Abstract
Urinary tract infections (UTIs) caused by Escherichia coli represent a significant public health concern due to the high virulence and antimicrobial resistance exhibited by these pathogens. This study aimed to analyze the phylogenetic diversity and antibiotic resistance profiles of Uropathogenic E. coli (UPEC) strains isolated from UTI patients in Algeria, focusing on virulence factors such as extended β-lactamase (ESBL) production, biofilm formation, and hemolytic activity. Phylogenetic grouping of 86 clinical imipenem resistant E. coli isolates showed the prevalence of group B2 (48.9%), followed by groups E (22.1%), unknown (12.8%), A (8.1%), and B1 (4.7%), and Clade I, D, Clade I, or Clade II (1.2%). The highest resistance rates were observed towards amoxicillin (86.04%), ticarcillin (82.55%), piperacillin (73.25%), nitrofurantoin (84.88%), and trimethoprim-sulfamethoxazole (51.16%). Notably, 69.8% of UPEC strains were multidrug-resistant (MDR) and 23.2% were extensively drug-resistant (XDR). Additionally, 48.9%, 42%, and 71% of strains demonstrated ESBL production, hemolytic activity, and weak biofilm production, respectively. Continuous monitoring and characterization of UPEC strains are essential to track the spread of the most resistant and virulent phylogenetic groups over time, facilitating rapid therapeutic decisions to treat infections and prevent the emergence of new resistant organisms, helping choose the most effective antibiotics and reducing treatment failure.
1. Introduction
Escherichia coli, a Gram-negative bacillus of the Enterobacterales family, is commonly found in the gastrointestinal tract of humans and various animals [1,2,3]. This organism is among the most significant and common species of the Escherichia genus in veterinary and medical fields [4] and is responsible for approximately 80–90% of infections [5]. Apart from intestinal diseases that E. coli can cause, the species possesses a high potential to cause extra-intestinal diseases, including urinary tract infections (UTIs), various intra-abdominal, pulmonary, skin, and soft tissue infections, neonatal meningitis, and bacteremia [2,6,7,8].
Phenotypic and genotypic characteristics are used to identify E. coli pathogenic strains or pathovars [1]. The definition of these pathotypes can be based on various criteria, such as the target organ, the infected host, the association with the targeted organs, the pathology caused by the strains, and the presence of a specific gene or genes alone or in combination [2]. In addition to the pathotype and pathovar, the classification of E. coli strains has been based on phylogenetic relationships [1,3].
Clermont et al. optimized a quadruplex polymerase chain reaction (PCR) to classify extracellular E. coli strains into eight phylogenetic groups: B2, B1, A, D, F, E, C, and clade I [1,7,9]. Commensal strains are primarily related to groups A and B1 and can be responsible for intestinal infections [10], while pathogenic strains primarily belong to groups B2 and D [4,5,9,11]. Moreover, the detection of phylogenetic groups plays an important role not only in understanding the populations of E. coli but also in clarifying the relationship between strains and diseases [12]. E. coli isolates can be distinguished in terms of characteristics such as patterns of antibiotic resistance, virulence genes, the use of sugars, and environmental characteristics [5,13]. Furthermore, several studies showed that regional variations in E. coli populations may exist due to differences in environmental factors, human population dynamics, and ecological conditions [14,15,16].
The global spread of multidrug-resistant (MDR) and extensively drug-resistant (XDR) E. coli strains has become a public health threat and a major concern worldwide [3]. Limited treatment options may complicate UTIs and their treatment and increase morbidity and mortality [3,17].
In this study, we analyzed 86 imipenem-resistant uropathogenic E. coli (UPEC) isolated from inpatients and outpatients with UPEC-associated urinary tract infections in northern Algerian populations. The isolates were analyzed for their phylogenetic groups, as well as antibiotic resistance patterns associated with virulence profiles such as extended β-lactamase production, biofilm formation, and hemolytic activity to characterize the resistance phenotype and to investigate the virulence factors associated with this type of resistance.
2. Results
2.1. Demographic Characteristics
A total of 86 urinary imipenem-resistant E. coli isolates were collected from several geographical locations around Algeria’s north-eastern province of Sétif: 20.9% (18 isolates) from the capital of the province (Sétif), 50% (43 isolates) from southwest of Sétif (Aïn Oulmène city), 14% (12 isolates) from extreme north (Bouandas city), 7% (six isolates) from the north (Tizi N’Bechar city), 8.1% (seven isolates) from the east (El-Eulma city). Of the total samples, 59.3% were collected from women and 40.7% from men (ratio of female/male: 51/35 = 1.45; p = 0.084), the ages of the patients ranged between 2 years and 92 years with a mean age of 36.19 years. Females were older (mean age: 39.4) than males mean age: 31.5), with statistical significance (p value < 0.001, t test). Most UPEC isolates were found in adults (p < 0.001), indicating they are more susceptible than other groups to UPEC UTIs. Table 1 shows the distribution of demographic characteristics of inpatients (hospitalized), or outpatients (day hospital visits) based on their gender and age.

Table 1.
Distribution of patients according to hospitalization (inpatients and outpatients), gender and, age (children ≤ 15 y, adults 15–64 y and elderly ≥ 65 y).
Regarding the age group distribution, female patients (44.1%) in the 15–64 age class were prevalent compared to males (12.7%), especially in outpatients. However, in the children age group (<15 years), there were more males (19.7%) than females (6.9%) in both inpatient and outpatient settings. The elderly included in the study were only outpatients and there was no significant gender difference. The statistical analyses (Fisher’s exact test, p < 0.0001) indicated a strong and significant correlation between the age group and gender distribution of the participants in this study.
2.2. Phylogenetic Grouping
Based on the quadruplex PCR assay, phylogenetic analysis of E. coli isolates showed that they mainly belonged to phylogroup B2 (48.9%) and E (22.1%), followed by A (8.1%), B1 (4.7%), and D, Clade I, Clade I or Clade II (1.2% for each one). Typing profiles that did not cluster with any of the known groups were frequently found (unknown groups; 12.8%), but no strain belonging to the phylogroup F was found (Figure 1). However, the statistical analyses did not reveal any significant correlation between the gender, the age of the patient, and the phylogenetic groups (p = 0.578 and 0.171, respectively).
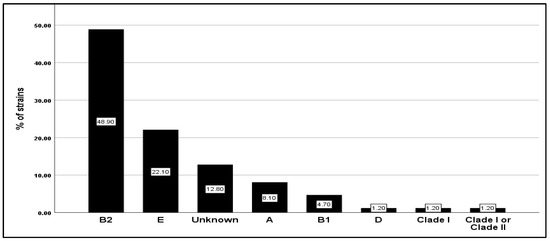
Figure 1.
Frequency distribution of the 86 E. coli clinical isolates in seven phylogroups (A, B1, B2, D, E, Unknown, Clade I, and Clade I or Clade II).
2.3. Antimicrobial Susceptibility
The 86 E. coli isolates showed different resistance profiles towards the 25 antibiotics tested. Apart from the resistance to imipenem (IMP), the highest resistance percentage was significantly (p < 0.0001) observed against β-lactam antibiotics: amoxicillin (AMX) (86.04%) and ticarcillin (TC) (82.55%), followed by piperacillin (PRL) (73.25%). The association of β-lactamase inhibitors with β-lactam antibiotics was also investigated; the association of the β-lactamase inhibitor tazobactam significantly reduced the resistance to piperacillin from 73% to 19%. However, the addition of the β-lactamase inhibitor clavulanic acid did not significantly reduce the resistant strains to amoxicillin (from 86% for amoxicillin to 62% for amoxicillin + clavulanic acid) and ticarcillin (from 83% for ticarcillin to 76% for ticarcillin + clavulanic acid). The lowest percentage of resistance was exhibited towards cephalosporin antibiotics (19.76–38.37%) (Figure 2).
