Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan
Abstract
1. Introduction
2. Results
2.1. Prevalence of B. cereus s. s. among the Samples Tested
2.2. Identification and Classification of B. cereus s. s. Isolates Using Matrix-Assisted Laser Desorption Ionization/Time of Flight Mass Spectrometry (MALDI-TOF MS)
2.3. Virulence Gene Distribution among B. cereus s. s. Isolates
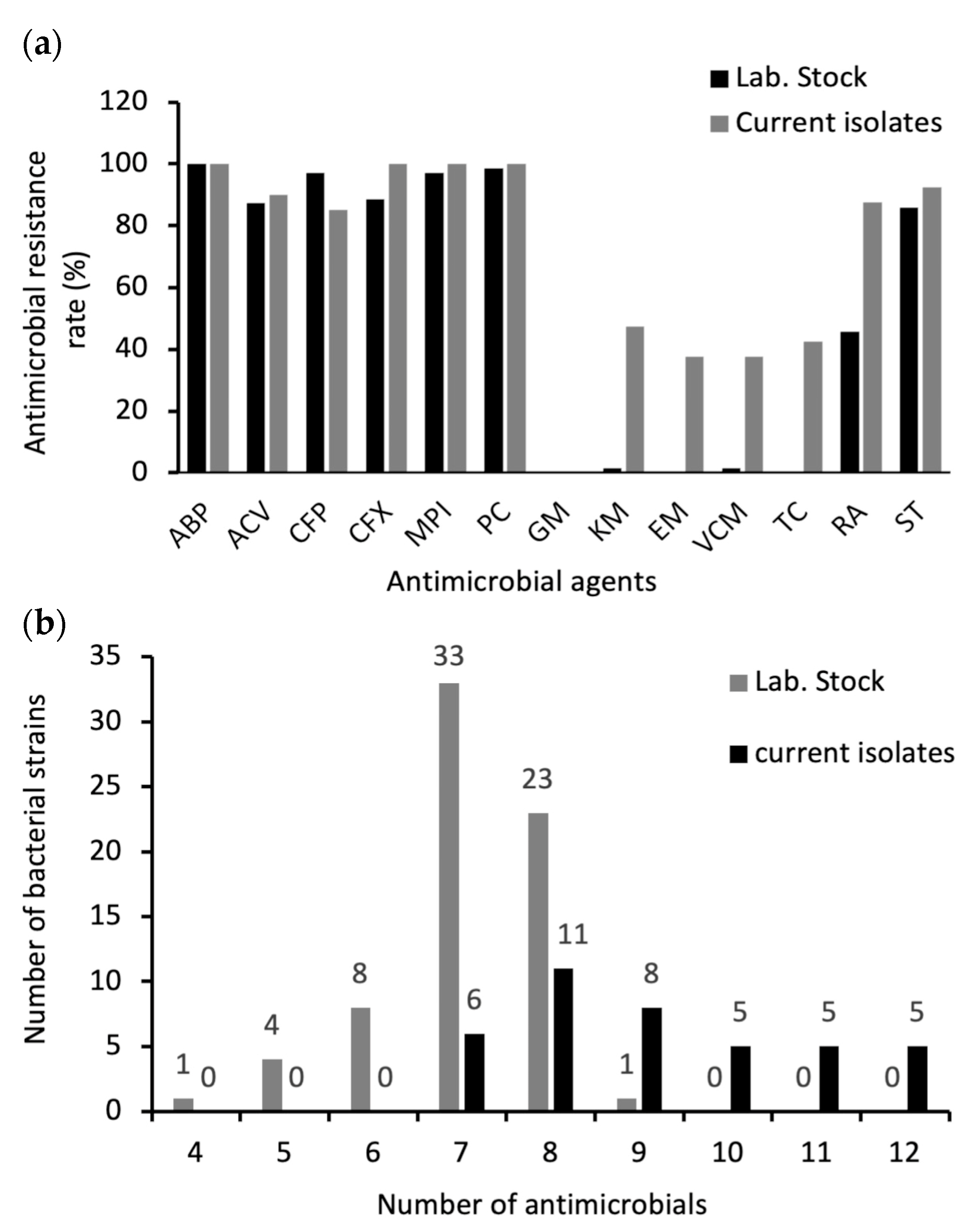
