Abstract
The increasing rates of morbidity and mortality owing to bacterial infections, particularly Staphylococcus aureus have necessitated finding solutions to face this issue. Thus, we elucidated the phytochemical constituents and antibacterial potential of Cleome droserifolia extract (CDE). Using LC-ESI-MS/MS, the main phytoconstituents of CDE were explored, which were kaempferol-3,7-O-bis-alpha-L-rhamnoside, isorhamnetin, cyanidin-3-glucoside, kaempferide, kaempferol-3-O-alpha-L-rhamnoside, caffeic acid, isoquercitrin, quinic acid, isocitrate, mannitol, apigenin, acacetin, and naringenin. The CDE exerted an antibacterial action on S. aureus isolates with minimum inhibitory concentrations ranging from 128 to 512 µg/mL. Also, CDE exhibited antibiofilm action using a crystal violet assay. A scanning electron microscope was employed to illuminate the effect of CDE on biofilm formation, and it considerably diminished S. aureus cell number in the biofilm. Moreover, qRT-PCR was performed to study the effect of CDE on biofilm gene expression (cna, fnbA, and icaA). The CDE revealed a downregulating effect on the studied biofilm genes in 43.48% of S. aureus isolates. Regarding the in vivo model, CDE significantly decreased the S. aureus burden in the liver and spleen of CDE-treated mice. Also, it significantly improved the mice’s survival and substantially decreased the inflammatory markers (interleukin one beta and interleukin six) in the studied tissues. Furthermore, CDE has improved the histology and tumor necrosis factor alpha immunohistochemistry in the liver and spleen of the CDE-treated group. Thus, CDE could be considered a promising candidate for future antimicrobial drug discovery studies.
1. Introduction
Cleome droserifolia (Forssk.) Delile Descr. is a small shrub that can grow to 60 cm in height. It grows naturally in Egyptian deserts like the Sinai Peninsula, stone soil, and rocky wadis. It became endangered due to extensive uprooting in Egypt’s Sinai and Eastern Deserts [1,2]. The genus Cleome (L.) DC. (Cleomacaea) [3] encompasses annual and perennial medicinal herbs or small shrubs [4,5,6,7]. Cleome droserifolia, known by local Bedouin people as Samwah, is employed in traditional medicine in Egypt as a hypoglycemic agent [8,9]. Different researchers analyzed the phytochemical content of C. droserifolia, exploring the existence of flavonoids [10], alkaloids, tannins, saponins, coumarins, catechins, sterols, glucosinolates, and terpenoids [1,9]. The essential oil profile of C. droserifolia was also studied, and the main compounds in the essential oil were (Z)-nerolidol and α-cadinol [11]. C. droserifolia exerted different biological effects as an antioxidant, antimicrobial [12], anticancer [13], allelopathic [11], and hypoglycaemic [14] properties.
Staphylococcus aureus is a common species of Staphylococci highly associated with multidrug resistance [15]. Such bacterial species can adapt to various environments and possess frequent virulence factors [16]. In addition, it is a common nosocomial pathogen that can trigger various diseases that range from mild severity to life-threatening ailments. The infections triggered by S. aureus are mild skin and soft tissue infections, bacteremia, osteomyelitis, endocarditis, and pneumonia [17].
S. aureus can exhibit resistance to antibiotics by various mechanisms, including decreasing the bacterial membrane permeability to the antibiotics, efflux, and excessive production of resistance enzymes like β-lactamases [18]. Multi-drug resistance (MDR) is a worldwide issue that has a deleterious effect on health care. S. aureus acquires resistance to antibiotics owing to persistent exposure to various antimicrobials. MDR S. aureus is resistant to multiple chemotherapeutic agents. Such MDR isolates have led to increasing global rates of mortality as well as morbidity in S. aureus-infected patients [19].
Biofilm is an important virulence factor of S. aureus, and it is defined as an extracellular complex structure that compromises a population of bacterial cells anchored to living or non-living surfaces [20]. The cells are surrounded by an extracellular polymer matrix formed by themselves as a protective tactic for bacterial survival to adapt to their surrounding environments [21].
Traditional antibiotics are excessively losing their potential to combat bacterial infections, particularly S. aureus [22]. Thus, novel treatment approaches should be elucidated to face such global concerns. Natural sources, like plants, are rich in many bioactive phytochemicals with various therapeutic activities [23]. Many studies have elucidated the potential antimicrobial action of plants and their bioactive constituents against viruses, fungi, and bacteria [24,25,26,27,28].
Here, we aimed to explore CDE’s potential antibacterial action in vitro and in vivo. Also, the phytoconstituents of this plant will be elucidated using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) to explore the active principles with different chemical entities which may have a role in developing new antibacterial agents against the MDR S. aureus isolates.
2. Results
2.1. Recognition of Different Phytochemical Contents of C. droserifolia by LC-ESI-MS/MS
It was revealed tentatively by this technique that C. droserifolia contains different phytochemical groups with variable biological effects. This plant contains dicarboxylic and tricarboxylic acids and their derivatives, hydroxy fatty acids, hydroxybenzoic acid derivatives, flavonols, flavones, flavanones, aurone O-glycosides, hydroxycinnamic acids, quinic acids and derivatives, flavonoid-3-O-glycosides, anthocyanidin-3-O-glycosides, anthocyanidin-5-O-glycosides, alkyl glucosinolate, methoxy phenols, and 4′-O-methylated flavonoids. The LC-ESI-MS/MS analysis disclosed that the predominant constituents are kaempferol-3,7-O-bis-alpha-L-rhamnoside, isorhamnetin, cyanidin-3-glucoside, 3, 5, 7-trihydroxy-4′-methoxyflavone (kaempferide), kaempferol-3-O-alpha-L-rhamnoside, caffeic acid, isoquercitrin, quinic acid, isocitrate, mannitol, apigenin, acacetin, and naringenin. Table 1 and Figure S1 in the Supplementary Materials demonstrate the tentatively recognized compounds supported by referenced data. The previously reported data revealed the presence of different phenolic compounds in C. droserifolia, such as kaempferol-3,7-dirhamnoside, isorharmnetin-3-O-gluco-7-O-rhamnoside, kaempferol-3-O-gluco-7-O-rhamnoside, quercetin-3-O-gluco-7-O-rhamnoside, kaempferol, artemitin [9], Isorhamnetin-3-O-β-d-glucoside, quercetin-3′-methoxy-3-O-(4″-acetyl rhamnoside)-7-O-α-rhamnoside, and kaempferol-4′-methoxy-3,7-dirhamnoside [29]. It was detected by RP-HPLC of the methanolic extract of the plant that the major phenolic compounds were benzoic acid, ellagic acid, rutin, o-coumaric acid, and naringenin. Moderate quantities of rosmarinic acid, p-hydroxybenzoic acid, resveratrol, kaempferol, quercetin, and ferulic acid were detected. The least abundant phenolic compounds were chlorogenic acid, caffeic acid, p-coumaric acid, syringic acid, and catechin [12].

Table 1.
The phytoconstituents of C. droserifolia methanol extract recognized by LC-ESI-MS/MS analysis.
2.2. Bacterial Isolates and Antibiotic Resistance
The clinical specimens from which the bacterial isolates were recovered include blood, wounds, sputum, and urine (Figure 1). The antibiotic susceptibility of the tested isolates is revealed in Figure 2.
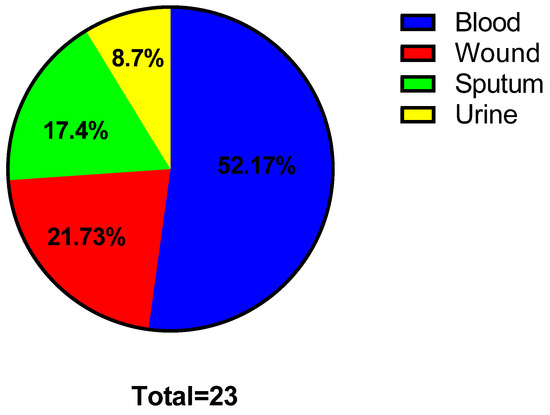
Figure 1.
Pie chart revealing the percentages of the clinical specimens.
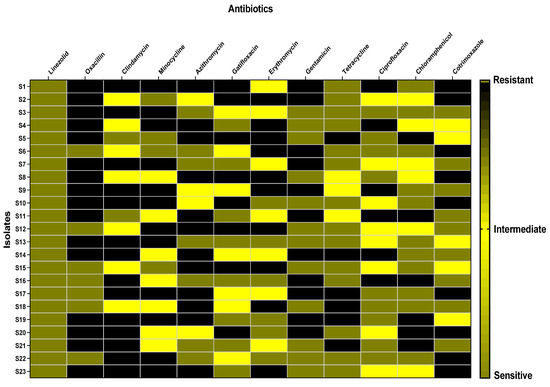
Figure 2.
