Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile
Abstract
1. Introduction
2. Results and Discussion
2.1. Antimicrobial-Resistance Levels
2.2. Bacterial Identification
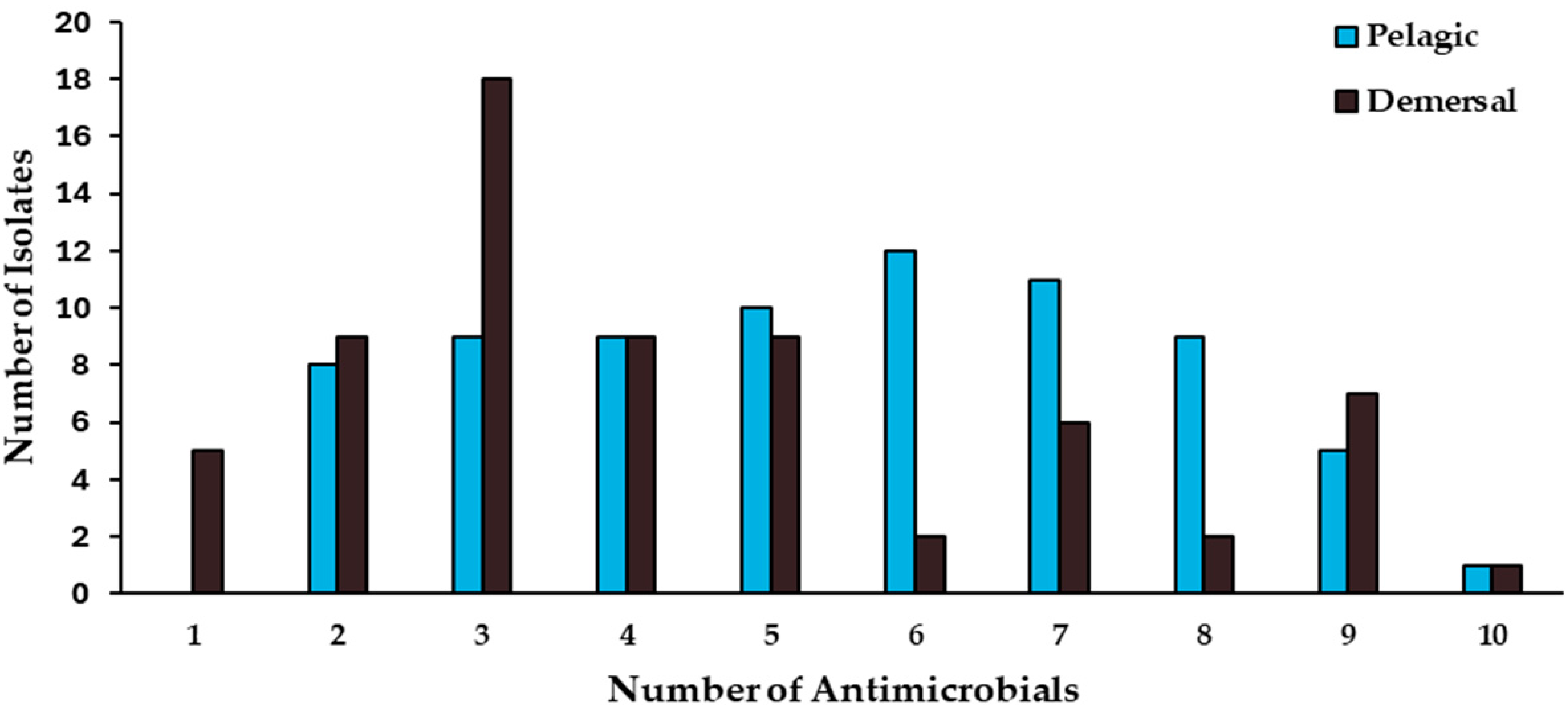
2.3. Antimicrobial-Resistance Patterns
2.4. ESBL and MBL Detection
2.5. Integron and Plasmid Detection
3. Materials and Methods
3.1. Sampling and Processing of Samples
3.2. Culturable Bacterial Counts
3.3. Bacterial Isolates
3.4. Bacterial Identification
3.5. Antimicrobial-Resistance Patterns
3.6. Detection of Extended-Spectrum-β-Lactamase (ESBL) Production
3.7. Screening for Class B Metallo-β-Lactamase (MBL) Production
3.8. Plasmid Content
3.9. Detection of Class 1 Integrons
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yang, D.; Jin, M.; Liu, W.; Zhao, X.; Li, C.; Zhao, T.; Wang, J.; Gao, Z.; Shen, Z. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017, 26, 5318–5333. [Google Scholar] [CrossRef] [PubMed]
- Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Carvalho, M.P.N.; Lincopan, N. Extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Mar. Pollut. Bull. 2018, 135, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D. Antimicrobial resistance in salmonid farming. In Antimicrobial Resistance in the Environment; Keen, P.L., Montforts, M.H.M.M., Eds.; Chapter 22; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 423–451. [Google Scholar]
- Domínguez, M.; Miranda, C.D.; Fuentes, O.; de la Fuente, M.; Godoy, F.A.; Bello-Toledo, H.; González-Rocha, G. Occurrence of transferable integrons and sul and dfr genes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Front. Microbiol. 2019, 10, 748. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Su, Y.; Deng, Y.; Guo, Z.; Cheng, C.; Ma, H.; Liu, G.; Xu, L.; Feng, J. Spatial and temporal variation of antibiotic resistance in marine fish cage-culture area of Guangdong, China. Environ. Pollut. 2019, 246, e463–e471. [Google Scholar] [CrossRef]
- Zhang, M.; Hou, L.; Zhu, Y.; Zhang, C.; Li, W.; Lai, X.; Yang, J.; Li, S.; Shu, H. Composition and distribution of bacterial communities and antibiotic resistance genes in fish of four mariculture systems. Environ. Pollut. 2022, 311, 119934. [Google Scholar] [CrossRef]
- Marti, E.; Huerta, B.; Rodríguez-Mozaz, S.; Barceló, D.; Marcé, R.; Balcázar, J.L. Abundance of antibiotic resistance genes and bacterial community composition in wild freshwater fish species. Chemosphere 2018, 196, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Jia, J.; Yue, X.; Guan, Y.; Zhu, L.; Wang, Z. River contamination shapes the microbiome and antibiotic resistance in sharpbelly (Hemiculter leucisculus). Environ. Pollut. 2021, 268, 115796. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-C.; Lin, Z.-J.; Shuai, X.-Y.; Zheng, J.; Meng, L.-X.; Zhu, L.; Sun, Y.-J.; Shang, W.-C.; Chen, H. Temporal variation and sharing of antibiotic resistance genes between water and wild fish gut in a peri-urban river. J. Environ. Sci. 2021, 103, 12–19. [Google Scholar] [CrossRef]
- Vivant, A.-L.; Marchand, E.; Janvier, B.; Berthe, T.; Guigon, E.; Grall, N.; Alliot, F.; Goutte, A.; Petit, F. Wild fish from a highly urbanized river (Orge, France) as vectors of culturable Enterobacterales resistant to antibiotics. Can. J. Microbiol. 2024, 70, 63–69. [Google Scholar] [CrossRef]