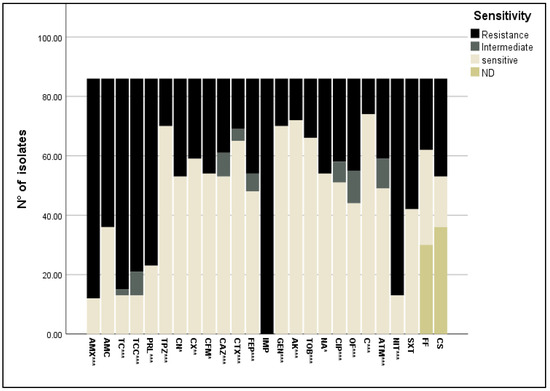
Figure 2.
Antibiotic resistance profile of E. coli isolates towards 25 antibiotics belonging to nine classes. *: p < 0.05; **: p < 0.01; ***: p < 0.0001. AMX: amoxicillin, AMC: amoxicillin + clavulanic acid, TC: ticarcillin, TCC; ticarcillin + clavulanic acid, PRL: piperacillin, TPZ: piperacillin + tazobactam, CN: cephalexin, CX: cefoxitin, CFM: cefixime, CAZ: ceftazidime, CTX: cefotaxime, FEP: cefipime, IMP: imipenem, GEN: gentamycin, AK: amikacin, TOB: tobramycin, NA: nalidixic acid, CIP: ciprofloxacin, OF: ofloxacin, C: chloramphenicol, ATM: aztreonam, NIT: nitrofurantoin, SXT: trimethoprim-sulfamethoxazole, FF: fosfomycin, CS: colistin, ND: not determined.
However, almost all isolates were resistant to nitrofurantoin (84.88%) and trimethoprim-sulfamethoxazole (51.16%). The resistance rate of isolates against fosfomycin was 27.9% and to colistin 38.4%, and lower percentages of nalidixic acid (37.2%), ofloxacin (36.04%), aztreonam (31.39%) and ciprofloxacin (32.55%) resistant strains were found (Figure 2).
Most of the isolates were susceptible to chloramphenicol, gentamycin, amikacin, and tobramycin (86.05%, 81.4%, 83.73%, and 76.75%, respectively) (Figure 2). A high number of E. coli isolates had a minimal inhibitory concentration (MIC) value > EUCAST resistance break point (Figure 3), especially for amoxicillin (AMC), piperacillin (PRL), piperacillin + tazobactam (TPZ), cephalexin (CN), cefixime (CFM), gentamycin (GEN), ciprofloxacin (CIP), and trimethoprim-sulfamethoxazole (SXT), with MIC values of >32 mg/L, 8 mg/L, 8 mg/L, 16 mg/L, 1 mg/L, 2 mg/L, and 0.5 mg/L, respectively. The rate of susceptibility was higher towards ceftazidime (CAZ), cefotaxime (CTX), tobramycin (TOB), nalidixic acid (NA), chloramphenicol (C), and aztreonam (ATM) (81.48%, 58.46%, 75.75%, 96.22%, and 58.44%, respectively).
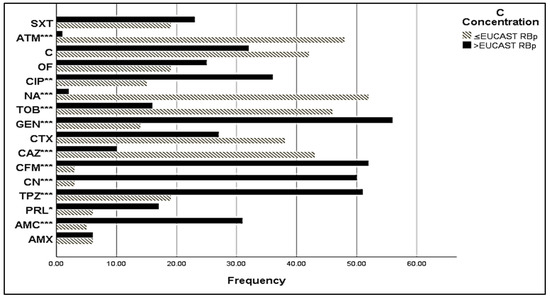
Figure 3.
MIC of sensitive E. coli isolates. C Concentration stands for Critical Concentration; EUCAST RBp stands for EUCAST Resistant Break point; *: signification (p < 0.05); **: signification (p < 0.01); ***: signification (p < 0.001). AMX: amoxicillin, AMC: amoxicillin + clavulanic acid, TC: ticarcillin, PRL: piperacillin, TPZ: piperacillin + tazobactam, CN: cephalexin, CFM: cefixime, CAZ: ceftazidime, CTX: cefotaxime, GEN: gentamycin, TOB: tobramycin, NA: nalidixic acid, CIP: ciprofloxacin, OF: ofloxacin, C: chloramphenicol, ATM: aztreonam, SXT: trimethoprim/sulfamethoxazole.
Different antibiotic susceptibility patterns were found among the phylogenetic groups. The most resistant was phylogroup B2 (resistance score: median 14; range 10–30) followed by phylogroup E (resistance score: median 8; range 5–16) and unknown state (resistance score: median 3; range 2–7) (ANOVA test, p = 0.001). The most susceptible phylogroups were phylogroup D and Clade I or Clade II. However, no significant correlation was seen between phylogenetic groups and antibiotic resistance; indeed, all phylogenetic groups had a variable resistance to penicillins (26–56.63%). The E groups presented with the highest percentage of resistance to cephalosporins. Moderate resistance of all E. coli groups was shown towards trimethoprim/sulfamethoxazole, while the aminoglycosides, especially amikacin, were the most effective antibiotics against all phylogenetic groups.
Further, 69.77% and 23.26% of the examined strains were MDR and XDR strains, respectively. The prevalence of MDR and XDR strains was very significant (p < 0.0001) in the adult group. According to phylogenetic groups, the prevalence of MDR strains was higher in phylogroup B1, unknown, and E (100%, 81.81%, and 73.68%, respectively). In contrast, only a few strains (6.98%) were resistant to less than three classes of antibiotics (R) (Table 2).

Table 2.
Distribution of phenotypic resistance (R, MDR, and XDR) among UTI strains and association to gender, age (adults, children, elderly), clinical status (inpatients and outpatients), phylogroups (A, B1, B2, Clade I or II, D, E, and unknown), ESBL production, and hemolytic activity.
2.4. Extended β-Lactamase (ESBL) Production and Haemolysin Activity
The production of ESBL is one of the main mechanisms by which bacteria resist lactam antibiotics. Out of the isolates tested, 48.83% were found to be ESBL-producers while 51.16% were not (Table 2). The ESBL-producing isolates showed greater resistance to β-lactam antibiotics than ESBL-negative, such as amoxicillin (100% vs. 73%, p = 0.0002), ticarcillin (98% vs. 68%, p = 0.0003), ticarcillin + clavulanic acid (96% vs. 57%), and piperacillin (98% vs. 50%, p < 0.0001), and there was a strong, positive, and significant correlation between ESBL production and cefixime (p = 0.017) and ceftazidime and cefotaxime (p < 0.0001) resistance. Most of the strains did not present hemolytic activity (48.83%), while 22.09% and 29.06% of the isolates featured α-hemolysin and β-hemolysin activity, respectively (Table 2). ESBL producing isolates, as well as strains that produced hemolysin (57.89% for α-hemolysin and 48% for β-hemolysin), were preferentially observed in phylogenetic group B2 more than in other phylogroups (Figure 4).
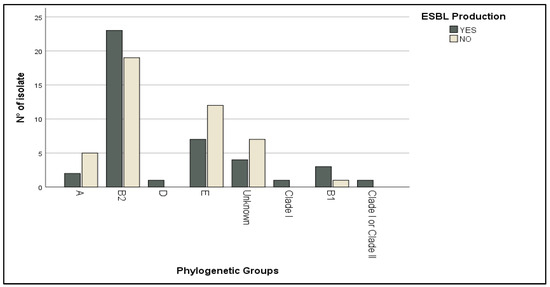
Figure 4.
Distribution of ESBL-producing strains according to phylogenetic groups.
2.5. Correlations between Biofilm Activity, Phylogroup, and Antibiotic Resistance
According to biofilm formation assay results (Table 3), most of the tested clinical strains were weak biofilm producers (71% with p value < 0.0001), while other strains did not produce biofilm at all (19%). Moderate (9%) and strong biofilm (1%) producers were in the minority. Biofilm-forming strains were mostly found in the phylogroup B2, although they were mostly weak producers (30%), while the only strong biofilm-forming strain belonged to phylogroup E (n = 1, 1.1%).

Table 3.