Heat map representing the susceptibility of S. aureus isolates to different antibiotics.
2.3. Susceptibility of S. aureus to CDE
The susceptibility of S. aureus to CDE was elucidated using the agar well diffusion method as a preliminary method to reveal whether CDE possesses antibacterial action (Figure 3 and Table S2). Then, the MICs were determined, as shown in Table S3.
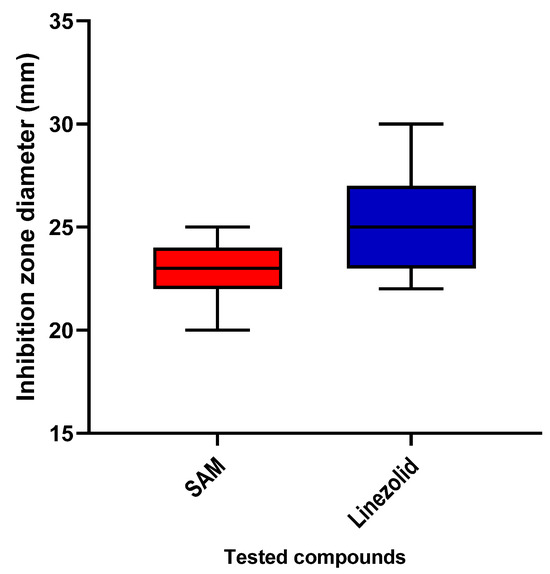
Figure 3.
The inhibition zone diameters of CDE against the tested isolates.
2.4. Determination of Antibiofilm Potential by Crystal Violet Assay, SEM, and qRT-PCR
CDE revealed antibiofilm potential through the semiquantitative method, crystal violet (Table 2), as it decreased the percentage of strong and moderate biofilm-forming isolates from 73.91% to 30.43%. Then, SEM was employed to elucidate the effect of CDE on the morphology of the biofilm (Figure 4). CDE has significantly decreased the number of cells in the formed biofilm.

Table 2.
Influence of CDE on the biofilm-forming ability of S. aureus isolates.
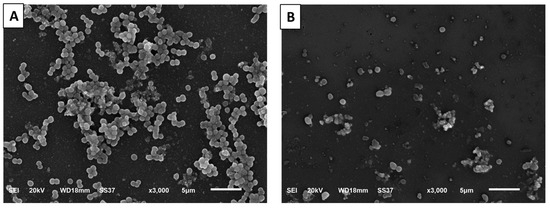
Figure 4.
SEM micrograph revealing the morphology of S. aureus biofilm formed on the surfaces of cover glass: (A) without (untreated isolates) and (B) with CDE (treated isolates).
qRT-PCR was utilized to reveal the potential of CDE on the expression level of biofilm genes. CDE was found to downregulate the biofilm-encoding genes in 43.48% of the isolates, as revealed in Figure 5.
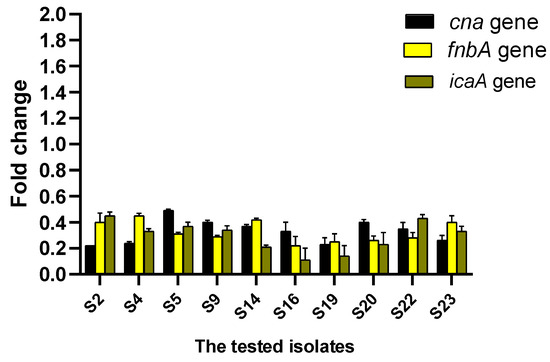
Figure 5.
Impact of CDE on the gene expression levels of the biofilm.
2.5. In Vivo Infection Model in Mice
A systemic infection was induced in mice, and the bacterial burden was detected in the liver and spleen in the experimental groups (Figure 6).
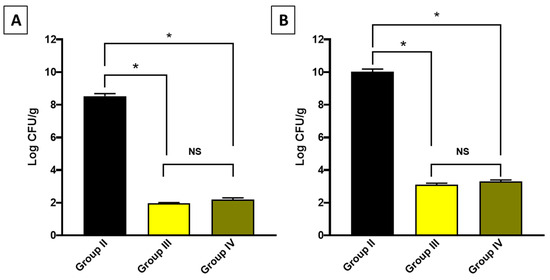
Figure 6.
Bacterial burden in (A) the liver and (B) the spleen. The symbol (*) denotes a significant difference (p < 0.05). The abbreviation (NS) denotes a non-significant difference (p > 0.05).
The survival curve was constructed as shown in Figure 7. No mice died in group I; in group II, two died after three days, one after five days, and another after one week. Also, two mice died after nine days. Regarding groups III and IV, one mouse died on the eighth and sixth days.
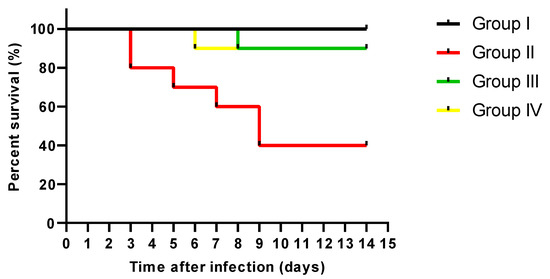
Figure 7.
Survival curve constructed for the experimental groups.
2.6. Histopathological and Immunohistochemical Investigations
The effect of CDE on the histological features of the liver and spleen is shown in Figure 8 and Figure 9. Also, the TNF-α immunohistochemical staining of the liver and spleen of the different experimental groups is shown in Figure 10 and Figure 11.
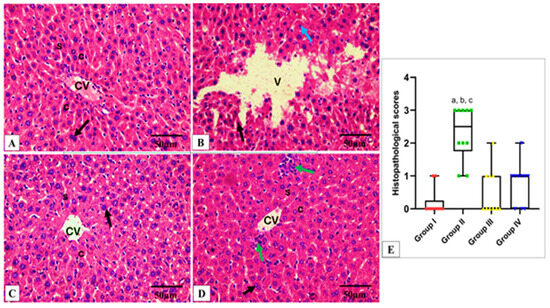
Figure 8.
Light microscopic H&E-stained images of hepatic sections of adult albino rats of all studied groups. (A) Group I shows normal hepatic architecture in the form of hepatic cords (c) radiating from the central vein (CV) and separated by the hepatic sinusoids (S), with pericentral zone and midzone hepatocytes. Polyhedral hepatocytes appear with rounded vesicular nuclei and granular eosinophilic cytoplasm (black arrow) separated by sinusoids (S) lined with endothelial cells and Kupffer cells. (B) Group II shows disturbed hepatic architecture, compressed hepatic sinusoids, multiple pyknotic nuclei (black arrow), multiple karyolitic nuclei (blue arrow), and diffuse vacuolar degeneration of hepatocytes with multiple large vacuoles (V) as well as ballooned hepatocytes. (C) Group III shows organized hepatic cords (C) around the central vein, resembling the normal hepatic structure of Group I. (D) Group IV shows marked improvement and a regaining of the normal hepatic structure. However, with this enhancement, inflammatory cells infiltrate the pericentral zone (green arrow). (E) Histopathological score of the hepatic tissue cross sections in all studied groups. A box plot was used to express the data. The bottom of the plot represents 25%, the middle represents the median, and the top represents 75% of the data. Significant difference at p ≤ 0.05, where (a) in comparison with group I, (b) in comparison with group III, and (c) in comparison with group IV using the Kruskal–Wallis test followed by Dunn’s pairwise comparison post-hoc test. (H&E × 400, scale bar = 50 μm).
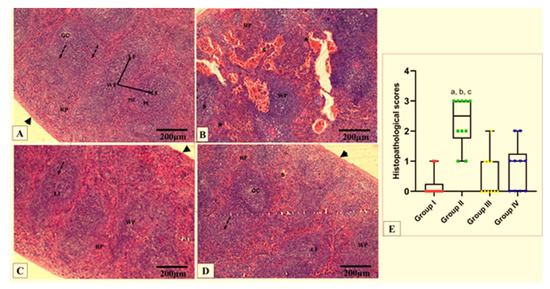
Figure 9.
