CrossRef]
- Hallaj-Nezhadi, S.; Hamdipour, R.; Shahrvirani, M.; Zare Tin, R.; Chapeland-Leclerc, F.; Ruprich-Robert, G.; Esnaashari, S.; Elyasi Far, B.; Dilmaghani, A. Antimicrobial activity of Bacillus sp. isolated strains of wild honey. BMC Complement. Med. Ther. 2022, 22, 78. [Google Scholar] [CrossRef] [PubMed]
- Shahali, A.; Soltani, R.; Akbari, V. Probiotic Lactobacillus and the potential risk of spreading antibiotic resistance: A systematic review. Res. Pharm. Sci. 2023, 18, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.M.B.; Shahidi, F.; Mortazavi, S.A.; Milani, E.; Eshaghi, Z. Potentially Probiotic Lactobacillus Strains from Traditional Kurdish Cheese. Probiot. Antimicrob. Prot. 2014, 6, 22–31. [Google Scholar] [CrossRef]
- Payne, J.; Bellmer, D.; Jadeja, R.; Muriana, P. The Potential of Bacillus Species as Probiotics in the Food Industry: A Review. Foods 2024, 13, 2444. [Google Scholar] [CrossRef] [PubMed]
- Golnari, M.; Bahrami, N.; Milanian, Z.; Rabbani Koreshgani, M.; Ali Asadollahi, M.; Shafiei, R.; Fatemi, S.S.A. Isolation and characterization of novel Bacillus strains with superior probiotic potential: Comparative analysis and safety evaluation. Sci. Rep. 2024, 14, 1457. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Suarez, J.M.; Gasparrini, M.; Forbes-Hernández, T.Y.; Mazzoni, L.; Giampieri, F. The Composition and Biological Activity of Honey: A Focus on Manuka Honey. Foods 2014, 21, 420–432. [Google Scholar] [CrossRef]
- Grabek-Lejko, D.; Miłek, M.; Sidor, E.; Puchalski, C.; Dżugan, M. Antiviral and Antibacterial Effect of Honey Enriched with Rubus spp. as a Functional Food with Enhanced Antioxidant Properties. Molecules 2022, 27, 4859. [Google Scholar] [CrossRef]
- Grabek-Lejko, D.; Hyrchel, T. The Antibacterial Properties of Polish Honey against Streptococcus mutans—A Causative Agent of Dental Caries. Antibiotics 2023, 12, 1640. [Google Scholar] [CrossRef]
- Luca, L.; Pauliuc, D.; Oroian, M. Honey microbiota, methods for determining the microbiological composition and the antimicrobial effect of honey—A review. Food Chem. X 2024, 23, 101524. [Google Scholar] [CrossRef]
- Seraglio, T.S.K.; Silva, B.; Bergamo, G.; Brugnerotto, P.; Gonzaga, L.V.; Fett, R.; Oliveira Costa, A.C. An overview of physicochemical characteristics and health-promoting properties of honeydew honey. Food Res. Int. 2019, 119, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Dżugan, M.; Ciszkowicz, E.; Tomczyk, M.; Miłek, M.; Lecka-Szlachta, K. Coniferous Honeydew Honey: Antibacterial Activity and Anti-Migration Properties against Breast Cancer Cell Line (MCF-7). Appl. Sci. 2024, 14, 710. [Google Scholar] [CrossRef]
- European Commission Regulation (EU) No 710/2010 of 6 August 2010 on entering a name in the register of protected designations of origin and protected geographical indications (“Podkarpacki Miód Spadziowy” (PDO)). Off. J. Eur. Communities 2010, 208, 1–2.