Correlations between the biofilm-forming ability of the E. coli isolates, phylogroups, and virulence factors: ESBL and hemolytic activity. (Only p values with statistical significance are shown in the table).
Weak biofilm production was more commonly found in ESBL-producing strains than in ESBL-negative (42% vs. 29%). There was a statistical correlation between ESBL production and biofilm production (p = 0.02) (Table 3).
In the present study, the production of biofilm and antibiotic resistance were analyzed (Figure 5). It was observed that strains possessing resistance to multiple classes of drugs (XDR) exhibited weak biofilm production (75%), or moderate (5%), or were completely unable to produce biofilm (20%). Regarding MDR, most tested isolates had a weak production of biofilm (72%). Furthermore, 50% of resistant strains (R) were not able to form biofilm or produced only weak biofilm (50%).
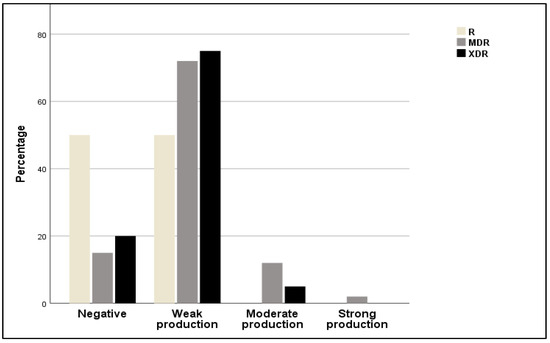
Figure 5.
Relative frequencies of resistant (R), multidrug-resistant (MDR), and extensively drug resistant (XDR) strains among non-producers and weak, moderate, and strong biofilm producers.
In addition, the optical density (OD) of biofilms was compared across the resistance profile (sensitive, intermediate, and resistant strains) for each of the antibiotics tested in this study (Figure 6). The analysis revealed that there was no difference in the OD value of biofilm activity between the resistant, sensitive, and intermediate strains to each of the antibiotics. In this case, the biofilm-forming ability was not associated with the resistance profile of strains; as an exception, the nitrofurantoin-resistant strains produced more biofilms than the sensitive strains with a statistically significant difference (p = 0. 012).
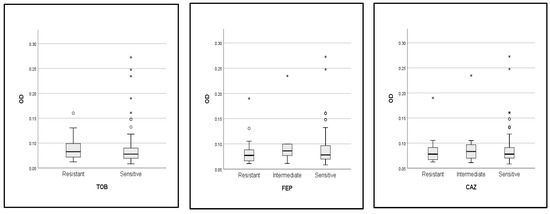
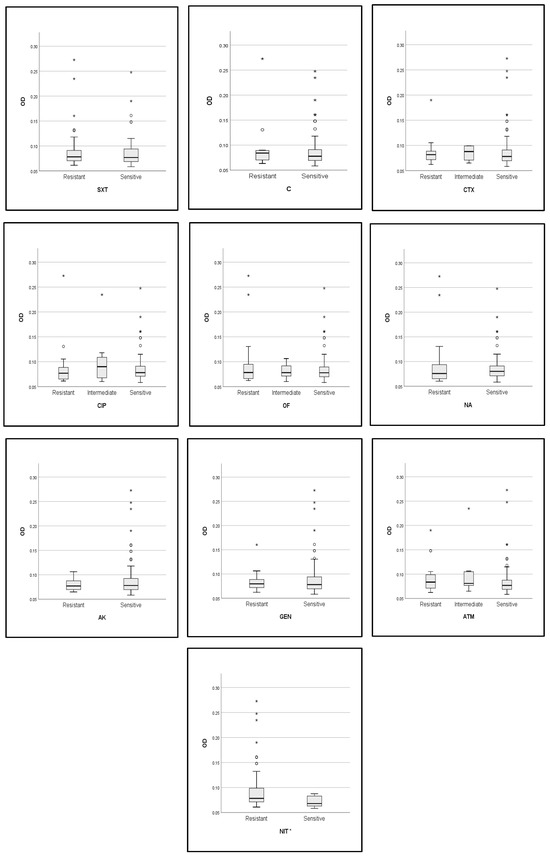
Figure 6.
Comparison of OD values of biofilm formation between resistant, intermediate, and susceptible strains. *: p value < 0.05, TOB: tobramycin, FEP: cefipime, CAZ: ceftazidime, SXT: trimethoprim-sulfamethoxazole, C: chloramphenicol, CTX: cefotaxime, CIP: ciprofloxacin, OF: ofloxacin, NA: nalidixic acid, AK: amikacin, GEN: gentamycin, ATM: aztreonam, NIT: nitrofurantoin.
Table 4 displays the percentage distribution of bacteria resistant to each tested antibiotic in weak, moderate, strong, and non-producing biofilm strains. It has been observed that cephalosporin resistance was significantly (p = 0.023, with correlation test) decreased for both moderate and strong biofilm-producing strains. On the other hand, weak or non-biofilm producing strains showed a higher percentage of resistance to cephalexin (CN), cefixime (CFM), ceftazidime (CAZ), and cefotaxime (CTX). These findings suggest that the ability to resist this group of antibiotics is inversely proportional to biofilm production. Moreover, with nitrofurantoin, the resistance percentage increased with the biofilm production profile from the non-producing strains to strong producers. In comparison to other resistance categories of antibiotic-resistant strains, there was no significant difference (p ˃ 0.05) in biofilm formation.

Table 4.
Percentage distribution of antibiotic-resistant strains based on biofilm formation ability.
3. Discussion
This study aimed to isolate imipenem-resistant UPECs from patients with UTIs and to characterize them by coupling biochemical and molecular approaches. We were able to confirm the high prevalence of E. coli in UTIs [18,19,20], with women being mostly affected, in accordance with previous studies [21,22]. This can be due to anatomical factors like a shorter urethra, making entering the urinary tract easier for bacteria [23,24]. Accordingly to previous research [25,26], UPEC isolates were more prevalent in adults than in the elderly, who can have weaker health conditions due to pathologies like diabetes or kidney stones, medications, and age-related changes in the immune system or the bladder [27]. Various socioeconomic factors, hygiene practices, diets, and lifestyles can also increase the frequency of UTIs, but they were not investigated in the current study.
Most strains causing extraintestinal infections are predominantly categorized into B2 and D groups [12], while commensal isolates are categorized into groups A and B1 [4]. Our findings align with numerous studies that have identified B2 group strains as the dominant type in UTIs. A study on 105 E. coli isolates from Slovenian patients with bacteremia of urinary tract origin showed that 51% belonged to group B2, 20% to group D, 15% to group A, and 13% to the B1 group [6]. Another study on 190 urinary E. coli isolates in Colombia showed that 46.8% of the isolates belonged to group B2 followed by D group with a percentage of 25.3% [28]. In a study on 228 UPEC in Egypt, 64.6% of the isolates belonged to phylogroup B2, and 18.9%, 10.7%, and 5.7% belonged to phylogenetic groups D, A, and B1, respectively [29]. Similarly in a study for phylogenetic typing urine samples in Korea, the prevalence of uropathogenic E. coli belonging to group B2 (77.7%) followed by group D (17.5%), B1 (3.4%), and A (1.4%) [25]. In a study on 113 uropathogenic E. coli isolates in Iran, 44.2% of the strains were classified into group B2, 31% into group D, 20.4% into group A, and 4.4% into group B1 [10]. Similarly, studies in India [4,30], Iran [31], and Egypt [32,33], demonstrated that most UPEC isolates from UTIs belonged to the B2 group. Among E. coli phylogenetic groups, the B2 phylogroup is believed to be more important than others. This phylogroup is associated with a high evolution of virulence capacity and characteristics, which may cause the spread and persistence of extraintestinal infections representing, therefore, a major public health concern [34,35].
Our analysis did not allow the classification of a small percentage of E. coli isolates (12.8%). This latter result can be dependent on the recombination of different or rare phylogroups resulting from the combination of the presence and absence of certain genes, as suggested by Boroumand et al. [5]. Phylogenetic group E also had a high prevalence among our strains, as found in a very recent study [36]. However, it should be noted that variations in the source of bacterial isolation, host health state, geographic locations, and genetic variables can affect the distributions and proportions of phylogenetic groupings.