Photomicrographs of H&E-stained sections of spleen in all studied groups. (A) Group I shows the normal histological organization of white pulp (WP) and red pulp (RP) surrounded by a capsule (arrowhead). The WP presents the central arteriole (dashed arrow) and lymphoid follicles (LF), with germinal centers (GC) and mantle regions (M), surrounded by a loosely distributed marginal zone (MZ). The RP presents lymphocytes, trabeculae, and sinusoids. (B) Group II shows a loss of normal architecture, shrunken WP, and broadened RP. Note congested, dilated splenic sinuses (S) in the RP. Many cells in the WP appear vacuolated. Thick fibrous trabeculae are observed (asterisk). (C) Group III shows a nearly normal appearance of the WP and RP splenic architecture. (D) Group IV shows a nearly normal outline of splenic architecture, but congested splenic sinuses are also seen. (E) Histopathological score of the splenic tissue cross sections in all studied groups. A box plot was used to express the data. The bottom of the plot represents 25%, the middle represents the median, and the top represents 75% of the data. Significant difference at p ≤ 0.05 (a) in comparison with group I, (b) in comparison with group III, and (c) in comparison with group IV using the Kruskal–Wallis test followed by Dunn’s pairwise comparison post-hoc test. (H&E × 100, scale bar = 200 μm).
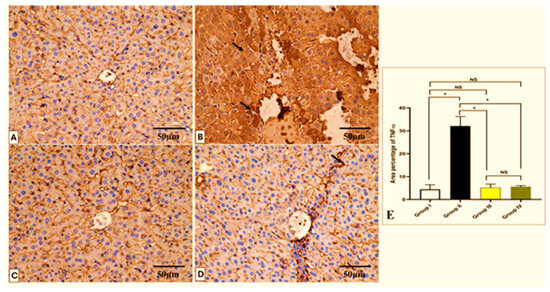
Figure 10.
Light microscopic TNF-α-stained pictures of a liver section in all studied groups. (A) Group I shows a negative TNF-α expression within the hepatocytes’ cytoplasm. (B) Group II shows a strong positive expression of TNF-α, which appears as brownish cytoplasm in hepatocytes (arrows). (C) Group III shows a negative expression of TNF-α. (D) Group IV shows a mild positive expression of TNF-α in a few hepatocytes (arrow) and a negative expression in most of the cells. (E) The area percentage of TNF-α in each group. Mean ± SD was used to represent the data. One-way ANOVA was used for the statistical comparison, and Tukey’s post-hoc test was used for multiple comparisons. The single asterisk indicates a significant change, and the abbreviation NS denotes a non-significant change (p < 0.05). (TNF-α × 400, scale bar = 50 μm).
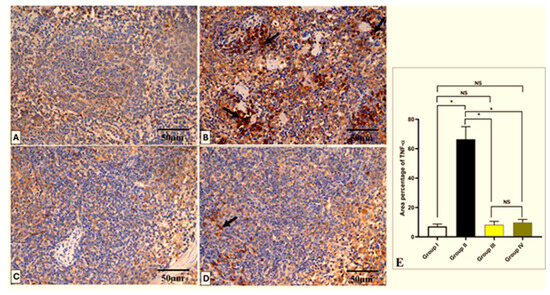
Figure 11.
Light microscopic TNF-α-stained pictures of the spleen in all studied groups. (A) Group I shows a negative expression of TNF-α within the cytoplasm of the cells. (B) Group II shows a strong positive expression of TNF-α, which appears as brownish cytoplasm in most cells (arrows). (C) Group III shows a negative expression of TNF-α. (D) Group IV shows few TNF-α-positive cells (arrow) and a negative expression in most cells. (E) The area percentage of TNF-α in each group. Mean ± SD is used to represent the data. One-way ANOVA was used for the statistical comparison, and Tukey’s post-hoc test was used for multiple comparisons. The single asterisk indicates a significant change, and the abbreviation NS denotes a non-significant change (p < 0.05). (TNF-α × 400, scale bar = 50 μm).
2.7. ELISA
Levels of IL-1β and IL-6 were detected in the liver and spleen tissues of the different groups (Table 3).

Table 3.
Effect of CDE on IL-1β and IL-6 levels in the liver and spleen of the experimental groups.
3. Materials and Methods
3.1. Collection, Drying, and Extraction of the Plant Material
The stems, flowers, and leaves of C. droserifolia shrubs were collected in March 2022 from the wadis around Sharm El-Sheikh and Dahab in South Sinai Governorate. The plant was dried and powdered to obtain 1.2 kg of dry weight. It was recognized by Prof. Dr. Hanafey Farouk Maswada, Professor of Plant Physiology, Agricultural Botany Department, Faculty of Agriculture, Tanta University, and depositing a voucher specimen (PG-A-00124) in the herbarium of the Department of Pharmacognosy, Faculty of Pharmacy, Tanta University. Methanol (Sigma Chemical Co., St. Louis, MO, USA) was used as an extracting solvent and was mixed with the powder. The extraction was performed thrice (4 L × 3 times), and then the concentration of the solvent was performed by a rotary evaporator under vacuum to obtain 45.3 g of dry residue from the plant’s extract.
3.2. Exploration of the Plant’s Phytoconstituents by LC-ESI-MS/MS
Different compounds in the extract were identified by the Proteomics and Metabolomics Unit, Children’s Cancer Hospital (57357), Basic Research Department, Cairo, Egypt. The crude extract was reconstituted in DI-Water:Methanol:Acetonitrile—50:25:25, and HPLC separation was accomplished by using in-line filter disks (0.5 µm × 3.0 mm, Phenomenex®, Torrance, CA, USA) and X select HSS T3 (2.5 µm, 2.1 × 150 mm, Waters®, Milford, MA, USA, 40 °C). The first is a pre-column, and the second is an analytical column. The mobile phases were composed of mobile phase A, which is composed of 5 mM ammonium formate buffer pH 8 with 1% methanol. Mobile phase B is 100% acetonitrile. Isocratic elution was done using 90% of solvent A and 10% of solvent B for one minute. After that a gradient elution from 90 to 10% solvent A and 10 to 90% of solvent B in twenty minutes was done, then elution with 90% of solvent B for four minutes was done then return to the initial condition (10% of acetonitrile) for three minutes. The flow rate was 0.3 mL/min. The instrument was coupled with Triple TOF 5600+ (Sciex®, Framingham, MA, USA) for IDA acquisition and Analyst TF 1.7.1 (Sciex®) for LC-Triple TOF control. Raw data files were loaded into MS-DIAL 3.52 for data-independent MS/MS deconvolution [69]. Compounds were recognized with >70% probability using an MS1 and MS2 tolerance of 0.2 mass units to be accepted as positive identifications. The ReSpect negative (1573 records) database was used as a reference database. PeakView 2.2 with the MasterView 1.1 package (AB SCIEX, Framingham, MA, USA) were used for feature or peaks extraction from the total ion chromatogram (TIC) based on that the signal-to-noise of features is more than ten, as well as their intensities of the sample-to blank should be more than three [37].
3.3. Bacteria
Twenty-three S. aureus isolates were from clinical specimens, including blood, wounds, sputum, and urine. They were identified by standard biochemical tests.
3.4. Antibiotic Susceptibility Testing
The antibiotic sensitivity of the S. aureus isolates was explored by the Kirby–Bauer disk diffusion technique. Mueller–Hinton agar (MHA) plates are utilized in this assay [70]. The following antibiotics were used: oxacillin (OX; 1 μg), erythromycin (E; 15 μg), gentamicin (GN; 10 μg), linezolid (LZD; 30 μg), clindamycin (DA; 2 μg), tetracycline (TE; 30 μg), cotrimoxazole (COT; 1.25/23.75 μg), minocycline (MI; 30 μg), gatifloxacin (GAT; 5 μg), chloramphenicol (C; 30 μg), azithromycin (AZM; 15 μg), and ciprofloxacin (CIP; 5 μg).
3.5. Antibacterial Action of C. droserifolia Methanol Extract
Antibacterial action was revealed by agar well diffusion in MHA plates [71]. The bacterial suspension (0.5 McFarland) was dispersed on the surface of the MHA plates. Three wells were performed. The first well-received CDE (2 mg/mL), the second received linezolid (positive control), and the third received dimethyl sulfoxide (DMSO, negative control). The appearance of inhibition zones revealed CDE’s antibacterial activity after incubating the plates at 37 °C for 24 h [72].
3.6. Determination of the Minimum Inhibitory Concentration (MIC) of CDE
The broth microdilution assay in MH broth was employed to estimate the MIC values of CDE against S. aureus isolates, as previously reported [72]. The MIC had the lowest concentration of CDE, and no growth was detected visually after overnight incubation at 37 °C [73].
3.7. Biofilm Inhibition
The effect of CDE on biofilm formation was investigated at 0.5 MIC values [74]. A tryptone soy broth (TSB) suspension was prepared from an over-night bacterial culture, and was adjusted to 106 CFU/mL in freshly prepared TSB. Then, 200 µL of the bacterial suspension was added to the microtitration plates and wells in the presence and absence of sub-MIC (0.5 MIC) of SAM and incubated at 37 °C for 48 h. The TSB was gently removed, and the wells were washed to remove any planktonic cells and subsequently left for air drying. Add 200 µL of 99% methanol for 20 min, and then the formed biofilm was stained with 200 µL of 1% crystal violet (CV) solution for 15 min. After washing the plate, 33% glacial acetic acid was utilized as a solvent for CV. Using a microtitration plate reader (Sunrise, Männedorf, Switzerland), the absorbance of the solubilized dye was measured at 570 nm [72].