- Pajor, M.; Worobo, R.W.; Milewski, S.; Szweda, P. The Antimicrobial Potential of Bacteria Isolated from Honey Samples Produced in the Apiaries Located in Pomeranian Voivodeship in Northern Poland. Int. J. Environ. Res. Public Health 2018, 15, 2002. [Google Scholar] [CrossRef]
- Lashani, E.; Davoodabadi, A.; Soltan Dallal, M.M. Some probiotic properties of Lactobacillus species isolated from honey and their antimicrobial activity against foodborne pathogens. Vet. Res. Forum. 2020, 11, 121–126. [Google Scholar] [CrossRef]
- Abadi, M.E.G.M.; Hosseini-Safa, A.; Habibi, S.; Dehghan, M.; Forouzani-Moghaddam, M.J.; Oshaghi, M. Isolation and characterization of the lactobacillus strain from honey and its probiotic properties. Iran. J. Microbiol. 2023, 15, 439–447. [Google Scholar] [CrossRef]
- Lee, N.K.; Kim, W.S.; Paik, H.D. Bacillus strains as human probiotics: Characterization, safety, microbiome, and probiotic carrier. Food Sci. Biotechnol. 2019, 28, 1297–1305. [Google Scholar] [CrossRef]
- FAO; WHO. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; Food and Nutrition Paper; World Health Organization: Geneva, Switzerland; Food Agriculture Organization of the United Nations: Rome, Italy, 2006. [Google Scholar]
- Daneshazari, R.; Rabbani Khorasgani, M.; Hosseini-Abari AKim, J.-H. Bacillus subtilis isolates from camel milk as probiotic candidates. Sci. Rep. 2023, 12, 3387. [Google Scholar] [CrossRef]
- Amoah, K.; Tan, B.; Zhang, S.; Chi, S.; Yang, Q.; Liu, H.; Yang, Y.; Zhang, H.; Dong, X. Host gut-derived Bacillus probiotics supplementation improves growth performance, serum and liver immunity, gut health, and resistive capacity against Vibrio harveyi infection in hybrid grouper (♀Epinephelus fuscoguttatus × ♂Epinephelus lanceolatus). Anim. Nutr. 2023, 14, 163–184. [Google Scholar] [CrossRef]
- Kim, B.J.; Hong, J.H.; Jeong, Y.S.; Jung, H.K. Evaluation of two Bacillus subtilis strains isolated from Korean fermented food as probiotics against loperamide-induced constipation in mice. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 797–806. [Google Scholar] [CrossRef]
- Duc, L.H.; Hong, H.A.; Barbosa, T.M.; Henriques, A.O.; Cutting, S.M. Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 2004, 70, 2161–2171. [Google Scholar] [CrossRef] [PubMed]
- Iurlina, M.O.; Fritz, R. Characterization of microorganisms in Argentinean honeys from different sources. Int. J. Food Microbiol. 2005, 105, 297–304. [Google Scholar] [CrossRef]
- Amin, Z.F.A.; Sabri, S.; Ismail, M.; Chan, K.W.; Ismail, N.; Mohd Esa, N.; Mohd Lila, M.A.; Zawawi, N. Probiotic Properties of Bacillus Strains Isolated from Stingless Bee (Heterotrigona itama) Honey Collected across Malaysia. Int. J. Environ. Res. Public Health 2020, 17, 278. [Google Scholar] [CrossRef]