In addition, this study investigated the antibiotic resistance profile of the UPEC isolates, since the spread of antibiotic-resistant strains is a major concern in clinical practice, particularly in developing countries. Due to its high levels of antibiotic resistance, the occurrence of virulence and resistance genes and frequent transmission between humans in different settings and between humans and animals [37,38], our study was focused on imipenem-resistant E. coli, since carbapenems are frequently used in hospital setting as first line drugs in the empirical treatment of several bacterial infections in Algeria. In this study, the 86 imipenem-resistant isolates displayed a high percentage of resistance to penicillins, similar to other studies: 78.6% penicillin-resistant urine strains in Uganda and 78.4% penicillin-resistant-UPEC in Mongolia, respectively [23,39]. A high percentage of resistance to nitrofurantoin (84.88%) was also found, while moderate percentages of resistant strains to amoxicillin + clavulanic acid, quinolones, trimethoprim/sulfamethoxazole, and fluroquinolones (32–37.1%) were observed. In Algeria, nitrofurantoin and trimethoprim-sulfamethoxazole are recommended as the first-line therapy, while β-lactams and fluoroquinolones are used as alternative agents in UTI therapy [20,40]. The resistance percentage of the cephalosporin class (19.76–38.37%) was similar to that obtained in Gabon (30–33%) and Rwanda (29.1%) [41,42] and can be linked to the spread and acquisition of the plasmid-borne ESBL genes [43]. The evidence that a lower percentage of isolates was resistant to aminoglycosides (16.2–23.25%), as reported in other studies carried out in Iran (16.7% and 21.8%) [44], could be explained by the limited use of this antibiotic in UTI treatment in developing countries. These results indicate a worrying trend of increased resistance to first-line treatments.
The antibiotic resistance profile of E. coli phylogroups showed that B2 groups were more resistant than the other phylogenetic groups. Our finding is consistent with several studies [5,12,45,46,47]. This can be explained by the fact that this phylogroup has a greater ability to exhibit characteristics associated with antibiotic resistance (antibiotic resistance genes), the coexistence of some virulence factors, followed by the acquisition of resistance [6]. On the contrary, many studies have proven that the phylogroup B2 is more sensitive than the other phylogroups (Iran, Taiwan) [48,49]. Social and environmental conditions and the therapy profile of patients may explain this difference. Most of our B2 strains were MDR (69.77%), similar to studies conducted in Egypt and Sri Lanka, featuring 65.17% and 60.3% of MDR strains, respectively [36,50]; this high similarity in percentage of MDR may be due to the similar inappropriate use of antibiotics and poor healthcare infrastructure and management in these developing countries. Several previous investigations have shown that MDR profiles are associated with less virulent strains and non-B2 phylogenetic groups [6].
In this study, as already reported [4,51,52], the majority of ESBL-producing and hemolytic strains belonged to B2 phylogroups. The B2 phylogroup‘s increased virulence has been correlated to its ability to persist in the gut microbiota, facilitating the accumulation of virulence and antibiotic resistance genes [9,53].
Biofilms represent a microbial characteristic that protect bacteria against hydrodynamic flow conditions, especially in UTIs and also against host defense mechanisms [54]. In this study, 81.39% of E. coli isolates analyzed showed considerable biofilm activity, with 24.8% of the isolates being classified as moderate to strong biofilm producers. This finding is consistent with Gunathilaka et al. (2024) (78% of tested strains were biofilm producers) [36]. Hashemizadeh et al. (2017) found that 74% of the tested strains were biofilm producers in inpatients and 83.4% in outpatients [55]. In another study, Maharjan et al. (2018) found that 21%, 14% and 11% of the strains tested were weak, moderate, and strong biofilm producers, respectively [56].
Several previous studies showed that the most of biofilm-forming strains belonged to phylogenetic group B2 [57,58], which is in accordance with our results. Virulence factors, toxin proteins, multi-drug resistance, and ESBL increased in UPEC and is related to phylogroup B2 [59]. The majority of biofilm-producing strains were MDR, confirming the results of a study conducted in Uganda in which 63% of E. coli urine isolates were biofilm formers [60]. Similar to our findings, Behzadi et al. (2020) found that there was a significant correlation between ESBL production and biofilm-formation [61].
We found an inverse relationship in 86 E. coli isolates between resistance to cephalosporins and biofilm production. Similar results were obtained by Gajdacs et al., who found an inverse relationship between resistance to cephalosporins and biofilm production, and the biofilm producers were less prevalent among third generation cephalosporin-resistant strains [62]. Cepas et al. found that there was an inverse association in biofilm formation ability and resistance to gentamicin and ceftazidime among E. coli strains [63].
4. Materials and Methods
4.1. Origin of Isolates and Bacterial Strains
This retrospective study was performed on 86 imipenem-resistant E. coli isolates collected from patients with UTIs (all symptomatic infections by uropathogenics involving any part of the urinary tree manifest in many symptoms [64])
After 3 years of collection (2021–2022–2023), 402 strains were isolated from different care territories (from the east, west, and north of Sétif province). Among them, 33.8% of the strains were resistant to imipenem, and 86 strains, collected from February to May 2023, were selected for this study from six medical diagnostic laboratories and three hospital laboratories. The eighty-six unique strains of E. coli were isolated from urine specimens and collected using standard sterile procedures (only positive cultures with count of 105 Colony Forming Units/mL were taken into consideration for this study). UPEC strains were isolated from both hospitalized and non-hospitalized patients of all age groups with a diagnosis of UTI. After collection, samples were cultured on standard media, including nutrient agar, MacConkey agar, and nutrient broth (TM-Media, Delhi, India) and incubated at 37 °C for 24 h to observe the colony morphology (shape, size, texture, edge and elevation, and opacity). Conventional microbiological methods like Gram staining, and biochemical characteristics, such as IMVIC (Indole test, methyl red test, Voges–Proskauer test, and citrate utilization test), catalase test, urease production, nitrate reduction, motility, triple sugar iron (TSI) test, and gas production were used for E. coli identification. Isolated strains were stored in nutrient broth (NB) with sterile glycerol at −20 °C.
4.2. Antimicrobial Susceptibility Testing
The Kirby–Bauer disk diffusion method was used for the evaluation of susceptibility in culture media of Muller–Hinton’s agar. Susceptibility testing was performed for 25 antimicrobial drugs (HiMedia Laboratories, Mumbai, India; BioMaxima, Lublin, Poland; BioScan Industrie, Setif, Algeria) including amoxicillin (AMX-25 µg), amoxicillin + clavulanic acid (AMC-20 µg and 10 µg), ticarcillin (TC-75 µg), ticarcillin + clavulanic acid (TCC-85 µg), piperacillin (PRL-30 µg), piperacillin + tazobactam (TPZ-110 µg), cephalexin (CN-30 µg), cefoxitin (CX-30 µg), cefixime (CFM-5 µg), ceftazidime (CAZ-30 µg), cefotaxime (CTX-30 µg), cefepime (FEP-30 µg), imipenem (IMP-10 µg), gentamicin (GEN-10 µg), amikacin (AK-30 µg), tobramycin (TOB-10 µg), nalidixic acid (NA-30 µg), ciprofloxacin (CIP-5 µg), ofloxacin (OF-5 µg), chloramphenicol (C-30 µg), aztreonam (ATM-30 µg), nitrofurantoin (NIT-300 µg), trimethoprim/sulfamethoxazole (SXT-1.25 µg and 23.75 µg), fosfomycin (FF-50 µg), and colistin (CS-10 µg). Antimicrobial susceptibility profiles were determined by interpreting the breakpoints recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guideline of 2022 [65]; the isolates were defined as susceptible, intermediate, or resistant. In our study R, MDR, and XDR have been used as acronyms (in all manuscripts): R (resistant) is defined as strains that resist less than three classes of antibiotics; MDR (multi-drug resistant) is defined as strains that resist three to six classes of antibiotics; and XDR (extensively-drug resistant) are strains that resist at least seven classes of antibiotics. Nine antibiotic classes were used: penicillins, cephalosporins, carbapenem, aminoglycosides, quinolones, phenicolate, monobactams, polymyxins, and other antibiotics, in particular, nitrofurantoin, trimethoprim/sulfamethoxazole, and fosfomycin. For all susceptible strains MIC determination was obtained through microtitration. The antimicrobial agents and dilution ranges tested for each are presented in (Table 5). The Muller–Hinton broth (TM-Media, Delhi, India), the prepared solutions with a double dilution of antibiotics, and the adjusted inoculum were distributed in wells of microtiter plates and incubated at 37 °C for 24 h. The results were compared with MIC clinical breakpoints published in the EUCAST guideline to determine the resistance profiles. E. coli ATCC 25922 was used as the quality control strain.