3.8. Scanning Electron Microscope (SEM)
The antibiofilm action of CDE on S. aureus bacteria was visualized under SEM, as previously explained (JEOL, Tokyo, Japan) [75].
3.9. Gene Expression Measurement Using qRT-PCR
The influence of CDE on the expression levels of the biofilm genes (cna, fnbA, and icaA) was elucidated using qRT-PCR. After growing the isolates in TSB in the presence and absence of sub-MICs of SAM, they were incubated overnight at 37 °C. After the incubation period, cells were harvested by centrifugation and immediately stored at −80 °C. The total RNA from S. aureus isolates was extracted and purified using TRIzol® reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer protocol. Reverse transcription was employed using the QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany). Then, the formed cDNA was amplified using Maximas SYBR Green/Fluorescein qPCR master mix (Thermo Fisher Scientific, Waltham, MA, USA). The average threshold cycle (CT) values were normalized to the housekeeping gene (16s rRNA). The relative gene expression of the treated isolates was compared to that of the untreated ones according to the 2−∆∆Ct method [76]. Primers are exposed in Table S1 [77,78].
3.10. In Vivo Assay
Forty male mice weighing 25–30 g and aged 6–8 weeks were obtained from the faculty of pharmacy at Tanta University, Egypt. They were grouped into four groups, each with ten mice. The first group was a normal control. The residual three groups were infected with 0.1 mL via intravenous injection of 1.5 × 107 colony-forming units (CFUs) of S. aureus [79]. The second group served as a positive control group (placebo), and the third group administered linezolid (160 mg/kg/24 h) orally as a standard drug. The fourth group administered CDE orally (200 mg/kg/24 h) [12]. The experimental procedures were approved by the ethics committee at the faculty of pharmacy, Tanta University (TP/RE/3/24 p-03).
After two weeks, mice from the diverse groups were euthanized. Liver and spleen samples were obtained from each group. The bacterial burden was determined in the liver and spleen after homogenization. A 1:10 serial dilution of the tissue homogenates in phosphate buffered saline was performed. Then, 100 μL of undiluted and each subsequent dilution were spread onto tryptic soya agar plates in duplicate using a glass spreader. The plates were then incubated at 37 °C overnight.
The number of colonies was counted on each plate, and the count of CFU/mL was determined as follows: CFU/mL = number of colonies × dilution factor/0.1 mL. CFU/g = CFU/mL × number of mL/g [80].
On the other hand, tissue samples (2 × 3 mm) were excised, fixed in buffered formalin (10%), treated as previously described [81], and finally stained with hematoxylin and eosin (H&E) and photographed using a light microscope. Also, tumor necrosis factor-alpha (TNF-α) monoclonal antibodies were utilized for staining the tissues. The immunostained tissues were then checked by a light microscope.
3.11. ELISA
The anti-inflammatory potential of CDE was illuminated by determining the levels of the inflammatory mediators, interleukin IL-1β and IL-6 in pg/mg protein in the liver and spleen tissues by an ELISA kit from Abcam Co., Waltham, MA, USA, following the manufacturer’s instructions.
3.12. Histopathological Examination
Formalin-fixed hepatic and splenic tissues were processed, and 5-µm-thick paraffin sections were stained with hematoxylin and eosin (H&E). Photomicrographs were taken at different magnification powers using a light microscope (Olympus, Tokyo, Japan) to assess the morphological changes [82]. Histopathological evaluation of the hepatic and splenic tissue damage was performed. A score of zero indicates the absence of tissue necrosis; a score of one indicates mild damage, 10–20% liver cell degeneration, necrosis, and 10–20% red blood cell depletion. A score of two indicates moderate degeneration in the form of 20–40% liver cell degeneration, necrosis, and 20–40% red blood cell depletion in the spleen. A score of three indicates severe degeneration in the form of >40% liver cell degeneration and necrosis and >40% red blood cell depletion in the spleen.
3.13. Immunohistochemistry
Six-micrometer tissue sections from the liver and spleen were subjected to immunohistochemical staining; they were first dewaxed, rehydrated by a diminishing alcohol series, and treated with 10% hydrogen peroxide in methanol for ten minutes. Following this, the sections were microwaved for ten minutes in 0.01 M sodium citrate buffer (pH 6.0), allowed to cool at room temperature, and then repeatedly washed with PBS for five minutes. After washing, antigens were recovered by autoclaving in citrate buffer for 11 min. Next, slices were incubated with primary antibodies for a whole night at 4 °C. Then, the tissues were treated for 30 min at room temperature with 3, 3-diaminobenzidine and a rabbit polyclonal TNF-α antibody. The tissue sections were mounted for visibility, washed in xylene, and subtly counterstained with hematoxylin. Slides were examined under a light microscope at a magnification of ×400 [83]. Using image analysis tools (Image J, 1.46a, NIH, Bethesda, MD, USA), morphometric analysis was carried out. At ×400 magnification, the mean area percentage of TNF-α protein expression for each of the experimental groups was evaluated in ten non-overlapping fields within each region.
3.14. Statistics
The assays were carried out three times and exposed as mean ± standard deviation (SD). ANOVA was utilized to reveal the significance of differences among the experimental groups by GraphPad software version 8.0 (GraphPad Software, LLC, Boston, MA, USA). Results of the histopathological scores were analyzed using Kruskal–Wallis test followed by Dunn’s multiple comparison test.
4. Discussion
Medicinal plants can act as a natural source of numerous therapeutic compounds that can be utilized safely to treat various diseases in humans and animals [84]. In recent decades, increasing attention has been paid to elucidating plants as an important source for numerous drugs, principally antimicrobials, to combat multidrug-resistant bacteria [85]. Here, the tested S. aureus isolates were from blood (52.17%), wounds (21.73%), sputum (17.4%), and urine (8.7%). Previous studies have documented that most recovered S. aureus isolates were from blood and wounds [86,87,88]. Regarding the susceptibility to antibiotics, the isolates tested in our study revealed multidrug resistance comparable to previous reports [89,90,91].
CDE revealed antibacterial action on S. aureus isolates with MIC values of 128–512 µg/mL. An earlier study described the antibacterial action of CDE on S. aureus NCTC 10788, Salmonella senftenberg ATCC 8400, Escherichia coli BA 12296, and Candida albicans ATCC MAY-2876 [12]. In addition, the antibacterial action of the essential oil obtained from Cleome species was previously reported on Gram-positive and Gram-negative bacterial species [92].
The phytochemical analysis by LC-ESI-MS/MS of C. droserifolia methanol extract tentatively identified 44 compounds belonging to different entities. Flavonoids, anthocyanin, and organic acids composed a major part of the extract, and it was found that the major constituents were kaempferol-3,7-O-bis-alpha-L-rhamnoside, isorhamnetin, cyanidin-3-glucoside, 3, 5, 7-trihydroxy-4′-methoxyflavone (kaempferide), kaempferol-3-O-alpha-L-rhamnoside, caffeic acid, isoquercitrin, quinic acid, isocitrate, mannitol, apigenin, acacetin, and naringenin. Kaempferol glycosides were reported to exert antimicrobial and anti-inflammatory effects [93,94]. Isorhamnetin, or 3′-methoxylated quercetin derivative, is a flavanol with antidiabetic, anti-inflammatory, and antimicrobial effects [95]. Flavonoids such as kaempferol derivatives, isorhamnetin, apigenin, acacetin, and naringenin are valuable groups of compounds with antimicrobial and anti-biofilm activities [96].
Wang et al. reported that cyanidin-3-glucoside exhibited antimicrobial and anti-inflammatory potential [97]. Cyanidin-3-glucoside inhibits the NF-κB pathway. Also, it was reported to inhibit the interferon-mediating inflammatory cascades and reduce the proinflammatory cytokines, such as interferon-γ, TNF-α, interleukin (IL)-5, IL-9, and IL-10 [95,98].
As biofilm is an important virulence factor for S. aureus, it enables it to resist multiple antibiotics by transferring the genes of resistance among the bacterial cells embedded in the biofilms and by hindering the penetration of the antibiotics across the biofilm [99,100]. Thus, antibiofilm agents are beneficial for managing MDR S. aureus infections [101]. Herein, CDE revealed antibiofilm action by crystal violet and SEM. Also, it revealed a downregulating effect on the biofilm-encoding genes (cna, fnbA, and icaA) using qRT-PCR in 43.48% of the isolates. Such genes encode intracellular adhesion molecules (icaA) and microbial surface components recognizing adhesive matrix molecules (fnbA and cna), which have a great role in biofilm formation [102].