2.4. Antimicrobial Susceptibility of B. cereus s. s. Isolates
2.5. Distribution of Antimicrobial Resistance Genes in B. cereus s. s. Isolates
2.6. Growth Profiles of B. cereus s. s. Isolates at 7 °C
2.7. Biofilm Formation of B. cereus s. s. Isolates
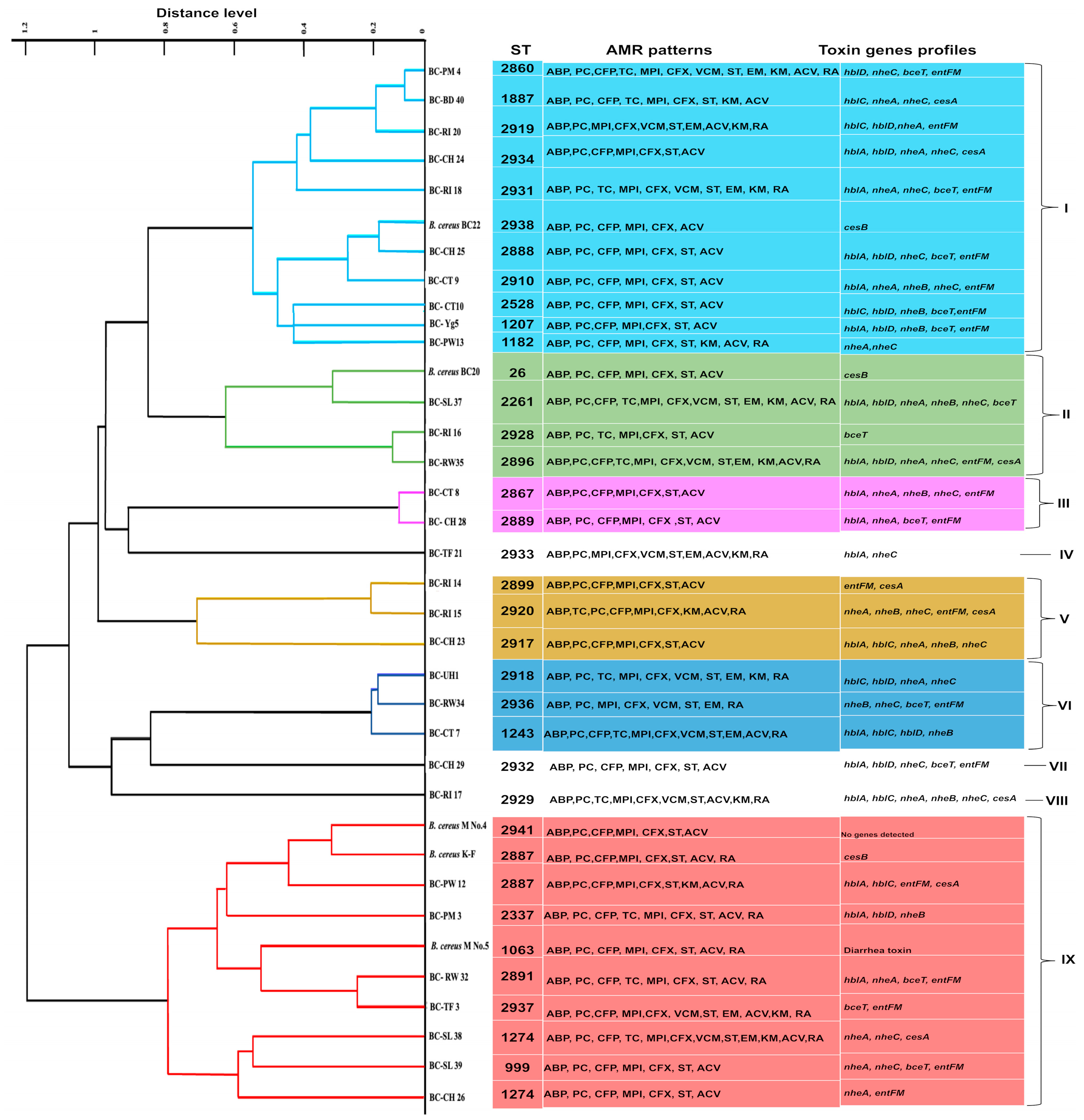
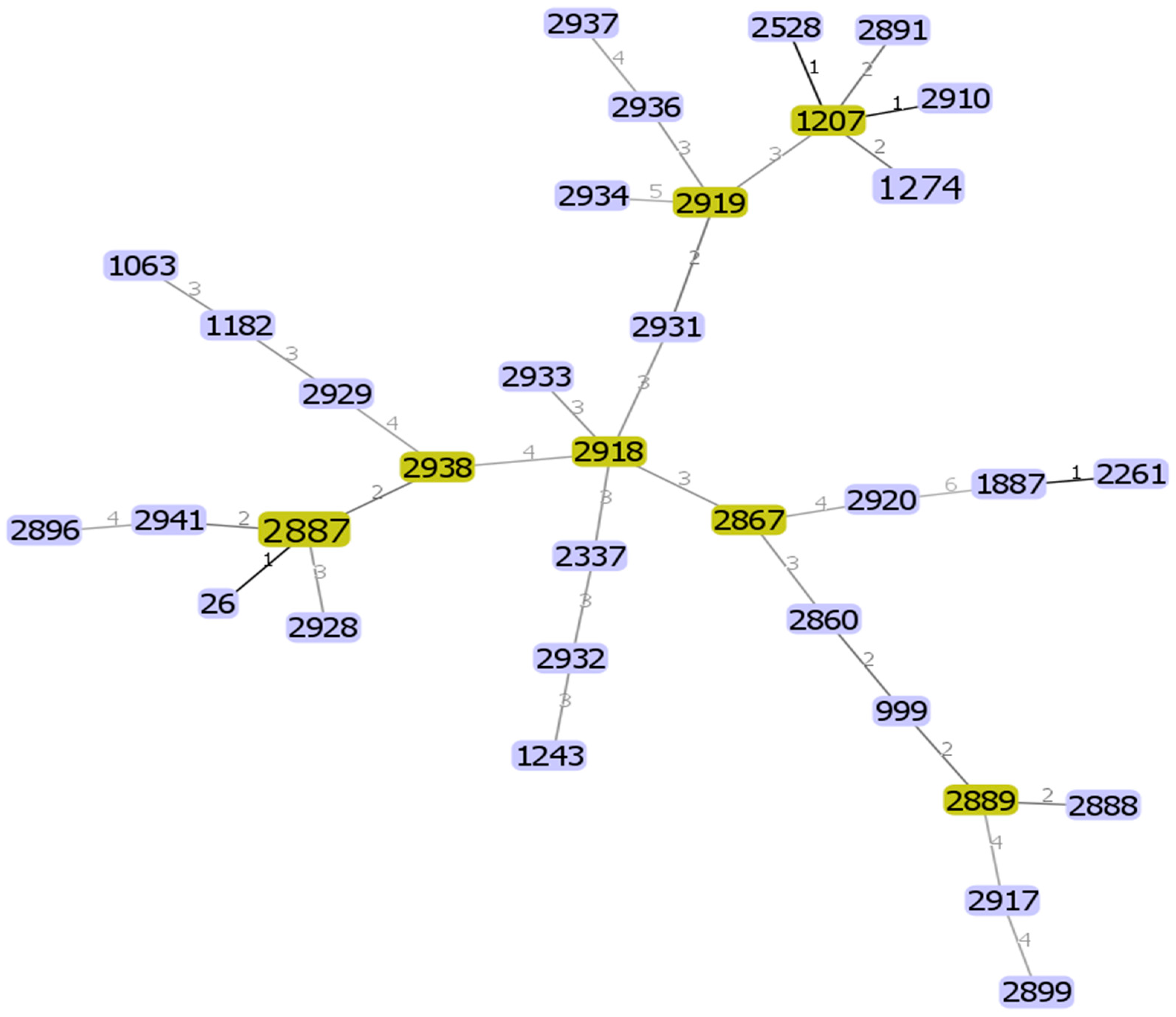
2.8. Genetic Diversity of B. cereus s. s. Isolates
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation and Identification of Bacterial Strains