Table 5.
Antibacterial agents and their dilution ranges used in the susceptibility test of E. coli isolates.
4.3. Detection of Extended Spectrum β-Lactamases Production
UPEC strains resistant to one or more third generation cephalosporins in the Kirby–Bauer disk diffusion test were screened for ESBL production through a confirmatory test. Confirmatory tests were performed using the double-disc synergy test [66]. Briefly, ceftazidime (30 µg), cefotaxime (30 µg), cefepime (30 µg), and aztreonam (30 µg) disks were placed at a 20 mm center-to-center distance of an amoxicillin + clavulanic acid (20 and 10 µg) disk. Samples were considered positive for ESBL when the inhibition zone around any of the cephalosporin discs increased in the direction of the disc containing clavulanic acid, promoting the appearance of either an enhanced or phantom zone. E. coli ATCC 25922 was used as the quality control strain.
4.4. DNA Extraction and Phylogenetic Grouping by Quadruplex PCR
Bacterial DNA was extracted from 86 E. coli isolates using the Direct PCR of Intact Bacteria (Colony PCR) method, described previously [67]. Briefly, after growing the strains in nutrient agar (Oxoid, Milano, Italy) overnight, 1–2 colonies were dissolved in 100 μL of sterile distilled water. The samples were vortexed for 10 s and then incubated at 99 °C for 15 min. Supernatants were collected after centrifugation at 10,000× g for 10 min, and pellets were discarded. A 1% agarose gel electrophoresis was conducted to evaluate the quality of the DNA, while the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, MA, USA) was used to assess DNA purity and concentration.
Molecular analyses were performed using the primers listed in Table 6. To characterize the 86 E. coli clinical strains within the seven phylogroups (A, B1, B2, C, D, E, and F), the method described in Clermont (2013) was used. Primers F1 and R12, described by Coy et al. (2014), were used to amplify the bacterial 16S rDNA gene (approximately 1500 bp fragment). PCR mixes contained 1 Unit of DreamTaq DNA Polymerase (Thermo Fisher), 10 pmol of the forward and reverse primers, 0.2 µM dNTPs in 1× buffer (Thermo Fisher), and 2 µL of DNA sample. The thermal profile used consisted of the initial denaturation step of 5 min at 95 °C, 35 cycles (30 s for denaturation at 95 °C, 30 s of primers annealing at the temperature reported in Table 2 and 30–90 s for extension at 72 °C based on the size of the amplification product), and a final step of extension for 5 min at 72 °C.

Table 6.
List of primers with amplicon size, annealing temperature, and the reference.
4.5. Biofilm Production
Biofilm production was assayed in microtiter plates, essentially as described by Stepanovic et al. (2007) [74], with a few adjustments. Briefly, cells were initially grown in brain-heart infusion broth medium BHIB (Liofilchem, Abruzzo, Italy) with glucose. Subsequently, cultures were diluted with fresh BHIB, and turbidity was adjusted to 0.5 McFarland Standard. The bacterial suspensions were incubated for 24 h at 37 °C in 96 well polystyrene microtiter plates. Unattached bacterial cells or planktonic bacteria were then removed from the culture medium by washing the plate with distilled water. Cells adhering to the plate walls were fixed and stained with crystal violet. The absorbance was measured with an ELISA reader (BioTek, El Dorado Hills, CA, USA) at 570 nm (OD570) to estimate the amount of biofilm formed. The experiments were performed in triplicate. The cut-off value (ODc) for judging whether the biofilm had formed was established as the mean absorbance value of the negative control well +3 standard deviation. Strains with a mean OD value ˃ ODc were considered to be biofilm producers. The interpretation of the results was as follows: OD ≤ ODc = not biofilm producer; ODc < OD ≤ 2×ODc = weak biofilm producer; 2×ODc < OD ≤ 4×ODc = moderate biofilm producer; 4×ODc < OD = strong biofilm producer. E. coli ATCC 25922 was used as the control organism [74].
4.6. Hemolysin Production
The production of hemolysin was tested on 5% human blood agar (type A and O) (BioScan Industrie, Setif, Algeria). E. coli strains were plated on blood agar plates and incubated at 37 °C for 18–24 h. Following the visualization of the plates, the bacterial strains were classified as α, β, and γ hemolytic: β hemolysis, when the toxin causes the complete lysis of the red blood cells (often referred to as true lysis) producing a clear, transparent area in the blood agar cultures; α hemolysis, when lysis does not occur but the hemoglobin of the red blood cells is reduced to methemoglobin and a brown/green colored area can be observed in blood agar cultures; and γ hemolysis, or non-hemolysis, when no damage to the cells is caused and no change in the agar plate is observed [75,76].
4.7. Statistical Analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences software, SPSS (version 26.0). Categories were compared using the chi-square test and Fisher’s exact test. Antibiotic resistance scores were compared between groups using one-way analysis of variance (ANOVA). The Kruskal–Wallis test was used to determine the association between resistance profiles and OD averages. The significance level was set at p < 0.05.
5. Conclusions
This study demonstrated that imipenem-resistant UPEC strains found in Algerian UTIs belonged mainly to phylogenetic groups B2 and E. Phylogenetic group B2 displayed heightened virulence attributes, including ESBL production, biofilm formation, and hemolysin activity. The majority of the examined isolates demonstrated a weak biofilm-forming capacity. In addition, our analysis showed that β-lactam antibiotics were ineffective against E. coli isolates, while aminoglycosides exhibited pronounced efficacy. Furthermore, MDR strains were weak biofilm producers. A variable relationship between antibiotic resistance and biofilm production was evidenced. In conclusion, this study confirms diversity and heterogeneity among imipenem-resistant UPEC strains and the complex association between biofilm production and antibiotic resistance profiles in UTIs caused by UPEC. The detection of the prevalence of phylogenetic groups, antibiotic resistance profiles, and virulence factors among urinary E. coli strains in Algeria will help in understanding the epidemiology of urine pathogens in the north-eastern of Algeria and developing the most appropriate treatment and prevention strategies for UTIs, to contain the spread of antimicrobial resistance and to avoid treatment failure in this geographical area.