Regarding the in vivo model, which was employed to simulate the human body [103], CDE revealed a promising effect in the studied infection model as it significantly decreased the bacterial count in the liver and spleen, indicating its antibacterial action in vivo. Also, it improved the histological features of the liver and spleen, manifested by regaining the normal hepatic structure and the splenic architecture.
As bacterial infections are among the causes that often trigger inflammation as a body response [104], we elucidated the consequence of CDE on the inflammatory markers in the liver and spleen using immunohistochemistry and ELISA. TNF-α is an inflammatory cytokine produced by macrophages as a response to acute inflammation [105,106]. Also, IL-1β is a proinflammatory cytokine that mediates numerous physiological responses, such as fever and lymphocyte activation [107,108]. IL-6 is an important pleiotropic cytokine in the inflammatory response [109,110,111]. Remarkably, CDE has significantly diminished these inflammatory mediators, which could have a role in its antibacterial potential. The experiment demonstrated a notable enhancement in the survival rate of rats infected with S. aureus when treated with CDE compared to the positive control group. CDE efficacy was comparable to that of the standard drug, linezolid. These results highlight the potential therapeutic value of CDE as an alternative treatment for S. aureus infections, warranting further investigation into the clinical relevance of these findings.
5. Conclusions
LC-ESI-MS/MS revealed that C. droserifolia methanol extract had variable phytochemicals with valuable biological activities. Flavonoid glycosides, anthocyanins, and other phenolic compounds were believed to cause multiple effects, such as antimicrobial, antibiofilm, and anti-inflammatory properties. The predominant compounds in CDE were kaempferol-3,7-O-bis-alpha-L-rhamnoside, isorhamnetin, cyanidin-3-glucoside, 3, 5, 7-trihydroxy-4′-methoxyflavone (kaempferide), kaempferol-3-O-alpha-L-rhamnoside, caffeic acid, isoquercitrin, quinic acid, isocitrate, mannitol, apigenin, acacetin, and naringenin. CDE exhibited a potent antibacterial action on S. aureus isolates with MICs that ranged from 128 to 512 µg/mL. It also revealed antibiofilm action using a crystal violet assay and SEM. This antibiofilm potential was further studied at the molecular level using qRT-PCR on the biofilm-encoding genes, and it revealed a downregulating action on the studied genes in 43.48% of the isolates. In the in vivo aspect, using ELISA and immunohistochemical studies, the CDE-treated group showed a significant improvement in the histological features, with a significant lessening in the inflammatory markers in the liver and spleen. Our work has shown that treatment with CDE can significantly improve the survival rate of S. aureus-infected rats, with efficacy similar to that of the standard drug linezolid, suggesting potential therapeutic value and prompting further exploration of its clinical relevance. Thus, it is important to perform future studies on CDE to reveal its potential activity on other bacterial species and to elucidate its action in clinical practice.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13050450/s1, Table S1. Sequences of the utilized primers. Table S2. Inhibition zone diameter of SAM, linezolid (positive control), and DMSA (negative control). Table S3. Minimum inhibitory concentrations (MICs) of SAM. Figure S1. Negative mode total ion chromatogram of LC-ESI-MS/MS of Cleome droserifolia methanol extract.
Author Contributions
J.A.: data curation, funding acquisition, software, validation, and writing—review and editing. W.A.N.: data curation, formal analysis, methodology, writing—original draft, and writing—review and editing. E.E.: data curation, formal analysis, methodology, writing—original draft, and writing—review and editing. I.A.H.: funding acquisition, investigation, resources, software, and writing—review and editing. H.S.H.: resources, software, validation, and writing—original draft. A.R.A.: funding acquisition, investigation, resources, and software. E.M.: data curation, project administration, resources, validation, and writing—review and editing. R.A.: data curation, investigation, software, validation, and writing—review and editing. S.I.: methodology, software, and writing—original draft. S.A.E.-S.: data curation, formal analysis, methodology, writing—original draft, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding form Researchers Supporting Project number (RSPD2024R583), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
The experimental procedures were approved by the ethics committee at the faculty of pharmacy, Tanta University (TP/RE/3/24 p-03).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
The authors are thankful to Researchers Supporting Project number (RSPD2024R583), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare absence of conflict of interests.
References
- Moustafa, A.A.; Mahmoud, M.A.K. Importance of Cleome droserifolia as an endangered medicinal plant species in the Sinai Peninsula and the need for its conservation. Adv. Med. Plant Res. 2023, 11, 43–51. [Google Scholar]
- Hegazy, A.K. Population ecology and implications for conservation of Cleome droserifolia: A threatened xerophyte. J. Arid Environ. 1990, 19, 269–282. [Google Scholar] [CrossRef]
- a Moustafa, A.R.; Alotaibi, M.O. Cleome droserifolia: An Egyptian natural heritage facing extinction. Asian J. Plant Sci. Res. 2019. [Google Scholar]
- Darbyshire, I.; Kordofani, M.; Farag, I.; Candiga, R.; Pickering, H. The Plants of Sudan and South Sudan: An Annotated Checklist; Royal Botanic Gardens: Kew, UK, 2015. [Google Scholar]
- Ghazanfar, S.A. An Annotated Catalogue of the Vascular Plants of Oman and Their Vernacular Names; Jardin Botanique National de Belgique: Meise, Belgium, 1992; Volume 2. [Google Scholar]
- Batanouny, K.; Aboutabl, E.; Shabana, M.; Soliman, F. Wild Medicinal Plants in Egypt; The Palm Press: Cairo, Egypt, 1999; Volume 154. [Google Scholar]
- Tackholm, V.; Boulos, L. Students’ Flora of Egypt; Cairo University: Cairo, Egypt, 1974. [Google Scholar]
- El-Askary, H.I. Terpenoids from Cleome droserifolia (Forssk.) Del. Molecules 2005, 10, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Helal, E.G.; Khattab, A.M.; Abou Aouf, N.; Abdallah, I.Z. Hypoglycemic and antioxidant effects of Cleome droserifolia (Samwah) in Alloxan-induced diabetic rats. Egypt. J. Hosp. Med. 2015, 58, 39–47. [Google Scholar] [CrossRef]
- Seif El-Din, A.; Darwish, F.; Abou-Donia, A. Flavonoids from Cleome droserifolia (Forssk.) Del. Growing in Egypt. Egypt. J. Pharm. Sci. 1987, 28, 313. [Google Scholar]
- Abd El-Gawad, A.M.; El-Amier, Y.A.; Bonanomi, G. Essential oil composition, antioxidant and allelopathic activities of Cleome droserifolia (Forssk.) Delile. Chem. Biodivers. 2018, 15, e1800392. [Google Scholar] [CrossRef] [PubMed]
- Hashem, N.M.; Shehata, M.G. Antioxidant and antimicrobial activity of Cleome droserifolia (Forssk.) Del. and its biological effects on redox status, immunity, and gut microflora. Animals 2021, 11, 1929. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, M.; Riad, A.; Soliman, R.A.; Elkhouly, A.M.; Nafae, H.; Gad, M.Z.; Motaal, A.A.; Youness, R.A. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell. Biochem. 2022, 477, 1281–1293. [Google Scholar] [CrossRef]