4.3. MALDI-TOF MS Analysis of B. cereus s. s. Isolates
4.4. Molecular Characterization and Toxin Gene Profiling of B. cereus s. s.
4.5. Antimicrobial Resistance Testing
4.5.1. Disk Diffusion Method
4.5.2. Molecular Identification of Genes Involved in Antibiotic Resistance
4.6. Discrimination of Psychrotrophic Strains of B. cereus s. s.
4.7. Biofilm Formation Assay
4.8. Multi-Locus Sequence Typing
4.8.1. DNA Extraction
4.8.2. Genes Amplification, Sequencing, and Determination
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naas, H.; Zurghani, M.; Garbaj, A.; Azwai, S.; Eshamah, H.; Gammoudi, F.; Abolghait, S.; Moawad, A.; Barbieri, I.; Eldaghayes, I. Bacillus cereus as an Emerging Public Health Concern in Libya: Isolation and Antibiogram from Food of Animal Origin. Libyan J. Med. Sci. 2018, 2, 56. [Google Scholar] [CrossRef]
- Berthold-Pluta, A.; Pluta, A.; Garbowska, M.; Stefańska, I. Prevalence and Toxicity Characterization of Bacillus cereus in Food Products from Poland. Foods 2019, 8, 269. [Google Scholar] [CrossRef]
- da Silva, N.B.; Baranyi, J.; Carciofi, B.A.M.; Ellouze, M. From Culture-Medium-Based Models to Applications to Food: Predicting the Growth of B. cereus in Reconstituted Infant Formulae. Front. Microbiol. 2017, 8, 01799. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Kim, H.J.; Jeong, M.; Koo, M. Enterotoxin Genes, Antibiotic Susceptibility, and Biofilm Formation of Low-Temperature-Tolerant Bacillus cereus Isolated from Green Leaf Lettuce in the Cold Chain. Foods 2020, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Rishi, E.; Rishi, P.; Sengupta, S.; Jambulingam, M.; Madhavan, H.N.; Gopal, L.; Therese, K.L. Acute Postoperative Bacillus cereus Endophthalmitis Mimicking Toxic Anterior Segment Syndrome. Ophthalmology 2013, 120, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Yagihara, Y.; Tatsuno, K.; Okazaki, M.; Okugawa, S.; Moriya, K. Clinical Characteristics and Antimicrobial Susceptibility of Bacillus cereus Blood Stream Infections. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and Pathogenesis of Bacillus cereus Infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, J.; Ornelis, V.F.; Madder, A.; Rajkovic, A. Bacillus cereus food intoxication and toxicoinfection. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3719–3761. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, D.; Fortunato, F.; Tafuri, S.; Cozza, V.; Chironna, M.; Germinario, C.; Prato, R. Lessons learnt from a birthday party: A Bacillus cereus outbreak, Bari, Italy, January 2012. Ann. Dell’istituto Super. Di Sanita 2013, 49, 391–394. [Google Scholar] [CrossRef]
- Delbrassinne, L.; Botteldoorn, N.; Andjelkovic, M.; Dierick, K.; Denayer, S. An emetic Bacillus cereus outbreak in a kindergarten: Detection and quantification of critical levels of cereulide toxin. Foodborne Pathog. Dis. 2015, 12, 84–87. [Google Scholar] [CrossRef]
- Bennett, S.D.; Walsh, K.A.; Gould, L.H. Foodborne Disease Outbreaks Caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus—United States, 1998–2008. Clin. Infect. Dis. 2013, 57, 425–433. [Google Scholar] [CrossRef]
- ECDC/EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef]
- Ishitobi, E. Bacillus cereus-associated food poisoning in milk. Food Hyg. Saf. Sci. 1991, 32, 139–143. [Google Scholar]
- Torii, T.; Ohkubo, Y. Distribution of Cereulide-Producing Bacillus cereus in Raw Milk in Hokkaido, Japan, and Evaluation of Cereulide Production. Int. Dairy J. 2023, 144, 105693. [Google Scholar] [CrossRef]
- Agata, N.; Ohta, M.; Mori, M.; Isobe, M. A Novel Dodecadepsipeptide, Cereulide, Is an Emetic Toxin of Bacillus cereus. FEMS Microbiol. Lett. 1995, 129, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Sasahara, T.; Hayashi, S.; Morisawa, Y.; Sakihama, T.; Yoshimura, A.; Hirai, Y. Bacillus cereus Bacteremia Outbreak Due to Contaminated Hospital Linens. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Barrie, D.; Hoffman, P.N.; Wilson, J.A.; Kramer, J.M. Contamination of Hospital Linen by Bacillus cereus. Epidemiol. Infect. 1994, 113, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Shigemi, H.; Suzuki, K.; Yasutomi, M.; Iwasaki, H.; Ohshima, Y. Successful Management of a Bacillus cereus Catheter-Related Bloodstream Infection Outbreak in the Pediatric Ward of Our Facility. J. Infect. Chemother. 2019, 25, 873–879. [Google Scholar] [CrossRef]
- Griffiths, M.W.; Schraft, H. Bacillus cereus Food Poisoning. In Foodborne Diseases, 3rd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 395–405. ISBN 9780123850072. [Google Scholar]
- de Noordhout, C.M.; Devleesschauwer, B.; Haagsma, J.A.; Havelaar, A.H.; Bertrand, S.; Vandenberg, O.; Quoilin, S.; Brandt, P.T.; Speybroeck, N. Burden of Salmonellosis, Campylobacteriosis and Listeriosis: A Time Series Analysis, Belgium, 2012 to 2020. Eurosurveillance 2017, 22, 6–18. [Google Scholar] [CrossRef]
- Mao, X.; Hu, J.; Liu, X. Epidemiological analysis of 1060 bacterial foodborne diseases in China in 2003–2007. Chin. J. Food Hyg. 2010, 22, 224–228. [Google Scholar]
- Frenzel, E.; Kranzler, M.; Stark, T.D.; Hofmann, T.; Ehling-Schulz, M. The endospore-forming pathogen Bacillus cereus exploits a small colony variant-based diversification strategy in response to aminoglycoside exposure. mBio 2015, 6, e01172-15. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Risks for public health related to the presence of Bacillus cereus and other Bacillus spp., including Bacillus thuringiensis, in foodstuffs. EFSA J. 2016, 14, 4524. [Google Scholar]
- Ibrahim, A.S.; Hafiz, N.M.; Saad, M.F. Prevalence of Bacillus cereus in dairy powders focusing on its toxigenic genes and antimicrobial resistance. Arch. Microbiol. 2022, 204, 339. [Google Scholar] [CrossRef] [PubMed]
- Decousser, J.-W.; Ramarao, N.; Duport, C.; Dorval, M.; Bourgeois-Nicolaos, N.; Guinebretière, M.-H.; Razafimahefa, H.; Doucet-Populaire, F. Bacillus cereus and severe intestinal infections in preterm neonates: Putative role of pooled breast milk. Am. J. Infect. Control 2013, 41, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Wendelboe, A.M.; Smelser, C.; Lucero, C.A.; McDonald, L.C. Cluster of necrotizing enterocolitis in a neonatal intensive care unit: New Mexico, 2007. Am. J. Infect. Control 2010, 38, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Fournier, S.; Faraut-Derouin, V.; Casseta, A.; Frange, P.; Doit, C.; Fortineau, N.; Romain, O.; Patkai, J.; de Chillaz, C.; Rigourd, V.; et al. Bactériémies à Bacillus cereus en réanimation néonatale à l’AP-HP en 2016. Bull. Epidemiol. Hebd. 2018, 25–26, 536–540. [Google Scholar]