Author Contributions
Investigation, A.K. and C.M.; resources, R.A. and N.B.; data curation, N.B.; writing—original draft preparation, A.K. and C.M.; writing—review and editing, G.M.G., R.A., and N.B.; supervision, R.A.; funding acquisition, N.B. and R.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been financially supported by the European Commission—NextGenerationEU, Project SUS-MIRRI.IT “Strengthening the MIRRI Italian Research Infrastructure for Sustainable Bioscience and Bioeconomy”, code n. IR0000005PO and by European Commission—NextGenerationEU, Piano Nazionale Resistenza e Resilienza (PNRR)—Missione 4 Componente 2 Investimento 1.4—Avviso N. 3138 del 16 dicembre 2021 rettificato con D.D. n.3175 del 18 dicembre 2021 del Ministero dell’Università e della Ricerca—CN5 “National Biodiversity Future Center”—NBFC—code n. CN00000033.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study by Ethics and Deontology Committee in University Ferhat Abbas of Setif-1- under the study registered with the number of the paper, UFAS1/09/03/2023/ETH-Deon-A-301, and title, Ethical Approval. In total, 86 urine samples were taken from the study’s recruited participants for various analyses. Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Coura, F.M.; Diniz, A.N.; Oliveira Junior, C.A.; Lage, A.P.; Lobato, F.C.F.; Heinemann, M.B.; Silva, R.O.S. Detection of Virulence Genes and the Phylogenetic Groups of Escherichia coli Isolated from Dogs in Brazil. Ciênc. Rural 2018, 48, e20170478. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The Population Genetics of Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Bozorgomid, A.; Chegene Lorestani, R.; Rostamian, M.; Nemati Zargaran, F.; Shahvaisi-Zadeh, Z.; Akya, A. Antibiotic Resistance, Virulence Factors, and Phylogenetic Groups of Escherichia coli Isolated from Hospital Wastewater: A Case Study in the West of Iran. Environ. Health Eng. Manag. 2023, 10, 131–139. [Google Scholar] [CrossRef]
- Saralaya, V.; Shenoy, S.; Baliga, S.; Hegde, A.; Adhikari, P.; Chakraborty, A. Characterization of Escherichia coli Phylogenetic Groups Associated with Extraintestinal Infections in South Indian Population. Ann. Med. Health Sci. Res. 2015, 5, 241. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Naghmachi, M.; Ghatee, M.A. Detection of Phylogenetic Groups and Drug Resistance Genes of Escherichia coli Causing Urinary Tract Infection in Southwest Iran. Jundishapur J. Microbiol. 2021, 14, e112547. [Google Scholar] [CrossRef]
- Rijavec, M.; Müller-Premru, M.; Zakotnik, B.; Žgur-Bertok, D. Virulence Factors and Biofilm Production among Escherichia coli Strains Causing Bacteraemia of Urinary Tract Origin. J. Med. Microbiol. 2008, 57, 1329–1334. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Nazari, S.; Farahani, O. Phylogenetic Analysis and Antimicrobial Resistance Profiles of Escherichia coli Strains Isolated from UTI-Suspected Patients. Iran. J. Public Health 2020, 49, 1743. [Google Scholar] [CrossRef] [PubMed]
- Nemattalab, M.; Rohani, M.; Evazalipour, M.; Hesari, Z. Formulation of Cinnamon (Cinnamomum Verum) Oil Loaded Solid Lipid Nanoparticles and Evaluation of Its Antibacterial Activity against Multi-Drug Resistant Escherichia coli. BMC Complement. Med. Ther. 2022, 22, 289. [Google Scholar] [CrossRef]
- Nowrouzian, F.L.; Clermont, O.; Edin, M.; Östblom, A.; Denamur, E.; Wold, A.E.; Adlerberth, I. Escherichia coli B2 Phylogenetic Subgroups in the Infant Gut Microbiota: Predominance of Uropathogenic Lineages in Swedish Infants and Enteropathogenic Lineages in Pakistani Infants. Appl. Environ. Microbiol. 2019, 85, e01681-19. [Google Scholar] [CrossRef]
- Bakhtiari, S.; Mahmoudi, H.; Khosravi Seftjani, S.; Amirzargar, M.A.; Ghiasvand, S.; Ghaffari, M.E.; Adabi, M. Antibiotic Resistance Pattern and Phylogenetic Groups of the Uropathogenic Escherichia coli Isolates from Urinary Tract Infections in Hamedan, West of Iran. Iran. J. Microbiol. 2020, 12, 388. [Google Scholar] [CrossRef]
- Baponi, S.; Taravati, A.; Dilmagani, M. Determinationof phylogenetic groups of Escherichia coli isolated fromhuman urine in Urmia city. Crescent J. Med. Biol. Sci. 2016, 3, 97–99. [Google Scholar]
- Halaji, M.; Fayyazi, A.; Rajabnia, M.; Zare, D.; Pournajaf, A.; Ranjbar, R. Phylogenetic Group Distribution of Uropathogenic Escherichia coli and Related Antimicrobial Resistance Pattern: A Meta-Analysis and Systematic Review. Front. Cell. Infect. Microbiol. 2022, 12, 790184. [Google Scholar] [CrossRef]
- Staji, H.; Khoshgoftar, J.; Javaheri Vayeghan, A.; Salimi Bejestani, M.R. Phylogenetic Grouping and Assessment of Virulence Genotypes, With Antibiotic Resistance Patterns, of Escherichia coli Strains Implicated in Female Urinary Tract Infections. Int. J. Enteric Pathog. 2016, 4, e31609. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal Gut Bacteria: Distribution of Enterococcus Species and Prevalence of Escherichia coli Phylogenetic Groups in Animals and Humans in Portugal. Ann. Microbiol. 2012, 62, 449–459. [Google Scholar] [CrossRef]
- Stoppe, N.D.C.; Silva, J.S.; Carlos, C.; Sato, M.I.Z.; Saraiva, A.M.; Ottoboni, L.M.M.; Torres, T.T. Worldwide Phylogenetic Group Patterns of Escherichia coli from Commensal Human and Wastewater Treatment Plant Isolates. Front. Microbiol. 2017, 8, 2512. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Talavera Rodríguez, A.; Roy, S.; Hossain, M.I.; Islam, M.A.; Lanza, V.F.; Julian, T.R. High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings. mSphere 2020, 5, e00704-19. [Google Scholar] [CrossRef] [PubMed]
- Fasugba, O.; Das, A.; Mnatzaganian, G.; Mitchell, B.G.; Collignon, P.; Gardner, A. Incidence of Single-Drug Resistant, Multidrug-Resistant and Extensively Drug-Resistant Escherichia coli Urinary Tract Infections: An Australian Laboratory-Based Retrospective Study. J. Glob. Antimicrob. Resist. 2019, 16, 254–259. [Google Scholar] [CrossRef]
- Al Nafeesah, A.; Al Fakeeh, K.; Chishti, S.; Hameed, T. E. coli. versus Non-E. coli. Urinary Tract Infections in Children: A Study from a Large Tertiary Care Center in Saudi Arabia. Int. J. Pediatr. Adolesc. Med. 2022, 9, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Jalil, M.B.; Al Atbee, M.Y.N. The Prevalence of Multiple Drug Resistance Escherichia coli and Klebsiella Pneumoniae Isolated from Patients with Urinary Tract Infections. J. Clin. Lab. Anal. 2022, 36, e24619. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, Z.; Zheng, L.; Gong, Z.; Li, Y.; Jin, Y.; Huang, Y.; Chi, M. Urinary Tract Infections Caused by Uropathogenic Escherichia Coli: Mechanisms of Infection and Treatment Options. Int. J. Mol. Sci. 2023, 24, 10537. [Google Scholar] [CrossRef]
- Mohsin, A.S.; Alsakini, A.H.; Ali, M.R. Outbreak of Drug Resistance Escherichia coli Phylogenetic F Group Associated Urinary Tract Infection. Iran. J. Microbiol. 2022, 14, 341. [Google Scholar] [CrossRef] [PubMed]
- Shea, A.E.; Stocki, J.A.; Himpsl, S.D.; Smith, S.N.; Mobley, H.L.T. Loss of an Intimin-Like Protein Encoded on a Uropathogenic E. coli. Pathogenicity Island Reduces Inflammation and Affects Interactions with the Urothelium. Infect. Immun. 2022, 90, e00275-21. [Google Scholar] [CrossRef] [PubMed]