- Nicola, W.; Ibrahim, K.; Mikhail, T.; Girgis, R.; Khadr, M. Role of the hypoglycemic plant extract cleome droserifolia in improving glucose and lipid metabolism and its relation to insulin resistance in fatty liver. Boll. Chim. Farm. 1996, 135, 507–517. [Google Scholar]
- Cave, R.; Misra, R.; Chen, J.; Wang, S.; Mkrtchyan, H.V. Whole genome sequencing revealed new molecular characteristics in multidrug resistant staphylococci recovered from high frequency touched surfaces in London. Sci. Rep. 2019, 9, 9637. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between virulence and resistance among gram-negative bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Cheung, G.Y.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Saha, M.; Sarkar, A. Review on multiple facets of drug resistance: A rising challenge in the 21st century. J. Xenobiotics 2021, 11, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Siddiqui, S.A.; Khan, S.; Mehdizadeh, M.; Bahmid, N.A.; Adli, D.N.; Walker, T.R.; Perestrelo, R.; Câmara, J.S. Phytochemicals and bioactive constituents in food packaging: A systematic review. Heliyon 2023, 9, e21196. [Google Scholar] [CrossRef]
- Bittner Fialová, S.; Rendeková, K.; Mučaji, P.; Nagy, M.; Slobodníková, L. Antibacterial activity of medicinal plants and their constituents in the context of skin and wound infections, considering European legislation and folk medicine—A review. Int. J. Mol. Sci. 2021, 22, 10746. [Google Scholar] [CrossRef]
- Attallah, N.G.; Al-Fakhrany, O.M.; Elekhnawy, E.; Hussein, I.A.; Shaldam, M.A.; Altwaijry, N.; Alqahtani, M.J.; Negm, W.A. Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches. Antibiotics 2022, 11, 993. [Google Scholar] [CrossRef] [PubMed]
- Alherz, F.A.; Negm, W.A.; Elekhnawy, E.; El-Masry, T.A.; Haggag, E.M.; Alqahtani, M.J.; Hussein, I.A. Silver Nanoparticles Prepared Using Encephalartos laurentianus De Wild Leaf Extract Have Inhibitory Activity against Candida albicans Clinical Isolates. J. Fungi 2022, 8, 1005. [Google Scholar] [CrossRef]
- Negm, W.A.; El-Aasr, M.; Attia, G.; Alqahtani, M.J.; Yassien, R.I.; Abo Kamer, A.; Elekhnawy, E. Promising Antifungal Activity of Encephalartos laurentianus de Wild against Candida albicans Clinical Isolates: In Vitro and In Vivo Effects on Renal Cortex of Adult Albino Rats. J. Fungi 2022, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, M.J.; Elekhnawy, E.; Negm, W.A.; Mahgoub, S.; Hussein, I.A. Encephalartos villosus Lem. Displays a strong in vivo and in vitro antifungal potential against Candida glabrata clinical isolates. J. Fungi 2022, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Motaal, A.A.; Ezzat, S.M.; Haddad, P. Determination of bioactive markers in Cleome droserifolia using cell-based bioassays for antidiabetic activity and isolation of two novel active compounds. Phytomedicine 2011, 19, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Jarukas, L.; Kamarauskaitė, J.; Marksa, M.; Trumbeckaitė, S.; Banienė, R.; Ivanauskas, L. Bio-based succinic acid sample preparation and derivatization procedure optimisation for gas chromatography-mass spectrometry analysis. Sci. Pharm. Sci. 2018, 4, 9–13. [Google Scholar] [CrossRef][Green Version]
- Vasilev, H.; Šmejkal, K.; Jusková, S.; Vaclavik, J.; Treml, J. Five New Tamarixetin Glycosides from Astragalus thracicus Griseb. Including Some Substituted with the Rare 3-Hydroxy-3-methylglutaric Acid and Their Collagenase Inhibitory Effects In Vitro. ACS Omega 2024, 9, 18023–18031. [Google Scholar] [CrossRef]
- Abela, L.; Spiegel, R.; Crowther, L.M.; Klein, A.; Steindl, K.; Papuc, S.M.; Joset, P.; Zehavi, Y.; Rauch, A.; Plecko, B. Plasma metabolomics reveals a diagnostic metabolic fingerprint for mitochondrial aconitase (ACO2) deficiency. PLoS ONE 2017, 12, e0176363. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, M.N.; Ahmad, S.; Tousif, M.I.; Ahmad, I.; Rao, H.; Ahmad, B.; Basit, A. Profiling of phytochemicals from aerial parts of Terminalia neotaliala using LC-ESI-MS2 and determination of antioxidant and enzyme inhibition activities. PLoS ONE 2022, 17, e0266094. [Google Scholar] [CrossRef]
- Surendra, S.V.; Mahalingam, B.L.; Velan, M. Degradation of monoaromatics by Bacillus pumilus MVSV3. Braz. Arch. Biol. Technol. 2017, 60, e16160319. [Google Scholar] [CrossRef]
- Mekky, R.H.; del Mar Contreras, M.; El-Gindi, M.R.; Abdel-Monem, A.R.; Abdel-Sattar, E.; Segura-Carretero, A. Profiling of phenolic and other compounds from Egyptian cultivars of chickpea (Cicer arietinum L.) and antioxidant activity: A comparative study. RSC Adv. 2015, 5, 17751–17767. [Google Scholar] [CrossRef]
- Mahrous, F.S.M.; Mohammed, H.; Sabour, R. LC-ESI-QTOF-MS/MS of Holoptelea integrifolia (Roxb.) Planch. leaves and In silico study of phenolic compounds’ antiviral activity against the HSV1 virus. Azhar Int. J. Pharm. Med. Sci. 2021, 1, 91–101. [Google Scholar] [CrossRef]
- Attallah, N.G.; Negm, W.A.; Elekhnawy, E.; Elmongy, E.I.; Altwaijry, N.; El-Haroun, H.; El-Masry, T.A.; El-Sherbeni, S.A. Elucidation of phytochemical content of Cupressus macrocarpa leaves: In Vitro and in vivo antibacterial effect against methicillin-resistant Staphylococcus aureus clinical isolates. Antibiotics 2021, 10, 890. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Schmeda-Hirschmann, G. Direct identification of phenolic constituents in Boldo Folium (Peumus boldus Mol.) infusions by high-performance liquid chromatography with diode array detection and electrospray ionization tandem mass spectrometry. J. Chromatogr. A 2010, 1217, 443–449. [Google Scholar] [CrossRef]
- Huang, G.; Liang, J.; Chen, X.; Lin, J.; Wei, J.; Huang, D.; Zhou, Y.; Sun, Z.; Zhao, L. Isolation and identification of chemical constituents from zhideke granules by ultra-performance liquid chromatography coupled with mass spectrometry. J. Anal. Methods Chem. 2020, 2020, 8889607. [Google Scholar] [CrossRef]
- Li, W.; Sun, Y.; Liang, W.; Fitzloff, J.F.; van Breemen, R.B. Identification of caffeic acid derivatives in Actea racemosa (Cimicifuga racemosa, black cohosh) by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 978–982. [Google Scholar] [CrossRef]
- Ferreira, P.S.; Victorelli, F.D.; Fonseca-Santos, B.; Chorilli, M. A review of analytical methods for p-coumaric acid in plant-based products, beverages, and biological matrices. Crit. Rev. Anal. Chem. 2019, 49, 21–31. [Google Scholar] [CrossRef]
- Andini, S.; Dekker, P.; Gruppen, H.; Araya-Cloutier, C.; Vincken, J.-P. Modulation of glucosinolate composition in Brassicaceae seeds by germination and fungal elicitation. J. Agric. Food Chem. 2019, 67, 12770–12779. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, M.; Ou, Y.; Zeng, B.; Lou, X.; Wang, M.; Zhao, C. Metabolic profile of esculin in rats by ultra high performance liquid chromatography combined with Fourier transform ion cyclotron resonance mass spectrometry. J. Chromatogr. B 2016, 1020, 120–128. [Google Scholar] [CrossRef]
- Jaiswal, R.; Müller, H.; Müller, A.; Karar, M.G.E.; Kuhnert, N. Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicera henryi L. (Caprifoliaceae) leaves by LC–MSn. Phytochemistry 2014, 108, 252–263. [Google Scholar] [CrossRef]
- Razgonova, M.; Zakharenko, A.; Pikula, K.; Manakov, Y.; Ercisli, S.; Derbush, I.; Kislin, E.; Seryodkin, I.; Sabitov, A.; Kalenik, T. LC-MS/MS Screening of Phenolic Compounds in Wild and Cultivated Grapes Vitis amurensis Rupr. Molecules 2021, 26, 3650. [Google Scholar] [CrossRef]
- El-Newary, S.A.; Abd Elkarim, A.S.; Abdelwahed, N.A.; Omer, E.A.; Elgamal, A.M.; ELsayed, W.M. Chenopodium murale Juice Shows Anti-Fungal Efficacy in Experimental Oral Candidiasis in Immunosuppressed Rats in Relation to Its Chemical Profile. Molecules 2023, 28, 4304. [Google Scholar] [CrossRef]