- Liao, S.-L.; Tsai, M.-H. Bacillus cereus bacteremia in a preterm infant caused by consumption of contaminated breastmilk. Pediatr. Neonatol. 2021, 62, 337–338. [Google Scholar] [CrossRef]
- Lotte, R.; Chevalier, A.; Boyer, L.; Ruimy, R. Bacillus cereus invasive infections in preterm neonates: An up-to-date review of the literature. Clin. Microbiol. Rev. 2022, 35, e00088-21. [Google Scholar] [CrossRef]
- Hwang, J.; Park, J. Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J. Dairy Sci. 2015, 98, 1652–1660. [Google Scholar] [CrossRef]
- Hassan, Z.H. Psychrotolerant Bacillus cereus: An Emerging Pathogen from Foodborne Diseases. Int. Food Res. J. 2022, 29, 496–509. [Google Scholar] [CrossRef]
- Gao, T.; Ding, Y.; Wu, Q.; Wang, J.; Zhang, J.; Yu, S.; Yu, P.; Liu, C.; Kong, L.; Feng, Z.; et al. Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus cereus Isolated from Pasteurized Milk in China. Front. Microbiol. 2018, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Xie, Q.; Yang, J.; Ma, L.; Feng, H. The Prevalence and Characterization of Bacillus cereus Isolated from Raw and Pasteurized Buffalo Milk in Southwestern China. J. Dairy Sci. 2021, 104, 3980–3989. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, X.; Zhan, L.; Chen, J.; Zhang, Y.; Zhang, J.; Chen, H.; Zhang, Z.; Zhang, Y.; Lu, Y.; et al. Multilocus Sequence Type Profiles of Bacillus cereus Isolates from Infant Formula in China. Food Microbiol. 2017, 62, 46–50. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Mukherjee, M.; Nicholas, D.C.; Mingle, L.A.; Dumas, N.B.; Cole, J.A.; Kovac, J. Characterization of Emetic and Diarrheal Bacillus cereus Strains from a 2016 Foodborne Outbreak Using Whole-Genome Sequencing: Addressing the Microbiological, Epidemiological, and Bioinformatic Challenges. Front. Microbiol. 2019, 10, 144. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Frenzel, E.; Gohar, M. Food–bacteria interplay: Pathometabolism of emetic Bacillus cereus. Front. Microbiol. 2015, 6, 704. [Google Scholar] [CrossRef]
- Inatsu, Y.; Chotiko, A.; Ananchaipattana, C. Contaminated Bacillus cereus in Lao and Thai fermented soybean “Tua Nao”. Jpn. Agric. Res. Q. JARQ 2020, 54, 47–51. [Google Scholar] [CrossRef]
- Shinagawa, K. Analytical Methods for Bacillus cereus and Other Bacillus Species. Int. J. Food Microbiol. 1990, 10, 125–141. [Google Scholar] [CrossRef]
- Huruse, A.; Ushigome, T.; Tsuruta, M.; Matayoshi, Y.; Ootsuka, H. An outbreak of Bacillus cereus. Infect. Immun. Child. 2004, 16, 151–155. [Google Scholar]
- Matsuoka, Y.; Araya, T.; Hujii, K. A case of B. cereus food poisoning caused by rice dumpling with sweet bean paste. Rep. Kumamoto Pref. Res. Instit. Environ. Sci. 2003, 9, 33–38. [Google Scholar]
- Vidal, A.M.C.; Junior, O.D.R.; de Abreu, I.L.; Bürger, K.P.; Cardoso, M.V.; Gonçalves, A.C.S.; Rossi, G.A.M.; D’Abreu, L.F. Detecção de Bacillus cereus Isolado Durante o Fluxograma de Produção Do Leite Tratado Por Ultra Alta Temperatura Através Do DNA Polimórfico Amplificado Ao Acaso Através Da Reação Em Cadeia Pela Polimerase. Cienc. Rural 2016, 46, 286–292. [Google Scholar] [CrossRef]
- Vyletělová, M.; Hanuš, O.; Páčová, Z.; Roubal, P.; Kopunecz, P. Frequency of Bacillus bacteria in raw cow’s milk and its relation to other hygienic parameters. Czech J. Anim. Sci. 2001, 46, 260–267. [Google Scholar]
- Sornchuer, P.; Tiengtip, R. Prevalence, virulence genes, and antimicrobial resistance of Bacillus cereus isolated from foodstuffs in Pathum Thani Province, Thailand. Pharm. Sci. Asia 2021, 48, 194–203. [Google Scholar] [CrossRef]
- Kashiwagi, M.; Harada, T.; Hiroi, M.; Ohata, K.; Kawamori, F.; Sugiyama, K. Investigation of Bacillus cereus producing emetic toxin in various foods. Bull. Shizuoka Inst. Environ. Hyg. 2005, 48, 11–14. [Google Scholar]
- Abdel Samad, R.; Al Disi, Z.; Mohammad Ashfaq, M.Y.; Wahib, S.M.; Zouari, N. The Use of Principle Component Analysis and MALDI-TOF MS for the Differentiation of Mineral-Forming: Virgibacillus and Bacillus Species Isolated from Sabkhas. RSC Adv. 2020, 10, 14606–14616. [Google Scholar] [CrossRef]
- Maung, A.T.; Abdelaziz, M.N.S.; Mohammadi, T.N.; Zhao, J.; EI-Telbany, M.; Nakayama, M.; Matsusita, K.; Masuda, Y.; Honjoh, K.; Miyamoto, T. Comparison of Prevalence, Characterization, Antimicrobial Resistance and Pathogenicity of Foodborne Listeria Monocytogenes in Recent 5 Years in Japan. Microb. Pathog. 2023, 183, 106333. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Smith, D.P.; Berrang, M.E.; Feldner, P.W.; Phillips, R.W.; Meinersmann, R.J. Detection of Bacillus cereus on Selected Retail Chicken Products. J. Food Prot. 2004, 67, 1770–1773. [Google Scholar] [CrossRef]
- Glasset, B.; Herbin, S.; Guillier, L.; Cadel-Six, S.; Vignaud, M.L.; Grout, J. Bacillus cereus-Induced food-borne outbreaks in France, 2007 to 2014: Epidemiology and genetic characterization. Euro Surveill. 2016, 21, 30413. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, J.; Zhan, L.; Chen, H.; Zhang, Z.; Huang, C.; Yue, M. Prevalence and Antimicrobial-Resistant Characterization of Bacillus cereus Isolated from Ready-to-Eat Rice Products in Eastern China. Front. Microbiol. 2022, 13, 964823. [Google Scholar] [CrossRef] [PubMed]
- Adame-Gómez, R.; Muñoz-Barrios, S.; Castro-Alarcón, N.; Leyva-Vázquez, M.A.; Toribio-Jiménez, J.; Ramírez-Peralta, A. Prevalence of the Strains of Bacillus cereus Group in Artisanal Mexican Cheese. Foodborne Pathog. Dis. 2020, 17, 8–14. [Google Scholar] [CrossRef]
- Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, Y.; Liao, X.; Zhang, J.; Wu, S.; Gu, Q.; Xue, L. Bacillus cereus Isolated from Vegetables in China: Incidence, Genetic Diversity, Virulence Genes, and Antimicrobial Resistance. Front. Microbiol. 2019, 10, 948. [Google Scholar] [CrossRef]
- Kim, J.B.; Kim, J.M.; Kim, C.H.; Seo, K.S.; Park, Y.B.; Choi, N.J.; Oh, D.H. Emetic Toxin Producing Bacillus cereus Korean Isolates Contain Genes Encoding Diarrheal-Related Enterotoxins. Int. J. Food Microbiol. 2010, 144, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.; Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, J.; Zhou, H.; Chen, M.; Zeng, H.; et al. Prevalence, Virulence Feature, Antibiotic Resistance and MLST Typing of Bacillus cereus Isolated from Retail Aquatic Products in China. Front. Microbiol. 2020, 11, 1513. [Google Scholar] [CrossRef]