- Bucekova, M.; Godocikova, J.; Kohutova, L.; Danchenko, M.; Barath, P.; Majtan, J. Antibacterial activity and bee-derived protein content of honey as important and suitable complementary tools for the assessment of honey quality. J. Food Compos. Anal. 2023, 123, 105610. [Google Scholar] [CrossRef]
- Snowdon, J.A.; Cliver, D.O. Microorganisms in honey. Int. J. Food Microbiol. 1996, 31, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, Z.; Khataybeh, B.; Al Ghzawi, A.M.; Ababneh, Q.; Nabusli, A.A. Molecular identification of major bacteria in honey and the effect of microwave treatment on its microbial quality and antibacterial activity. AIMS Agric. Food 2022, 7, 594–613. [Google Scholar] [CrossRef]
- Sowa, P.; Grabek-Lejko, D.; Wesołowska, M.; Swacha, S.; Dżugan, M. Hydrogen peroxide-dependent antibacterial action of Melilotus albus honey. Lett. Appl. Microbiol. 2017, 65, 82–89. [Google Scholar] [CrossRef]
- Tsadila, C.; Nikolaidis, M.; Dimitriou, T.G.; Kafantaris, I.; Amoutzias, G.D.; Pournaras, S.; Mossialos, D. Antibacterial Activity and Characterization of Bacteria Isolated from Diverse Types of Greek Honey against Nosocomial and Foodborne Pathogens. Appl. Sci. 2021, 11, 5801. [Google Scholar] [CrossRef]
- López, A.C.; Alippi, A.M. Feasibility of using RFLP of PCR-amplified 16S rRNA gene(s) for rapid differentiation of isolates of aerobic spore-forming bacteria from honey. J. Microbiol. Methods 2019, 165, 105690. [Google Scholar] [CrossRef]
- Alippi, A.M.; Abrahamovich, E. HiCrome Bacillus agar for presumptive identification of Bacillus and related species isolated from honey samples. Int. J. Food Microbiol. 2019, 305, 108245. [Google Scholar] [CrossRef]
- Balzan, S.; Carraro, L.; Merlanti, R.; Lucatello, L.; Capolongo, F.; Fontana, F.; Cardazzo, B. Microbial metabarcoding highlights different bacterial and fungal populations in honey samples from local beekeepers and market in North-Eastern Italy. Int. J. Food Microbiol. 2020, 334, 108806. [Google Scholar] [CrossRef]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Y.; Gu, L.; Yu, J.; Liu, Q.; Zhang, R.; Liang, G.; Chen, H.; Gu, F.; Liu, H.; et al. Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine. Microorganisms 2024, 12, 672. [Google Scholar] [CrossRef] [PubMed]
- Amenu, D.; Bacha, K. Probiotic potential and safety analysis of lactic acid bacteria isolated from Ethiopian traditional fermented foods and beverages. Ann. Microbiol. 2023, 73, 37. [Google Scholar] [CrossRef]
- Andrews, J.M.; Wise, R. Susceptibility testing of Bacillus species. J. Antimicrob. Chemother. 2002, 49, 1040–1042. [Google Scholar] [CrossRef]