- Feketeová, L.; Barlow, C.K.; Benton, T.M.; Rochfort, S.J.; Richard, A. The formation and fragmentation of flavonoid radical anions. Int. J. Mass Spectrom. 2011, 301, 174–183. [Google Scholar] [CrossRef]
- Schütz, K.; Persike, M.; Carle, R.; Schieber, A. Characterization and quantification of anthocyanins in selected artichoke (Cynara scolymus L.) cultivars by HPLC–DAD–ESI–MSn. Anal. Bioanal. Chem. 2006, 384, 1511–1517. [Google Scholar] [CrossRef]
- Sánchez-Ilárduya, M.; Sánchez-Fernández, C.; Viloria-Bernal, M.; López-Márquez, D.; Berrueta, L.; Gallo, B.; Vicente, F. Mass spectrometry fragmentation pattern of coloured flavanol-anthocyanin and anthocyanin-flavanol derivatives in aged red wines of Rioja. Aust. J. Grape Wine Res. 2012, 18, 203–214. [Google Scholar] [CrossRef]
- Abdl Aziz, F.T.; Temraz, A.S.; Hassan, M.A. Metabolites profiling by LC-ESI-MS/MS technique and in-vitro antioxidant activity of Bauhinia madagascariensis Desv. and Bauhinia purpurea L. aerial parts cultivated in Egypt: A comparative study. Azhar Int. J. Pharm. Med. Sci. 2024, 4, 169–188. [Google Scholar] [CrossRef]
- Siger, A.; Czubinski, J.; Dwiecki, K.; Kachlicki, P.; Nogala-Kalucka, M. Identification and antioxidant activity of sinapic acid derivatives in Brassica napus L. seed meal extracts. Eur. J. Lipid Sci. Technol. 2013, 115, 1130–1138. [Google Scholar] [CrossRef]
- Asen, S.; Plimmer, J.R. 4,6,4′-Trihydroxyaurone and other flavonoids from Limonium. Phytochemistry 1972, 11, 2601–2603. [Google Scholar] [CrossRef]
- Mizuno, T.; Uchiyama, N.; Tanaka, S.; Nakane, T.; Devkota, H.P.; Fujikawa, K.; Kawahara, N.; Iwashina, T. Flavonoids from Sedum japonicum subsp. oryzifolium (Crassulaceae). Molecules 2022, 27, 7632. [Google Scholar] [CrossRef]
- Hu, Q.; Sun, Y.; Mu, X.; Wang, Y.; Tang, H. Reliable quantification of citrate isomers and isobars with direct-infusion tandem mass spectrometry. Talanta 2023, 259, 124477. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, P.; Brantner, A.; Wang, H.; Shu, X.; Yang, J.; Si, N.; Han, L.; Zhao, H.; Bian, B. Chemical profiling analysis of Maca using UHPLC-ESI-Orbitrap MS coupled with UHPLC-ESI-QqQ MS and the neuroprotective study on its active ingredients. Sci. Rep. 2017, 7, 44660. [Google Scholar] [CrossRef]
- Ablajan, K.; Tuoheti, A. Fragmentation characteristics and isomeric differentiation of flavonol O-rhamnosides using negative ion electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 451–460. [Google Scholar] [CrossRef]
- Rini Vijayan, K.P.; Raghu, A. Tentative characterization of phenolic compounds in three species of the genus Embelia by liquid chromatography coupled with mass spectrometry analysis. Spectrosc. Lett. 2019, 52, 653–670. [Google Scholar] [CrossRef]
- Brito, A.; Areche, C.; Sepúlveda, B.; Kennelly, E.J.; Simirgiotis, M.J. Anthocyanin characterization, total phenolic quantification and antioxidant features of some Chilean edible berry extracts. Molecules 2014, 19, 10936–10955. [Google Scholar] [CrossRef]
- Abd-El-Aziz, N.M.; Hifnawy, M.S.; Lotfy, R.A.; Younis, I.Y. LC/MS/MS and GC/MS/MS metabolic profiling of Leontodon hispidulus, in vitro and in silico anticancer activity evaluation targeting hexokinase 2 enzyme. Sci. Rep. 2024, 14, 6872. [Google Scholar] [CrossRef]
- Kaur, J.; Dhiman, V.; Bhadada, S.; Katare, O.; Ghoshal, G. LC/MS guided identification of metabolites of different extracts of Cissus quadrangularis. Food Chem. Adv. 2022, 1, 100084. [Google Scholar] [CrossRef]
- Brito, A.; Ramirez, J.E.; Areche, C.; Sepúlveda, B.; Simirgiotis, M.J. HPLC-UV-MS profiles of phenolic compounds and antioxidant activity of fruits from three citrus species consumed in Northern Chile. Molecules 2014, 19, 17400–17421. [Google Scholar] [CrossRef]
- Choi, S.S.; Lee, S.H.; Lee, K.A. A Comparative Study of Hesperetin, Hesperidin and Hesperidin Glucoside: Antioxidant, Anti-Inflammatory, and Antibacterial Activities In Vitro. Antioxidants 2022, 11, 1618. [Google Scholar] [CrossRef]
- Wu, W.; Liu, Z.; Song, F.; Liu, S. Structural analysis of selected characteristic flavones by electrospray tandem mass spectrometry. Anal. Sci. 2004, 20, 1103–1105. [Google Scholar] [CrossRef]
- Fabre, N.; Rustan, I.; de Hoffmann, E.; Quetin-Leclercq, J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Huang, Y.; Wen, X.; Wu, Y.; Zhao, Y.; Ni, Y. Extraction, purification, and hydrolysis behavior of apigenin-7-O-Glucoside from Chrysanthemum Morifolium tea. Molecules 2018, 23, 2933. [Google Scholar] [CrossRef] [PubMed]
- Prasain, J.K.; Jones, K.; Kirk, M.; Wilson, L.; Smith-Johnson, M.; Weaver, C.; Barnes, S. Profiling and quantification of isoflavonoids in kudzu dietary supplements by high-performance liquid chromatography and electrospray ionization tandem mass spectrometry. J. Agric. Food Chem. 2003, 51, 4213–4218. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.A.; dos Santos, L.R.; de Freitas, J.S.; de Sousa, R.P.; de Farias, R.R.S.; Vieira Júnior, G.M.; Rai, M.; Chaves, M.H. First report of flavonoids from leaves of Machaerium acutifolium by DI-ESI-MS/MS. Arab. J. Chem. 2022, 15, 103765. [Google Scholar] [CrossRef]
- Sayed, D.; Afifi, A.; Temraz, A.; Ahmed, A. Metabolic Profiling of Mimusops elengi Linn. Leaves extract and in silico anti-inflammatory assessment targeting NLRP3 inflammasome. Arab. J. Chem. 2023, 16, 104753. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Shibabaw, A.; Abebe, T.; Mihret, A. Antimicrobial susceptibility pattern of nasal Staphylococcus aureus among Dessie Referral Hospital health care workers, Dessie, Northeast Ethiopia. Int. J. Infect. Dis. 2014, 25, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, B.; El-Masry, T.A.; Elekhnawy, E.; El-Kadem, A.H.; Saleh, A.; Negm, W.A.; Abdelkader, D.H. Aqueous core epigallocatechin gallate PLGA nanocapsules: Characterization, antibacterial activity against uropathogens, and in vivo reno-protective effect in cisplatin induced nephrotoxicity. Drug Deliv. 2022, 29, 1848–1862. [Google Scholar] [CrossRef]
- Abbas, H.A.; Atallah, H.; El-Sayed, M.A.; El-Ganiny, A.M. Diclofenac mitigates virulence of multidrug-resistant Staphylococcus aureus. Arch. Microbiol. 2020, 202, 2751–2760. [Google Scholar] [CrossRef]
- Binsuwaidan, R.; Sultan, A.A.; Negm, W.A.; Attallah, N.G.; Alqahtani, M.J.; Hussein, I.A.; Shaldam, M.A.; El-Sherbeni, S.A.; Elekhnawy, E. Bilosomes as nanoplatform for oral delivery and modulated in vivo antimicrobial activity of lycopene. Pharmaceuticals 2022, 15, 1043. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Khayat, M.T.; Abbas, H.A.; Ibrahim, T.S.; Khayyat, A.N.; Alharbi, M.; Darwish, K.M.; Elhady, S.S.; Khafagy, E.-S.; Safo, M.K.; Hegazy, W.A. Anti-Quorum Sensing Activities of Gliptins against Pseudomonas aeruginosa and Staphylococcus aureus. Biomedicines 2022, 10, 1169. [Google Scholar] [CrossRef]
- Schmittgen, T.D. Real-Time Quantitative PCR; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Nejabatdoust, A.; Zamani, H.; Salehzadeh, A. Functionalization of ZnO nanoparticles by glutamic acid and conjugation with thiosemicarbazide alters expression of efflux pump genes in multiple drug-resistant Staphylococcus aureus strains. Microb. Drug Resist. 2019, 25, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Mastoor, S.; Nazim, F.; Rizwan-ul-Hasan, S.; Ahmed, K.; Khan, S.; Ali, S.N.; Abidi, S.H. Analysis of the antimicrobial and anti-biofilm activity of natural compounds and their analogues against Staphylococcus aureus isolates. Molecules 2022, 27, 6874. [Google Scholar] [CrossRef]