- Torki Baghbadorani, S.; Rahimi, E.; Shakerian, A. Investigation of Virulence and Antibiotic-Resistance of Bacillus cereus Isolated from Various Spices. Can. J. Infect. Dis. Med. Microbiol. 2023, 2023, 8390778. [Google Scholar] [CrossRef]
- Lee, N.; Kim, M.D.; Chang, H.J.; Choi, S.W.; Chun, H.S. Genetic Diversity, Antimicrobial Resistance, Toxin Gene Profiles, and Toxin Production Ability of Bacillus cereus Isolates from Doenjang, a Korean Fermented Soybean Paste. J. Food Saf. 2017, 37, e12363. [Google Scholar] [CrossRef]
- Owusu-Kwarteng, J.; Wuni, A.; Akabanda, F.; Tano-Debrah, K.; Jespersen, L. Prevalence, Virulence Factor Genes and Antibiotic Resistance of Bacillus cereus Sensu Lato Isolated from Dairy Farms and Traditional Dairy Products. BMC Microbiol. 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Biesta-Peters, E.G.; Dissel, S.; Reij, M.W.; Zwietering, M.H.; In’t Veld, P.H. Characterization and Exposure Assessment of Emetic Bacillus cereus and Cereulide Production in Food Products on the Dutch Market. J. Food Prot. 2016, 79, 230–238. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, Q.; Göker, M.; Meier-Kolthoff, J.P.; Wang, M.; Sun, Y.; Wang, L.; Shao, Z. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 2015, 5, 14082. [Google Scholar] [CrossRef]
- Agarwal, V.; Tiwari, A.; Varadwaj, P. An Extensive Review on β-lactamase Enzymes and Their Inhibitors. Curr. Med. Chem. 2023, 30, 783–808. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-B.; Kim, J.-B.; Shin, S.-W.; Kim, J.-C.; Cho, S.-H.; Lee, B.-K.; Ahn, J.; Kim, J.-M.; Deog-Hwan Oh, A. Prevalence, Genetic Diversity, and Antibiotic Susceptibility of Bacillus cereus Strains Isolated from Rice and Cereals Collected in Korea. J. Food Prot. 2009, 72, 612–617. [Google Scholar] [CrossRef]
- Lee, N.; Sun, J.M.; Kwon, K.Y.; Kim, H.J.; Koo, M.; Chun, H.S. Genetic Diversity, Antimicrobial Resistance, and Toxigenic Profiles of Bacillus cereus Strains Isolated from Sunsik. J. Food Prot. 2012, 75, 225–230. [Google Scholar] [CrossRef]
- Jawad, N.; Sahilah, A.M.; Abdullah, A.; Mutalib, S.A. Antimicrobial Resistance Pattern of Bacillus cereus Strains Isolated from Fried Rice Samples. Int. J. ChemTech Res. 2016, 9, 160–167. [Google Scholar]
- Navaneethan, Y.; Effarizah, M.E. Prevalence, Toxigenic Profiles, Multidrug Resistance, and Biofilm Formation of Bacillus cereus Isolated from Ready-to Eat Cooked Rice in Penang, Malaysia. Food Control 2021, 121, 107553. [Google Scholar] [CrossRef]
- Schlegelová, J.; Brychta, J.; Klimova, E.; Nápravníková, E.; Babak, V. The prevalence of and resistance to antimicrobial agents of Bacillus cereus isolates from foodstuffs. Veterinární Med. 2003, 48, 331–338. [Google Scholar] [CrossRef]
- Gdoura-Ben Amor, M.; Jan, S.; Baron, F.; Grosset, N.; Culot, A.; Gdoura, R.; Gautier, M.; Techer, C. Toxigenic Potential and Antimicrobial Susceptibility of Bacillus cereus Group Bacteria Isolated from Tunisian Foodstuffs. BMC Microbiol. 2019, 19, 196. [Google Scholar] [CrossRef]
- Sasano, H.; Yoshizawa, T.; Suzuki, M.; Fukui, Y.; Arakawa, R.; Tamura, N.; Naito, T. A Case of Persistent Bacillus cereus Bacteremia Responding to a Combination of Vancomycin and Gentamicin. Case Rep. Infect. Dis. 2022, 2022, 8725102. [Google Scholar] [CrossRef] [PubMed]
- Thery, M.; Cousin, V.L.; Tissieres, P.; Enault, M.; Morin, L. Multi-organ failure caused by lasagnas: A case report of Bacillus cereus food poisoning. Front. Pediatr. 2022, 10, 978250. [Google Scholar] [CrossRef]
- Nacharaju, D.; Tapley, A.; Karmarkar, E.N.; Fang, F.C.; Johnson, N.J. Vancomycin-resistant Bacillus cereus pneumonia and bacteremia after nonfatal freshwater drowning. Ann. Intern. Med. Clin. Cases 2024, 3, e240226. [Google Scholar] [CrossRef]
- Abdelaziz, M.N.; Maung, A.T.; El-Telbany, M.; Lwin, S.Z.; Mohammadi, T.N.; Zayda, M.; Wang, C.; Damaso, C.H.; Lin, Y.; Masuda, Y.; et al. Applications of bacteriophage in combination with nisin for controlling multidrug-resistant Bacillus cereus in broth and various food matrices. Food Res. Int. 2024, 91, 114685. [Google Scholar] [CrossRef]
- Rather, M.A.; Aulakh, R.S.; Gill, J.P.S.; Mir, A.Q.; Hassan, M.N. Detection and sequencing of plasmid-encoded tetracycline resistance determinants (tetA and tetB) from food-borne Bacillus cereus isolates. Asian Pac. J. Trop. Med. 2012, 5, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Iwabuchi, E.; Hasegawa, M.; Esaki, H.; Muramatsu, M.; Hirayama, N.; Hirai, K. Prevalence and molecular epidemiological characterization of antimicrobial-resistant Escherichia coli isolates from Japanese black beef cattle. J. Food Prot. 2013, 76, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.B.; Angrasan, M.; Chandel, N.; Rajamohan, G. Genome sequence and comparative analysis of Bacillus cereus BC04, reveals genetic diversity and alterations for antimicrobial resistance. Funct. Integr. Genom. 2018, 18, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Khishigtuya, T.; Matsuyama, H.; Suzuki, K.; Watanabe, T.; Nishiyama, M. Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Beef Cattle and Dairy Cows in a Livestock Farm in Yamagata, Japan. Microorganisms 2024, 12, 1342. [Google Scholar] [CrossRef]
- Suzuki, Y.; Hiroki, H.; Xie, H.; Nishiyama, M.; Sakamoto, S.H.; Uemura, R.; Nukazawa, K.; Ogura, Y.; Watanabe, T.; Kobayashi, I. Antibiotic-resistant Escherichia coli isolated from dairy cows and their surrounding environment on a livestock farm practicing prudent antimicrobial use. Int. J. Hyg. Environ. Health 2022, 240, 113930. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Asai, T. Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J. Biomed. Biotechnol. 2010, 2010, 180682. [Google Scholar] [CrossRef]
- Tamura, T. Trends in antimicrobial agents for veterinary use and control measures of antimicrobial resistance. J. Jpn. Vet. Med. Assoc. 2003, 56, 685–691. [Google Scholar] [CrossRef][Green Version]
- AMR Clinical Reference Center. Nippon AMR One Health Report (NAOR 2018). 2018. Available online: https://amr-onehealth.ncgm.go.jp/en/ (accessed on 1 June 2023).