- Mills, M.; Lee, S.; Mollenkopf, D.; Wittum, T.; Sullivan, S.M.P.; Lee, J. Comparison of environmental microbiomes in an antibiotic resistance-polluted urban river highlights periphyton and fish gut communities as reservoirs of concern. Sci. Total Environ. 2022, 851, 158042. [Google Scholar] [CrossRef] [PubMed]
- Varol, M.; Rasit, M. Environmental contaminants in fish species from a large dam reservoir and their potential risks to human health. Ecotoxicol. Environ. Saf. 2019, 169, 507–515. [Google Scholar] [CrossRef]
- Griboff, J.; Carrizo, J.C.; Bonansea, R.I.; Valdés, M.E.; Wunderlin, D.A.; Amé, M.V. Multiantibiotic residues in commercial fish from Argentina. The presence of mixtures of antibiotics in edible fish, a challenge to health risk assessment. Food Chem. 2020, 332, 127380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yu, K.; Li, A.; Wang, Y.; Pan, C.; Huang, X. Antibiotics in coral reef fishes from the South China Sea: Occurrence, distribution, bioaccumulation, and dietary exposure risk to human. Sci. Total Environ. 2020, 704, 135288. [Google Scholar] [CrossRef]
- Carrizo, J.C.; Griboff, J.; Bonansea, R.I.; Nimptsch, J.; Valdés, M.E.; Wunderlin, D.A.; Amé, M.V. Different antibiotic profiles in wild and farmed Chilean salmonids. Which is the main source for antibiotic in fish? Sci. Total Environ. 2021, 800, 149516. [Google Scholar] [CrossRef] [PubMed]
- Bueno, I.; Verdugo, C.; Jimenez-Lopez, O.; Alvarez, P.P.; Gonzalez-Rocha, G.; Lima, C.A.; Travis, D.A.; Wass, B.; Zhang, Q.; Ishii, S.; et al. Role of wastewater treatment plants on environmental abundance of antimicrobial resistance genes in chilean rivers. Int. J. Hyg. Environ. Health 2020, 223, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Ballash, G.A.; Baesu, A.; Lee, S.; Mills, M.C.; Mollenkopf, D.F.; Sullivan, S.M.P.; Lee, J.; Bayen, S.; Wittum, T.E. Fish as sentinels of antimicrobial resistant bacteria, epidemic carbapenemase genes, and antibiotics in surface water. PLoS ONE 2022, 17, e0272806. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Zemelman, R. Antibiotic Resistant Bacteria in Fish from the Concepción Bay, Chile. Mar. Pollut. Bull. 2001, 42, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Ahlstrom, C.A. Antibiotic resistance bacteria in wildlife: Perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildl. Dis. 2020, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Dagot, C.; Chesneau, O.; Bibbal, D.; Labanowski, J.; Vialette, M.; Bouchard, D.; Martin-Laurent, F.; Calsat, L.; Nazaret, S.; et al. Environmental contamination in a high-income country (France) by antibiotics, antibiotic-resistant bacteria, and antibiotic resistance genes: Status and possible causes. Environ. Int. 2022, 159, 107047. [Google Scholar] [CrossRef]
- Bollache, L.; Bardet, E.; Depret, G.; Motreuil, S.; Neuwirth, C.; Moreau, J.; Hartmann, A. Dissemination of CTX-M-producing Escherichia coli in freshwater fishes from a French watershed (burgundy). Front. Microbiol. 2019, 9, 3239. [Google Scholar] [CrossRef] [PubMed]
- Kerry, J.; Hiney, M.; Coyne, R.; Cazabon, D.; NicGabhainn, S.; Smith, P. Frequency and distribution of resistance to oxytetracycline in micro-organisms isolated from marine fish farm sediments following therapeutic use of oxytetracycline. Aquaculture 1994, 123, 43–54. [Google Scholar] [CrossRef]