- Kabugo, D.; Kizito, S.; Ashok, D.D.; Kiwanuka, A.G.; Nabimba, R.; Namunana, S.; Kabaka, R.M.; Achan, B.; Najjuka, F.C. Factors Associated with Community-Acquired Urinary Tract Infections among Adults Attending Assessment Centre, Mulago Hospital Uganda. Afr. Health Sci. 2017, 16, 1131. [Google Scholar] [CrossRef] [PubMed]
- Dadi, B.R.; Abebe, T.; Zhang, L.; Mihret, A.; Abebe, W.; Amogne, W. Distribution of Virulence Genes and Phylogenetics of Uropathogenic Escherichia coli among Urinary Tract Infection Patients in Addis Ababa, Ethiopia. BMC Infect. Dis. 2020, 20, 108. [Google Scholar] [CrossRef]
- Hyun, M.; Lee, J.Y.; Kim, H.A. Differences of Virulence Factors, and Antimicrobial Susceptibility According to Phylogenetic Group in Uropathogenic Escherichia coli Strains Isolated from Korean Patients. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Millán, Y.; Araque, M.; Ramírez, A. Distribución de Grupos Filogenéticos, Factores de Virulencia y Susceptibilidad Antimicrobiana En Cepas de Escherichia coli Uropatógena. Rev. Chil. Infectol. 2020, 37, 117–123. [Google Scholar] [CrossRef]
- Ahmed, A.E.; Abdelkarim, S.; Zenida, M.; Baiti, M.A.H.; Alhazmi, A.A.Y.; Alfaifi, B.A.H.; Majrabi, R.Q.M.; Khormi, N.Q.M.; Hakami, A.A.A.; Alqaari, R.A.M.; et al. Prevalence and Associated Risk Factors of Urinary Tract Infection among Diabetic Patients: A Cross-Sectional Study. Healthcare 2023, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- Baldiris-Avila, R.; Montes-Robledo, A.; Buelvas-Montes, Y. Phylogenetic Classification, Biofilm-Forming Capacity, Virulence Factors, and Antimicrobial Resistance in Uropathogenic Escherichia coli (UPEC). Curr. Microbiol. 2020, 77, 3361–3370. [Google Scholar] [CrossRef]
- Elsayed Gawad, W.; Mohamed Helmy, O.; Mostafa Tawakkol, W.; Mohamed Hashem, A. Antimicrobial Resistance, Biofilm Formation, and Phylogenetic Grouping of Uropathogenic Escherichia coli Isolates in Egypt: The Role of Efflux Pump-Mediated Resistance. Jundishapur J. Microbiol. 2018, 11, e14444. [Google Scholar] [CrossRef]
- Agarwal, J.; Mishra, B.; Srivastava, S.; Srivastava, R. Genotypic Characteristics and Biofilm Formation among Escherichia coli Isolates from Indian Women with Acute Cystitis. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 183–187. [Google Scholar] [CrossRef]
- Moez, N.M.; Mashouf, R.Y.; Sedighi, I.; Shokoohizadeh, L.; Taheri, M. Phylogroup Classification and Investigation the Relationships between Phylogroups and Antibiotic Resistance Patterns of Uropathogenic E. coli. Isolated from Pediatric Urinary Tract Infection. Gene Rep. 2020, 20, 100758. [Google Scholar] [CrossRef]
- Hassuna, N.A.; Khairalla, A.S.; Farahat, E.M.; Hammad, A.M.; Abdel-Fattah, M. Molecular Characterization of Extended-Spectrum β Lactamase- Producing E. coli. Recovered from Community-Acquired Urinary Tract Infections in Upper Egypt. Sci. Rep. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Farahat, E.M.; Hassuna, N.A.; Hammad, A.M.; Fattah, M.A.; Khairalla, A.S. Distribution of Integrons and Phylogenetic Groups among Escherichia coli Causing Community-Acquired Urinary Tract Infection in Upper Egypt. Can. J. Microbiol. 2021, 67, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.; Najar Peerayeh, S.; Mohabati Mobarez, A.; Bakhshi, B.; Karbalaei, M.; Abdolvand, Z. Phylogenetic Groups/B2 Subgroup Distributions, Serogrouping and Identification of Virulence Factors in Extended-Spectrum Cephalosporin-Resistant Escherichia coli Strains Isolated from the Stool of Healthy Children Under 10 Years Old. Arch. Pediatr. Infect. Dis. 2022, 10, e118889. [Google Scholar] [CrossRef]
- Hogins, J.; Xuan, Z.; Zimmern, P.E.; Reitzer, L. The Distinct Transcriptome of Virulence-Associated Phylogenetic Group B2 Escherichia coli. Microbiol. Spectr. 2023, 11, e02085-23. [Google Scholar] [CrossRef]
- Gunathilaka, G.A.D.K.K.; Dewasmika, W.A.P.M.; Sandaruwan, U.M.; Neelawala, N.G.D.A.K.; Madhumali, G.E.D.; Dissanayake, B.N.; Priyantha, M.A.R.; Prasada, D.V.P.; Dissanayake, D.R.A. Biofilm-Forming Ability, Antibiotic Resistance and Phylogeny of Escherichia coli Isolated from Extra Intestinal Infections of Humans, Dogs, and Chickens. Comp. Immunol. Microbiol. Infect. Dis. 2024, 105, 102123. [Google Scholar] [CrossRef]
- Suay-García, B.; Pérez-Gracia, M.T. Present and Future of Carbapenem-Resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lv, C.; Li, M.; Rahman, T.; Chang, Y.-F.; Guo, X.; Song, Z.; Zhao, Y.; Li, Q.; Ni, P.; et al. Carbapenem-Resistant Escherichia coli Exhibit Diverse Spatiotemporal Epidemiological Characteristics across the Globe. Commun. Biol. 2024, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Munkhdelger, Y.; Gunregjav, N.; Dorjpurev, A.; Juniichiro, N.; Sarantuya, J. Detection of Virulence Genes, Phylogenetic Group and Antibiotic Resistance of Uropathogenic Escherichia coli in Mongolia. J. Infect. Dev. Ctries. 2017, 11, 51–57. [Google Scholar] [CrossRef]
- Tewawong, N.; Kowaboot, S.; Pimainog, Y.; Watanagul, N.; Thongmee, T.; Poovorawan, Y. Distribution of Phylogenetic Groups, Adhesin Genes, Biofilm Formation, and Antimicrobial Resistance of Uropathogenic Escherichia coli Isolated from Hospitalized Patients in Thailand. PeerJ 2020, 8, e10453. [Google Scholar] [CrossRef]
- Mouanga Ndzime, Y.; Onanga, R.; Kassa Kassa, R.F.; Bignoumba, M.; Mbehang Nguema, P.P.; Gafou, A.; Lendamba, R.W.; Mbombe Moghoa, K.; Bisseye, C. Epidemiology of Community Origin Escherichia coli and Klebsiella Pneumoniae Uropathogenic Strains Resistant to Antibiotics in Franceville, Gabon. Infect. Drug Resist. 2021, 14, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Muvunyi, C.M.; Masaisa, F.; Musemakweri, A.; Muhirwa, G.; Claeys, G.W.; Bayingana, C.; Mutesa, L. Decreased Susceptibility to Commonly Used Antimicrobial Agents in Bacterial Pathogens Isolated from Urinary Tract Infections in Rwanda: Need for New Antimicrobial Guidelines. Am. J. Trop. Med. Hyg. 2011, 84, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Jeanvoine, A.; Bouxom, H.; Leroy, J.; Gbaguidi-Haore, H.; Bertrand, X.; Slekovec, C. Resistance to Third-Generation Cephalosporins in Escherichia coli in the French Community: The Times They Are a-Changin’? Int. J. Antimicrob. Agents 2020, 55, 105909. [Google Scholar] [CrossRef] [PubMed]
- Naziri, Z.; Derakhshandeh, A.; Soltani Borchaloee, A.; Poormaleknia, M.; Azimzadeh, N. Treatment Failure in Urinary Tract Infections: A Warning Witness for Virulent Multi-Drug Resistant ESBL- Producing Escherichia Coli. Infect. Drug Resist. 2020, 13, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Iranpour, D.; Hassanpour, M.; Ansari, H.; Tajbakhsh, S.; Khamisipour, G.; Najafi, A. Phylogenetic Groups of Escherichia coli Strains from Patients with Urinary Tract Infection in Iran Based on the New Clermont Phylotyping Method. BioMed Res. Int. 2015, 2015, 846219. [Google Scholar] [CrossRef] [PubMed]