- Asai, T. Antimicrobial resistance monitoring program in food-producing animals in Japan. J. Vet. Epidemiol. 2008, 12, 93–98. [Google Scholar] [CrossRef]
- Huang, Y.; Flint, S.H.; Palmer, J.S. Bacillus cereus Spores and Toxins—The Potential Role of Biofilms. Food Microbiol. 2020, 90, 103493. [Google Scholar] [CrossRef]
- Samaržija, D.; Zamberlin, Š.; Pogačić, T. Psychrotrophic bacteria and their negative effects on milk and dairy products quality. Mljekarstvo 2012, 62, 77–95. [Google Scholar]
- Shemesh, M.; Ostrov, I. Role of Bacillus species in biofilm persistence and emerging antibiofilm strategies in the dairy industry. J. Sci. Food Agric. 2020, 100, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- Cardazzo, B.; Negrisolo, E.; Carraro, L.; Alberghini, L.; Patarnello, T.; Giaccone, V. Multiple-Locus Sequence Typing and Analysis of Toxin Genes in Bacillus cereus Food-Borne Isolates. Appl. Environ. Microbiol. 2008, 74, 850–860. [Google Scholar] [CrossRef]
- Bianco, A.; Normanno, G.; Capozzi, L.; Del Sambro, L.; Di Fato, L.; Miccolupo, A.; Di Taranto, P.; Caruso, M.; Petruzzi, F.; Ali, A.; et al. High Genetic Diversity and Virulence Potential in Bacillus cereus Sensu Lato Isolated from Milk and Cheeses in Apulia Region, Southern Italy. Foods 2023, 12, 1548. [Google Scholar] [CrossRef]
- Chaves, J.Q.; Pires, E.S.; Vivoni, A.M. Genetic Diversity, Antimicrobial Resistance and Toxigenic Profiles of Bacillus cereus Isolated from Food in Brazil over Three Decades. Int. J. Food Microbiol. 2011, 147, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Cho, S.H.; Kang, S.H.; Park, Y.B.; Yoon, M.H.; Lee, J.B.; No, W.S.; Kim, J.B. Prevalence, Genetic Diversity, and Antibiotic Resistance of Bacillus cereus Isolated from Korean Fermented Soybean Products. J. Food Sci. 2015, 80, M123–M128. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Kamikado, H.; Sasaki, C.; Sadakari, K.; Honjoh, K.; Iio, M. An attempt to identify Bacillus cereus by PCR. Biocontrol Sci. 2004, 9, 69–75. [Google Scholar] [CrossRef]
- GB4789.14-2014; Food Microbiological Examination: Bacillus cereus Testing. China Food and Drug Administration: Beijing, China, 2014.
- U.S. Food and Drug Administration (USFDA). Bacteriological Analytical Manual Online (BAM Online): Chapter 14: Bacillus cereus. 2012. Available online: https://www.fda.gov/food/laboratory-methods-food/bacteriological-analytical-manual-bam (accessed on 1 April 2022).