- Bush, K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010, 13, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A. β-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a025239. [Google Scholar] [CrossRef]
- Khurana, H.; Singh, D.N.; Singh, A.; Singh, Y.; Lal, R.; Negi, R.K. Gut microbiome of endangered Tor putitora (Ham.) as a reservoir of antibiotic resistance genes and pathogens associated with fish health. BMC Microbiol. 2020, 20, 249. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Singh, B.; Billekallu Thammegowda, N.K.; Singh, N.K. Shotgun metagenomics offers novel insights into taxonomic compositions, metabolic pathways and antibiotic resistance genes in fish gut microbiome. Arch. Microbiol. 2019, 201, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Bruijn, I.; Liu, Y.; Wiegertjes, G.F.; Raaijmakers, J.M. Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 2018, 94, fix161. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Torres, C.; Barros, J.; Somalo, S.; Igrejas, G.; Poeta, P. Gilthead seabream (Sparus aurata) as carriers of SHV-12 and TEM-52 extended-spectrum betalactamases-containing Escherichia coli isolates. Foodborne Pathog. Dis. 2011, 8, 1139–1141. [Google Scholar] [CrossRef] [PubMed]
- Brahmi, S.; Touati, A.; Dunyach-Remy, C.; Sotto, A.; Pantel, A.; Lavigne, J.P. High prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in wild fish from the Mediterranean Sea in Algeria. Microb. Drug Resist. 2018, 24, 290–298. [Google Scholar] [CrossRef]
- Bourdonnais, E.; Le Bris, C.; Brauge, T.; Midelet, G. Tracking antimicrobial resistance indicator genes in wild flatfish from the English Channel and the North Sea area: A one health concern. Environ. Pollut. 2024, 343, 123274. [Google Scholar] [CrossRef]
- Barraud, O.; Laval, L.; Le Devendec, L.; Larvor, E.; Chauvin, C.; Jouy, E.; Le Bouquin, S.; Vanrobaeys, Y.; Thuillier, B.; Lamy, B.; et al. Integrons from Aeromonas isolates collected from fish: A global indicator of antimicrobial resistance and anthropic pollution. Aquaculture 2023, 576, 739768. [Google Scholar] [CrossRef]
- Jia, J.; Gomes-Silva, G.; Plath, M.; Pereira, B.B.; Vieira, C.U.; Wang, Z. Shifts in bacterial communities and antibiotic resistance genes in surface water and gut microbiota of guppies (Poecilia reticulata) in the upper Rio Uberabinha, Brazil. Ecotoxicol. Environ. Saf. 2021, 211, 111955. [Google Scholar] [CrossRef] [PubMed]
- Higuera-Llantén, S.; Vásquez-Ponce, F.; Barrientos-Espinoza, B.; Mardones, F.O.; Marshall, S.H.; Olivares-Pacheco, J. Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes. PLoS ONE 2018, 13, e0203641. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Rojas, R. Occurrence of florfenicol resistance in bacteria associated with two Chilean salmon farms with different history of antibacterial usage. Aquaculture 2007, 266, 39–46. [Google Scholar] [CrossRef]