- Calhau, V.; Domingues, S.; Ribeiro, G.; Mendonça, N.; Da Silva, G.J. Interplay between Pathogenicity Island Carriage, Resistance Profile and Plasmid Acquisition in Uropathogenic Escherichia Coli. J. Med. Microbiol. 2015, 64, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, S.; Cui, L.; Yang, H.; Yi, L. Phylogenetic Group, Biofilm Formation and Drug Resistance of Escherichia coli Isolated from Goose and Fish in Henan, China. Indian J. Anim. Res. 2023, 57, 1375–1379. [Google Scholar] [CrossRef]
- Norouzian, H.; Katouli, M.; Shahrokhi, N.; Sabeti, S.; Pooya, M.; Bouzari, S. The Relationship between Phylogenetic Groups and Antibiotic Susceptibility Patterns of Escherichia coli Strains Isolated from Feces and Urine of Patients with Acute or Recurrent Urinary Tract Infection. Iran. J. Microbiol. 2019, 11, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-C.; Fan, Y.-H.; Zhang, Y.-Z.; Bregente, C.J.B.; Lin, W.-H.; Chen, C.-A.; Lin, T.-P.; Kao, C.-Y. Characterization of Uropathogenic Escherichia coli Phylogroups Associated with Antimicrobial Resistance, Virulence Factor Distribution, and Virulence-Related Phenotypes. Infect. Genet. Evol. 2023, 114, 105493. [Google Scholar] [CrossRef]
- A Kadry, A.; M Al-Kashef, N.; M El-Ganiny, A. Distribution of Genes Encoding Adhesins and Biofilm Formation Capacity among Uropathogenic Escherichia coli Isolates in Relation to the Antimicrobial Resistance. Afr. Health Sci. 2020, 20, 238–247. [Google Scholar] [CrossRef]
- Poeta, P.; Radhouani, H.; Pinto, L.; Martinho, A.; Rego, V.; Rodrigues, R.; Gonçalves, A.; Rodrigues, J.; Estepa, V.; Torres, C.; et al. Wild Boars as Reservoirs of Extended-spectrum Beta-lactamase (ESBL) Producing Escherichia coli of Different Phylogenetic Groups. J. Basic Microbiol. 2009, 49, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Mirkalantari, S.; Masjedian, F.; Irajian, G.; Siddig, E.E.; Fattahi, A. Determination of the Frequency of β-Lactamase Genes (Bla SHV, Bla TEM, Bla CTX-M) and Phylogenetic Groups among ESBL-Producing Uropathogenic Escherichia coli Isolated from Outpatients. J. Lab. Med. 2020, 44, 27–33. [Google Scholar] [CrossRef]
- Rahimifard, B.; Soheili, V.; Hashemitabar, G.; Askari Badouei, M. Possible Relationship of Novel Phylogenetic Structure With Antimicrobial Resistance, Biofilm Formation, and Hemolytic Activity in Uropathogenic Escherichia coli (UPEC). Int. J. Enteric Pathog. 2022, 10, 98–104. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for Combating Bacterial Biofilms: A Focus on Anti-Biofilm Agents and Their Mechanisms of Action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Hashemizadeh, Z.; Kalantar-Neyestanaki, D.; Mansouri, S. Association between Virulence Profile, Biofilm Formation and Phylogenetic Groups of Escherichia coli Causing Urinary Tract Infection and the Commensal Gut Microbiota: A Comparative Analysis. Microb. Pathog. 2017, 110, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, G.; Khadka, P.; Siddhi Shilpakar, G.; Chapagain, G.; Dhungana, G.R. Catheter-Associated Urinary Tract Infection and Obstinate Biofilm Producers. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 7624857. [Google Scholar] [CrossRef]
- Soto, S.M.; Smithson, A.; Martinez, J.A.; Horcajada, J.P.; Mensa, J.; Vila, J. Biofilm Formation in Uropathogenic Escherichia coli Strains: Relationship With Prostatitis, Urovirulence Factors and Antimicrobial Resistance. J. Urol. 2007, 177, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Javed, S.; Mirani, Z.A.; Pirzada, Z.A. Phylogenetic Group B2 Expressed Significant Biofilm Formation among Drug Resistant Uropathogenic Escherichia Coli. Libyan J. Med. 2021, 16, 1845444. [Google Scholar] [CrossRef]
- Matinfar, S.; Ahmadi, M.; Sisakht, A.M.; Sadeghi, J.; Javedansirat, S. Phylogenetic and Antibiotics Resistance in Extended-Spectrum B-Lactamase (ESBL) Uropathogenic Escherichia Coli: An Update Review. Gene Rep. 2021, 23, 101168. [Google Scholar] [CrossRef]
- Katongole, P.; Nalubega, F.; Florence, N.C.; Asiimwe, B.; Andia, I. Biofilm Formation, Antimicrobial Susceptibility and Virulence Genes of Uropathogenic Escherichia coli Isolated from Clinical Isolates in Uganda. BMC Infect. Dis. 2020, 20, 453. [Google Scholar] [CrossRef]
- Behzadi, P.; Urbán, E.; Gajdács, M. Association between Biofilm-Production and Antibiotic Resistance in Uropathogenic Escherichia coli (UPEC): An In Vitro Study. Diseases 2020, 8, 17. [Google Scholar] [CrossRef]
- Gajdács, M.; Kárpáti, K.; Nagy, Á.L.; Gugolya, M.; Stájer, A.; Burián, K. Association between Biofilm-Production and Antibiotic Resistance in Escherichia coli Isolates: A Laboratory-Based Case Study and a Literature Review. Acta Microbiol. Immunol. Hung. 2021, 68, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship Between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Chlebicki, M. Urinary Tract Infections in Adults. Singapore Med. J. 2016, 57, 485–490. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. 2022. Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2022/05/CASFM2022_V1.0.pdf (accessed on 7 August 2024).
- Kaur, J. Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella Pneumoniae. J. Clin. Diagn. Res. 2013, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Woodman, M.E.; Savage, C.R.; Arnold, W.K.; Stevenson, B. Direct PCR of Intact Bacteria (Colony PCR). Curr. Protoc. Microbiol. 2016, 42, A-3D. [Google Scholar] [CrossRef] [PubMed]
- Coy, M.R.; Hoffmann, M.; Kingdom Gibbard, H.N.; Kuhns, E.H.; Pelz-Stelinski, K.S.; Stelinski, L.L. Nested-Quantitative PCR Approach with Improved Sensitivity for the Detection of Low Titer Levels of Candidatus Liberibacter Asiaticus in the Asian Citrus Psyllid, Diaphorina Citri Kuwayama. J. Microbiol. Methods 2014, 102, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-typing Method Revisited: Improvement of Specificity and Detection of New Phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Characterization of an Anonymous Molecular Marker Strongly Linked to Escherichia coli Strains Causing Neonatal Meningitis. J. Clin. Microbiol. 2004, 42, 1770–1772. [Google Scholar] [CrossRef][Green Version]
- Lescat, M.; Clermont, O.; Woerther, P.L.; Glodt, J.; Dion, S.; Skurnik, D.; Djossou, F.; Dupont, C.; Perroz, G.; Picard, B.; et al. Commensal Escherichia coli Strains in Guiana Reveal a High Genetic Diversity with Host-dependant Population Structure. Environ. Microbiol. Rep. 2013, 5, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Lescat, M.; O’Brien, C.L.; Gordon, D.M.; Tenaillon, O.; Denamur, E. Evidence for a Human-specific Escherichia coli Clone. Environ. Microbiol. 2008, 10, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Buxton, R. Blood Agar Plates and Hemolysis Protocols. 2005. Available online: https://asm.org/getattachment/7ec0de2b-bb16-4f6e-ba07-2aea25a43e76/protocol-28 (accessed on 7 August 2024).
- Mogrovejo-Arias, D.C.; Brill, F.H.H.; Wagner, D. Potentially Pathogenic Bacteria Isolated from Diverse Habitats in Spitsbergen, Svalbard. Environ. Earth Sci. 2020, 79, 109. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).