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry: A Fundamental Shift in the Routine Practice of Clinical Microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef]
- Yamada, S.; Ohashi, E.; Agata, N.; Venkateswaran, K. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice. Appl. Environ. Microbiol. 1999, 65, 1483–1490. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Vukov, N.; Schulz, A.; Shaheen, R.; Andersson, M.; Märtlbauer, E.; Scherer, S. Identification and Partial Characterization of the Nonribosomal Peptide Synthetase Gene Responsible for Cereulide Production in Emetic Bacillus cereus. Appl. Environ. Microbiol. 2005, 71, 105–113. [Google Scholar] [CrossRef]
- Horwood, P.F.; Burgess, G.W.; Jane Oakey, H. Evidence for Non-Ribosomal Peptide Synthetase Production of Cereulide (the Emetic Toxin) in Bacillus cereus. FEMS Microbiol. Lett. 2004, 236, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute CLSI. M100 Performance Standards for Antimicrobial; CLSI: Wayne, PE, USA, 2021; Volume 31, ISBN 9781684400669. [Google Scholar]
- De Jesus, R.; Dedeles, G. Data on Quantitation of Bacillus cereus Sensu Lato Biofilms by Microtiter Plate Biofilm Formation Assay. Data Brief. 2020, 28, 104951. [Google Scholar] [CrossRef]
- Hussain, M.S.; Oh, D.H. Impact of the Isolation Source on the Biofilm Formation Characteristics of Bacillus cereus. J. Microbiol. Biotechnol. 2018, 28, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Miyako, K.; Nakamura, J.; Hatano, S. Production and Partial Purification of HEp-2 Cell-Vacuolation Factor Derived from Bacillus cereus. Jpn. J. Food Microbiol. 1994, 11, 113–117. [Google Scholar] [CrossRef]
- Hayrapetyan, H.; Muller, L.; Tempelaars, M.; Abee, T.; Nierop Groot, M. Comparative Analysis of Biofilm Formation by Bacillus cereus Reference Strains and Undomesticated Food Isolates and the Effect of Free Iron. Int. J. Food Microbiol. 2015, 200, 72–79. [Google Scholar] [CrossRef]
- Faille, C.; Bénézech, T.; Midelet-Bourdin, G.; Lequette, Y.; Clarisse, M.; Ronse, G.; Ronse, A.; Slomianny, C. Sporulation of Bacillus Spp. within Biofilms: A Potential Source of Contamination in Food Processing Environments. Food Microbiol. 2014, 40, 64–74. [Google Scholar] [CrossRef]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016, 44, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Kuo, W.-S.; Chak, K.-F. Identification of Novel Cry-Type Genes from Bacillus Thuringiensis Strains on the Basis of Restriction Fragment Length Polymorphism of the PCR-Amplified DNA. Appl. Environ. Microbiol. 1996, 62, 369–377. [Google Scholar] [CrossRef]
- Bundy, J.G.; Willey, T.L.; Castell, R.S.; Ellar, D.J.; Brindle, K.M. Discrimination of Pathogenic Clinical Isolates and Laboratory Strains of Bacillus cereus by NMR-Based Metabolomic Profiling. FEMS Microbiol. Lett. 2005, 242, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Lee, N.A.; Woo, G.J.; Park, J.H. Prevalence of Bacillus cereus group in rice and distribution of enterotoxin genes. Food Sci. Biotechnol. 2006, 15, 232–237. [Google Scholar]
- Rowan, N.J.; Caldow, G.; Gemmell, C.G.; Hunter, I.S. Production of diarrheal enterotoxins and other potential virulence factors by veterinary isolates of Bacillus species associated with nongastrointestinal infections. Appl. Environ. Microbiol. 2003, 69, 2372–2376. [Google Scholar] [CrossRef] [PubMed]
- Melnick, R.L.; Testen, A.L.; Poleatewich, A.M.; Backman, P.A.; Bailey, B.A. Detection and Expression of Enterotoxin Genes in Endophytic Strains of Bacillus cereus. Lett. Appl. Microbiol. 2012, 54, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Guinebretière, M.H.; Broussolle, V.; Nguyen-The, C. Enterotoxigenic Profiles of Food-Poisoning and Food-Borne Bacillus cereus Strains. J. Clin. Microbiol. 2002, 40, 3053–3056. [Google Scholar] [CrossRef] [PubMed]
- Ghelardi, E.; Celandroni, F.; Salvetti, S.; Barsotti, C.; Baggiani, A.; Senesi, S. Identification and Characterization of Toxigenic Bacillus cereus Isolates Responsible for Two Food-Poisoning Outbreaks. FEMS Microbiol. Lett. 2002, 208, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Blanch, J.F.; Sánchez, G.; Garay, E.; Aznar, R. Development of a Real-Time PCR Assay for Detection and Quantification of Enterotoxigenic Members of Bacillus cereus Group in Food Samples. Int. J. Food Microbiol. 2009, 135, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lund, T.; De Buyser, M.L.; Granum, P.E. A New Cytotoxin from Bacillus cereus That May Cause Necrotic Enteritis. Mol. Microbiol. 2000, 38, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Adimpong, D.B.; Sørensen, K.I.; Thorsen, L.; Stuer-Lauridsen, B.; Abdelgadir, W.S.; Nielsen, D.S.; Derkx, P.M.; Jespersen, L. Antimicrobial susceptibility of Bacillus strains isolated from primary starters for African traditional bread production and characterization of the bacitracin operon and bacitracin biosynthesis. Appl. Environ. Microbiol. 2012, 78, 7903–7914. [Google Scholar] [CrossRef]
- Mirzaie, S.; Faghiri, I.; Askari Badouei, M.; Madani, S.A. Molecular detection and occurrence of vancomycin resistance genes (van A, B, C1, C2/C3) among Enterococcus species isolated from farm ostriches. Vet. Med. Sci. 2023, 9, 226–233. [Google Scholar] [CrossRef]
| Tested Sample | No. of Positive Samples by the Method (%) | Total Samples Collected | ||
|---|---|---|---|---|
| XBC | BACARA | PCR | ||
| Milk and milk products | 40 | 22 | 18 | 89 |
| Raw chicken meat | 21 | 9 | 9 | 50 |
| Tofu | 3 | 3 | 3 | 5 |
| RTE cooked rice | 11 | 7 | 6 | 20 |
| Soil and bedding | 13 | 8 | 4 | 15 |
| Total No. (%) | 88 (49.1%) | 50 (27.3%) | 40 (22.3%) | 179 |