- Owusu-Kwarteng, J.; Wuni, A.; Akabanda, F.; Tano-Debrah, K.; Jespersen, L. Prevalence, virulence factor genes, and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]



| Honey Sample | S. aureus | B. cereus | E. coli | Y. enterocolitica |
|---|---|---|---|---|
| 1 | 15 | 11 | 5 | 0 |
| 2 | 11 | 18 | 3 | 3 |
| 3 | 12 | 12 | 6 | 0 |
| 4 | 8 | 2 | 4 | 2 |
| 5 | 8 | 5 | 3 | 2 |
| 6 | 4 | 1 | 0 | 0 |
| 7 | 11 | 7 | 5 | 1 |
| 8 | 6 | 14 | 4 | 1 |
| 9 | 5 | 6 | 1 | 0 |
| 10 | 9 | 10 | 0 | 0 |
| Total | 89 | 86 | 31 | 9 |
| % | 47.09 | 45.5 | 16.4 | 4.76 |
| Isolates | Survival Rates, % | |
|---|---|---|
| Acid Tolerance (pH = 2.0) | Bile Tolerance (0.3%) | |
| 1 | 59.90 | 79.06 |
| 2 | 47.45 | 81.06 |
| 3 | 49.50 | 85.77 |
| 4 | 0.00 | 42.01 |
| 5 | 66.95 | 85.99 |
| 6 | 0.00 | 0.00 |
| 7 | 40.44 | 74.09 |
| 8 | 0.00 | 50.35 |
| 9 | 0.00 | 0.00 |
| 10 | 37.93 | 44.87 |
| 11 | 36.36 | 62.47 |
| 12 | 0.00 | 0.00 |
| 13 | 58.49 | 70.98 |
| 14 | 52.26 | 63.64 |
| 15 | 63.46 | 78.05 |
| 16 | 76.66 | 107.97 |
| 17 | 60.67 | 96.59 |
| 18 | 54.42 | 72.32 |
| Isolate | Diameter of Inhibition Zone (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S10 | FOX30 | AX25 | AM10 | TE30 | CN10 | E15 | C30 | DO30 | DA2 | |
| 1 | 23.3 ± 0.6 (S) | 17.0 ± 1.0 (MS) | 14.0 ± 1.5 (R) | 32.3 ± 1.2 (S) | 24.5 ± 1.0 (S) | 28.3 ± 1.5 (S) | 26.0 ± 1.0 (S) | 25.3 ± 0.6 (S) | 28.3 ± 1.5 (S) | 26.3 ± 0.6 (S) |
| 2 | 24.7 ± 0.6 (S) | 16.3 ± 0.6 (MS) | 16.0 ± 1.0 (MS) | 32.7 ± 1.2 (S) | 24.7 ± 0.6 (S) | 28.7 ± 1.2 (S) | 30.0 ± 0.0 (S) | 24.0 ± 1.0 (S) | 28.3 ± 1.4 (S) | 23.3 ± 1.2 (S) |
| 3 | 19.7 ± 0.6 (MS) | 13.3 ± 0.6 (R) | 14.0 ± 1.0 (R) | 24.3 ± 0.6 (S) | 22 ± 1.0 (S) | 21.0 ± 1.0 (S) | 23.3 ± 0.6 (S) | 25.3 ± 1.2 (S) | 27.3 ± 1.5 (S) | 24.0 ± 1.0 (S) |
| 4 | 27.0 ± 1.0 (S) | 18.0 ± 1.0 (MS) | 15.0 ± 1.0 (R) | 30.3 ± 2.1 (S) | 21.7 ± 0.6 (S) | 30.7 ± 0.6 (S) | 28.3 ± 1.5 (S) | 25.0 ± 1.0 (S) | 30.3 ± 1.5 (S) | 27.3 ± 0.6 (S) |
| 5 | 20.0 ± 0.0 (MS) | 15.0 ± 2.0 (R) | 17.7 ± 0.6 (MS) | 23.3 ± 1.5 (S) | 23.0 ± 2.0 (S) | 24.3 ± 0.6 (S) | 29.3 ± 1.5 (S) | 19.7 ± 2.1 (MS) | 26.0 ± 1.0 (S) | 10.0 ± 0.0 (R) |
| 6 | 25.3 ± 2.1 (S) | NI (R) | NI (R) | 7.0 ± 2.0 (R) | 25.7 ± 1.5 (S) | 25.3 ± 0.6 (S) | 30.0 ± 2.0 (S) | 26.7 ± 0.6 (S) | 22 ± 1.0 (S) | 17.0 ± 1.0 (MS) |
| 7 | 22 (S) | 14.3 ± 1.5 (R) | 11.0 ± 2.0 (R) | 25.0 ± 2.0 (S) | 26.3 ± 1.2 (S) | 25.0 ± 1.0 (S) | 28.7 ± 1.2 (S) | NI (R) | 27.0 ± 2.0 (S) | 19.7 ± 0.6 (MS) |
| 8 | 16.0 ± 1.5 (MS) | 22.3 ± 2.1 (S) | 15.3 ± 0.6 (R) | 27.3 ± 0.6 (S) | 19.7 ± 0.6 (MS) | 30.0 ± 2.0 (S) | 30.3 ± 1.2 (S) | 33.0 ± 2.0 (S) | 24.0 ± 1.0 (S) | 25.3 ± 0.6 (S) |