- Zhang, S.; Qu, X.; Tang, H.; Wang, Y.; Yang, H.; Yuan, W.; Yue, B. Diclofenac Resensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactams and Prevents Implant Infections. Adv. Sci. 2021, 8, 2100681. [Google Scholar] [CrossRef] [PubMed]
- Stackhouse, R.R.; Faith, N.G.; Kaspar, C.W.; Czuprynski, C.J.; Wong, A.C.L. Survival and virulence of Salmonella enterica serovar enteritidis filaments induced by reduced water activity. Appl. Environ. Microbiol. 2012, 78, 2213–2220. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Albogami, S.M.; Jean-Marc, S.; Nadwa, E.H.; Hafiz, A.A.; A. Negm, W.; Kamal, M.; Al-Jouboury, M.; Elekhnawy, E.; et al. Potential Therapeutic Benefits of Metformin Alone and in Combination with Sitagliptin in the Management of Type 2 Diabetes Patients with COVID-19. Pharmaceuticals 2022, 15, 1361. [Google Scholar] [CrossRef]
- Bancroft, J.D.; Gamble, M. Theory and Practice of Histological Techniques; Elsevier Health Sciences: Philadelphia, PA, USA, 2008. [Google Scholar]
- Miotto, A.; Lins, T.A.; Montero, E.; Oshima, C.; Alonso, L.G.; Perfeito, J.A. Immunohistochemical analysis of the COX-2 marker in acute pulmonary injury in rats. Ital. J. Anat. Embryol. Arch. Ital. Anat. Embriol. 2009, 114, 193–199. [Google Scholar]
- Anand, U.; Jacobo-Herrera, N.; Altemimi, A.; Lakhssassi, N. A comprehensive review on medicinal plants as antimicrobial therapeutics: Potential avenues of biocompatible drug discovery. Metabolites 2019, 9, 258. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Min, C.; Wang, H.; Xia, F.; Tang, M.; Li, J.; Hu, Y.; Dou, Q.; Zou, M. Characteristics of Staphylococcus aureus small colony variants isolated from wound specimen of a tertiary care hospital in China. J. Clin. Lab. Anal. 2022, 36, e24121. [Google Scholar] [CrossRef]
- Sapkota, J.; Sharma, M.; Jha, B.; Bhatt, C.P. Prevalence of Staphylococcus aureus isolated from clinical samples in a tertiary care hospital: A descriptive cross-sectional study. JNMA J. Nepal. Med. Assoc. 2019, 57, 398. [Google Scholar] [CrossRef]
- Idrees, M.M.; Saeed, K.; Shahid, M.A.; Akhtar, M.; Qammar, K.; Hassan, J.; Khaliq, T.; Saeed, A. Prevalence of mecA- and mecC-associated methicillin-resistant Staphylococcus aureus in clinical specimens, Punjab, Pakistan. Biomedicines 2023, 11, 878. [Google Scholar] [CrossRef]
- Chew, C.H.; Yeo, C.C.; Che Hamzah, A.M.; Al-Trad, E.a.I.; Jones, S.U.; Chua, K.H.; Puah, S.M. Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus Associated with Hospitalized Newborn Infants. Diagnostics 2023, 13, 1050. [Google Scholar] [CrossRef]
- Steinig, E.J.; Duchene, S.; Robinson, D.A.; Monecke, S.; Yokoyama, M.; Laabei, M.; Slickers, P.; Andersson, P.; Williamson, D.; Kearns, A. Evolution and global transmission of a multidrug-resistant, community-associated methicillin-resistant Staphylococcus aureus lineage from the Indian subcontinent. mBio 2019, 10, 1–20. [Google Scholar] [CrossRef]
- Raut, S.; Bajracharya, K.; Adhikari, J.; Pant, S.S.; Adhikari, B. Prevalence of methicillin resistant Staphylococcus aureus in Lumbini medical college and teaching hospital, Palpa, Western Nepal. BMC Res. Notes 2017, 10, 187. [Google Scholar] [CrossRef]
- Muhaidat, R.; Al-Qudah, M.A.; Samir, O.; Jacob, J.H.; Hussein, E.; Al-Tarawneh, I.N.; Bsoul, E.; Orabi, S.T.A. Phytochemical investigation and in vitro antibacterial activity of essential oils from Cleome droserifolia (Forssk.) Delile and C. trinervia Fresen. (Cleomaceae). S. Afr. J. Bot. 2015, 99, 21–28. [Google Scholar] [CrossRef]
- Tatsimo, S.J.N.; Tamokou, J.d.D.; Havyarimana, L.; Csupor, D.; Forgo, P.; Hohmann, J.; Kuiate, J.-R.; Tane, P. Antimicrobial and antioxidant activity of kaempferol rhamnoside derivatives from Bryophyllum pinnatum. BMC Res. Notes 2012, 5, 158. [Google Scholar] [CrossRef]
- Rho, H.S.; Ghimeray, A.K.; Yoo, D.S.; Ahn, S.M.; Kwon, S.S.; Lee, K.H.; Cho, D.H.; Cho, J.Y. Kaempferol and kaempferol rhamnosides with depigmenting and anti-inflammatory properties. Molecules 2011, 16, 3338–3344. [Google Scholar] [CrossRef]
- Kalai, F.Z.; Boulaaba, M.; Ferdousi, F.; Isoda, H. Effects of isorhamnetin on diabetes and its associated complications: A review of in vitro and in vivo studies and a post hoc transcriptome analysis of involved molecular pathways. Int. J. Mol. Sci. 2022, 23, 704. [Google Scholar] [CrossRef] [PubMed]
- Faleye, O.S.; Lee, J.-H.; Lee, J. Selected flavonoids exhibit antibiofilm and antibacterial effects against Vibrio by disrupting membrane integrity, virulence and metabolic activities. Biofilm 2023, 6, 100165. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Wang, Y.; Zheng, F.; Qu, M.; Huang, Z.; Yan, J.; Bao, F.; Li, X.; Sun, C. Cyanidin-3-O-glucoside extracted from the Chinese bayberry (Myrica rubra Sieb. et Zucc.) alleviates antibiotic-associated diarrhea by regulating gut microbiota and down-regulating inflammatory factors in NF-κB pathway. Front. Nutr. 2022, 9, 970530. [Google Scholar] [CrossRef]
- Frountzas, M.; Karanikki, E.; Toutouza, O.; Sotirakis, D.; Schizas, D.; Theofilis, P.; Tousoulis, D.; Toutouzas, K.G. Exploring the Impact of Cyanidin-3-Glucoside on Inflammatory Bowel Diseases: Investigating New Mechanisms for Emerging Interventions. Int. J. Mol. Sci. 2023, 24, 9399. [Google Scholar] [CrossRef]
- Kalia, V.C.; Patel, S.K.; Lee, J.-K. Bacterial biofilm inhibitors: An overview. Ecotoxicol. Environ. Saf. 2023, 264, 115389. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Sedarat, Z.; Taylor-Robinson, A.W. Biofilm formation by pathogenic bacteria: Applying a Staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens 2022, 11, 388. [Google Scholar] [CrossRef]
- Haddad, O.; Merghni, A.; Elargoubi, A.; Rhim, H.; Kadri, Y.; Mastouri, M. Comparative study of virulence factors among methicillin resistant Staphylococcus aureus clinical isolates. BMC Infect. Dis. 2018, 18, 560. [Google Scholar] [CrossRef]
- Vlachou, M.; Karalis, V. An in vitro–in vivo simulation approach for the prediction of bioequivalence. Materials 2021, 14, 555. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Jang, D.-i.; Lee, A.-H.; Shin, H.-Y.; Song, H.-R.; Park, J.-H.; Kang, T.-B.; Lee, S.-R.; Yang, S.-H. The role of tumor necrosis factor alpha (TNF-α) in autoimmune disease and current TNF-α inhibitors in therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef] [PubMed]
- Alomair, B.M.; Al-Kuraishy, H.M.; Al-Buhadily, A.K.; Al-Gareeb, A.I.; De Waard, M.; Elekhnawy, E.; Batiha, G.E.-S. Is sitagliptin effective for SARS-CoV-2 infection: False or true prophecy? Inflammopharmacology 2022, 30, 2411–2415. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, N.; Kurata, M.; Yamamoto, T.; Morikawa, S.; Masumoto, J. The role of interleukin-1 in general pathology. Inflamm. Regen. 2019, 39, 12. [Google Scholar] [CrossRef] [PubMed]
- Batiha, G.E.-S.; Al-Gareeb, A.I.; Elekhnawy, E.; Al-Kuraishy, H.M. Potential role of lipoxin in the management of COVID-19: A narrative review. Inflammopharmacology 2022, 30, 1993–2001. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Nadwa, E.H.; Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Elekhnawy, E.; Albogami, S.M.; Alorabi, M.; Batiha, G.E.-S.; De Waard, M. Cholinergic dysfunction in COVID-19: Frantic search and hoping for the best. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2023, 396, 453–468. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Elekhnawy, E.; Batiha, G.E.-S. Dipyridamole and adenosinergic pathway in COVID-19: A juice or holy grail. Egypt. J. Med. Hum. Genet. 2022, 23, 140. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).