| Virulence Genes | Number of Isolates (%) Positive for Target Gene (s) |
|---|---|
| NHE gene complexes | |
| nheA | 22 (55%) |
| nheB | 13 (32.5%) |
| nheC | 25 (62.5%) |
| nheA + nheB + nheC | 7 (17.5%) |
| HBl gene complexes | |
| hblA | 20 (50%) |
| hblC | 12 (30%) |
| hblD | 17 (42.5%) |
| hblA + hblC + hblD | 1 (2.5%) |
| bceT | 17 (42.5%) |
| entFM | 23(57.5%) |
| Ces A | 7 (17.5%) |
| cytK | 0 (not detected) |
| Category | Antimicrobial Agents | B. cereus s. s. Strains No. (%) | |||||
|---|---|---|---|---|---|---|---|
| Current Isolates (n = 40) | Lab. Stock (n = 70) | ||||||
| Resistance | Intermediate | Susceptible | Resistance | Intermediate | Susceptible | ||
| β lactam Antibiotics | Ampicillin (ABP) 10 µg | 40 (100%) | 0 (0) | 0 (0) | 70 (100%) | 0 (0) | 0 (0) |
| Amoxicillin-Clavulanic acid (ACV) 20 µg/10 µg | 36 (90%) | 0 (0) | 4 (10%) | 61 (87.1%) | 0 (0) | 9 (12.8%) | |
| Cefepime (CFP) 30 µg | 34 (85%) | 6 (15%) | 0 (0) | 68 (97.1%) | 2 (2.8%) | 0 (0) | |
| Cefoxitin (CFX) 30 µg | 40 (100%) | 0 (0) | 0 (0) | 62 (88.5%) | 0 (0) | 8 (11.4%) | |
| Oxacillin (MPI) 1 µg | 40 (100%) | 0 (0) | 0 (0) | 68 (97.1%) | 1 (1.4%) | 1 (1.42%) | |
| Penicillin (PC) 10 units | 40 (100%) | 0 (0) | 0 (0) | 69 (98.5%) | 0 (0) | 1 (1.42%) | |
| Aminoglycosides | Gentamicin (GM) 10 µg | 0 (0) | 0 (0) | 40 (100%) | 0 (0) | 0 (0) | 70 (100%) |
| Kanamycin (KM) 30 µg | 19 (47.5%) | 5 (12.5%) | 16 (40%) | 1 (1.4%) | 2 (2.8%) | 67 (95.7%) | |
| Macrolides | Erythromycin (EM) 15 µg | 15 (37.5%) | 1 (2.5%) | 24 (60%) | 0 (0) | 11 (15.7%) | 59(84.2%) |
| Glycopeptides | Vancomycin (VCM) 30 µg | 15 (37.5%) | 1 (2.5%) | 24 (60%) | 1 (1.4%) | 0 (0) | 69 (98.5%) |
| Tetracyclines | Tetracycline (TC) 30 µg | 17 (42.5%) | 5 (12.5%) | 18 (45%) | 0 (0) | 2 (2.8%) | 68 (97.1%) |
| Rifamycin | Rifampicin (RA) 5 µg | 35 (87.5%) | 2 (5%) | 3 (7.5%) | 32 (45.7%) | 26 (37.1%) | 12 (17.1%) |
| Folic acid Inhibitors | Trimethoprim-Sulfamethoxazole (ST) 1.25 µg–23.75 µg | 37 (92.5%) | 3 (7.5%) | 0 (0) | 60 (85.7%) | 7 (10%) | 3 (4.2%) |
| Isolates | Total No. | No. of Positive Isolates (% in Resistant Isolates) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tetracycline | Erythromycin | Vancomycin | |||||||||||
| Resistant Strain No. | tetA | Resistant Strain No. | erm | Resistant Strain No. | vanA | vanB | vanR | vanS | vanY | vanW | vanH | ||
| Lab. stock | 70 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 |
| Current | 40 | 17 | 9 (52.9) | 15 | 12 (80) | 15 | 0 | 0 | 12 (80) | 3 (20) | 12(80) | 14 (93.3) | 0 |
| OD600 at 7th Day | Total | ||||
|---|---|---|---|---|---|
| 0.2–0.3 | 0.4–0.5 | 0.6–0.7 | 0.8–0.9 | ||
| No. of strains grown | 8 | 13 | 9 | 10 | 40 |
| Percentage | 20% | 32.5% | 22.5% | 25% | 100% |
| Strain Code | SBF Index ± SD | Biofilm Forming Strength | Strain Code | SBF Index ± SD | Biofilm Forming Strength |
|---|---|---|---|---|---|
| BC-UH 1 | 0.31 ± 0.02 | weak | BC-TF 21 | 1.59 ± 0.10 | strong |
| BC-UH 2 | 0.26 ± 0.04 | weak | BC-TF 3 | 1.16 ± 0.07 | strong |
| BC-PM 3 | 0.89 ± 0.01 | moderate | BC-CH 23 | 1.75 ± 0.04 | strong |
| BC-PM 4 | 0.52 ± 0.02 | moderate | BC-CH 24 | 0.45 ± 0.02 | weak |
| BC-Yg5 | 0.61 ± 0.02 | moderate | BC-CH 25 | 0.73 ± 0.03 | moderate |
| BC-YG 6 | 0.33 ± 0.01 | weak | BC-CH 26 | 0.86 ± 0.04 | moderate |
| BC-CT 7 | 1.37 ± 0.12 | strong | BC-CH 27 | 1.10 ± 0.03 | strong |
| BC-CT 8 | 0.45 ± 0.02 | weak | BC-CH 28 | 0.88 ± 0.01 | moderate |
| BC-CT 9 | 0.44 ± 0.03 | moderate | BC-CH 29 | 1.21 ± 0.02 | strong |
| BC-CT 10 | 0.8 ± 0.02 | moderate | BC-CH 30 | 1.01 ± 0.03 | moderate |
| BC-CT 11 | 0.70 ± 0.02 | moderate | BC-CH 31 | 1.10 ± 0.09 | strong |
| BC-PW 12 | 0.54 ± 0.01 | moderate | BC-RW 32 | 0.71 ± 0.02 | moderate |
| BC-PW13 | 0.65 ± 0.03 | moderate | BC-RW 33 | 0.71 ± 0.01 | moderate |
| BC-RI 14 | 0.44 ± 0.04 | weak | BC-RW34 | 1.21 ± 0.02 | strong |
| BC-RI 15 | 0.66 ± 0.01 | moderate | BC-RW35 | 1.85 ± 0.01 | strong |
| BC-RI 16 | 0.30 ± 0.01 | weak | BC-RW 36 | 1.43 ± 0.03 | strong |
| BC-RI 17 | 0.98 ± 0.00 | moderate | BC-SL 37 | 0.61 ± 0.03 | moderate |
| BC-RI 18 | 1.13 ± 0.09 | strong | BC-SL 38 | 1.23 ± 0.04 | strong |
| BC-TF 19 | 1.64 ± 0.02 | strong | BC-SL 39 | 1.01 ± 0.02 | strong |
| BC-RI 20 | 0.74 ± 0.02 | moderate | BC-BD 40 | 0.60 ± 0.01 | moderate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelaziz, M.N.S.; Zayda, M.G.; Maung, A.T.; El-Telbany, M.; Mohammadi, T.N.; Lwin, S.Z.C.; Linn, K.Z.; Wang, C.; Yuan, L.; Masuda, Y.; et al. Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan. Antibiotics 2024, 13, 774. https://doi.org/10.3390/antibiotics13080774
Abdelaziz MNS, Zayda MG, Maung AT, El-Telbany M, Mohammadi TN, Lwin SZC, Linn KZ, Wang C, Yuan L, Masuda Y, et al. Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan. Antibiotics. 2024; 13(8):774. https://doi.org/10.3390/antibiotics13080774
Chicago/Turabian StyleAbdelaziz, Marwa Nabil Sayed, Mahmoud Gamaleldin Zayda, Aye Thida Maung, Mohamed El-Telbany, Tahir Noor Mohammadi, Su Zar Chi Lwin, Khin Zar Linn, Chen Wang, Lu Yuan, Yoshimitsu Masuda, and et al. 2024. "Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan" Antibiotics 13, no. 8: 774. https://doi.org/10.3390/antibiotics13080774
APA StyleAbdelaziz, M. N. S., Zayda, M. G., Maung, A. T., El-Telbany, M., Mohammadi, T. N., Lwin, S. Z. C., Linn, K. Z., Wang, C., Yuan, L., Masuda, Y., Honjoh, K.-i., & Miyamoto, T. (2024). Genetic Characterization, Antibiotic Resistance, and Virulence Genes Profiling of Bacillus cereus Strains from Various Foods in Japan. Antibiotics, 13(8), 774. https://doi.org/10.3390/antibiotics13080774