| 9 | 20.0 ± 1.0 (MS) | 15.7 ± 0.6 (R) | 12.5 ± 0.5 (R) | 28.3 ± 2.0 (S) | 27.0 ± 1.0 (S) | 29.0 ± 2.0 (S) | 29.7 ± 0.6 (S) | 25.3 ± 0.6 (S) | 30.0 ± 2.0 (S) | 25.3 ± 3.1 (S) |
| 10 | 22.7 ± 1.2 (S) | 19.0 ± 2.0 (MS) | 20.3 ± 0.6 (MS) | 31.3 ± 1.5 (S) | 30.2 ± 1.3 (S) | 23.3 ± 0.6 (S) | 30.0 ± 2.0 (S) | 26.7 ± 0.6 (S) | 32.3 ± 1.2 (S) | 30.3 ± 1.5 (S) |
| 11 | 26.7 ± 1.5 (S) | 16.0 ± 2.0 (MS) | 15.3 ± 0.6 (R) | 30.7 ± 1.5 (S) | 27.0 ± 1.0 (S) | 30.3 ± 2.1 (S) | 32.7 ± 1.2 (S) | 23.3 ± 0.6 (S) | 30.0 ± 1.0 (S) | 27.7 ± 1.2 (S) |
| 12 | 18.0 ± 2.0 (MS) | NI (R) | NI (R) | NI (R) | 25.7 ± 0.6 (S) | 25.0 ± 1.0 (S) | 32.3 ± 1.2 (S) | 30.7 ± 0.6 (S) | 25.3 ± 1.5 (S) | 25.7 ± 0.6 (S) |
| 13 | 14.0 ± 0.0 (R) | 17.3 ± 0.6 (MS) | 13.0 ± 2.0 (R) | 26.7 ± 1.5 (S) | 15.0 ± 1.0 (R) | 30.7 ± 1.2 (S) | 33.3 ± 0.6 (S) | 32.0 ± 2.0 (S) | 27.3 ± 1.6 (S) | 23.7 ± 1.2 (S) |
| 14 | 18.7 ± 0.6 (MS) | 18.0 ± 2.0 (MS) | 15.3 ± 0.6 (R) | 31.0 ± 2.0 (S) | 25.7 ± 0.6 (S) | 27.0 ± 1.0 (S) | 30.7 ± 1.2 (S) | 24.0 ± 2.0 (S) | 28.3 ± 0.6 (S) | 22.0 ± 0.0 (S) |
| 15 | 16.0 ± 1.0 (MS) | 16.0 ± 1.0 (MS) | 19.7 ± 1.5 (MS) | 22.0 ± 2.0 (S) | 13.0 ± 0.0 (R) | 25.3 ± 0.6 (S) | 30.0 ± 2.0 (S) | 28.3 ± 1.5 (S) | 16.0 ± 2.0 (MS) | 27.7 ± 0.6 (S) |
| 16 | 20.7 ± 1.5 (MS) | 13.0 ± 2.0 (R) | 9.0 ± 1.0 (R) | 24.7 ± 0.6 (S) | 20.0 ± 1.0 (MS) | 25.0 ± 2.0 (S) | 25.3 ± 0.6 (S) | 24.0 ± 1.0 (S) | 30.0 ± 2.0 (S) | 19.0 ± 1.0 (MS) |
| 17 | 17.0 ± 1.0 (MS) | 16.3 ± 1.5 (MS) | 14.0 ± 1.0 (R) | 25.3 ± 0.6 (S) | 12.0 ± 1.0 (R) | 27.0 ± 1.0 (S) | 33.3 ± 0.6 (S) | 32.0 ± 2.0 (S) | 18.3 ± 1.2 (MS) | 25.3 ± 1.2 (S) |
| 18 | 22.3 ± 2.1 (S) | 17.0 ± 1.0 (MS) | 16.0 ± 0.0 (MS) | 28.3 ± 0.6 (S) | 30.7 ± 0.6 (S) | 25.3 ± 1.5 (S) | 31.0 ± 2.0 (S) | 28.7 ± 0.6 (S) | 31.3 ± 1.2 (S) | 24.0 ± 1.0 (S) |
| B. c. | 20.0 ± 2.0 (MS) | 10.0 ± 1.0 (R) | - | NI (R) | 24.3 ± 0.6 (S) | 22.0 ± 1.0 (S) | 30.7 ± 1.2 (S) | 22.0 ± 2.0 (S) | 27.3 ± 0.6 (S) | 21.0 ± 1.0 (S) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grabek-Lejko, D.; Worek, M. Honeydew Honey as a Reservoir of Bacteria with Antibacterial and Probiotic Properties. Antibiotics 2024, 13, 855. https://doi.org/10.3390/antibiotics13090855
Grabek-Lejko D, Worek M. Honeydew Honey as a Reservoir of Bacteria with Antibacterial and Probiotic Properties. Antibiotics. 2024; 13(9):855. https://doi.org/10.3390/antibiotics13090855
Chicago/Turabian StyleGrabek-Lejko, Dorota, and Mariusz Worek. 2024. "Honeydew Honey as a Reservoir of Bacteria with Antibacterial and Probiotic Properties" Antibiotics 13, no. 9: 855. https://doi.org/10.3390/antibiotics13090855
APA StyleGrabek-Lejko, D., & Worek, M. (2024). Honeydew Honey as a Reservoir of Bacteria with Antibacterial and Probiotic Properties. Antibiotics, 13(9), 855. https://doi.org/10.3390/antibiotics13090855