- Miranda, C.D.; Rojas, R.; Garrido, M.; Geisse, J.; González, G. Role of shellfish hatchery as reservoir of antimicrobial resistant bacteria. Mar. Pollut. Bull. 2013, 74, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Opazo, R.; Ortúzar, F.; Navarrete, P.; Espejo, R.; Romero, J. Reduction of soybean meal non-starch polysaccharides and α-Galactosides by solid-state fermentation using cellulolytic bacteria obtained from different environments. PLoS ONE 2012, 7, e44783. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Antimicrobial Disk Susceptibility Testing of Bacteria Isolated from Aquatic Animals; Approved Guideline VET03-A; Number 23; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2006; Volume 26. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Test, 12th ed.; Approved Standard, M02-A12; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2015. [Google Scholar]
- Hinton, M.; Hedges, A.J.; Linton, A.H. The ecology of Escherichia coli in market claves fed a milk-substitute diet. J. Appl. Bacteriol. 1985, 58, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Ogochukwu, E.C.; Regina, A.N.; Nkpeh, U.S.; Macmagnus, A.I.; Grace, E.; Ikenna, U.; Peter, E.C. Detection of ESBL, AmpC, and MBL-Producing Chryseobacterium indologenes Isolates Recovered from Hospital Environment in a Tertiary Health Care Facility. Am. J. Infect. Dis. Microbiol. 2021, 9, 129–135. [Google Scholar] [CrossRef]
- De Gheldre, Y.; Avesani, V.; Berhin, C.; Delmée, M.; Glupczynski, Y. Evaluation of Oxoid combination discs for detection of extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2003, 52, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Dey, T.K.; Lindahl, J.F.; Lundkvist, Å.; Grace, D.; Deka, R.P.; Shome, R.; Bandyopadhyay, S.; Goyal, N.K.; Sharma, G.; Shome, B.R. Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India. Antibiotics 2023, 12, 1449. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- El Aila, N.A.; Al Laham, N.A.; Ayesh, B.M. Prevalence of extended spectrum beta lactamase and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Gram negative bacilli isolates from pediatric patient population in Gaza strip. BMC Infect. Dis. 2023, 23, 99. [Google Scholar] [CrossRef] [PubMed]
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum b-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 90–103. [Google Scholar] [CrossRef] [PubMed]
- Niumsup, P.R.; Tansawai, U.; Boonkerd, N.; Polwichai, P.; Dejsirilert, S. Dissemination of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli in Thai hospitals. J. Infect. Chemother. 2008, 14, 404–408. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing Disk Susceptibility Test; Twenty-first informational supplement, CLSI document M100-S21; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2011. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI document M100-S27; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Aibinu, I.; Nwanneka, T.; Odugbemi, T. Occurrence of ESBL and MBL in clinical isolates of Pseudomonas aeruginosa from Lagos, Nigeria. J. Am. Sci. 2007, 3, 81–85. [Google Scholar]
- Owlia, P.; Saderi, H.; Karimi, Z.; Akhavi Rad, S.M.B.; Bahar, M.A. Phenotypic detection of metallo-β-lactamase producing Pseudomonas aeruginosa strains isolated from burned patients. Iran. J. Pathol. 2008, 3, 20–24. [Google Scholar]
- Ejikeugwu, P.C.; Ugwu, C.M.; Iroha, I.R.; Eze, P.; Gugu, T.H. Phenotypic detection of metallo-β-Lactamase (MBL) enzyme in Enugu, Southeast Nigeria. Am. J. Biol. Chem. Pharm. Sci. 2014, 2, 1–6. [Google Scholar]
- Aguirre-Quiñonero, A.; Martínez-Martínez, L. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J. Infect. Chemother. 2017, 23, 1–11. [Google Scholar] [CrossRef][Green Version]
- Egbule, O.S. Antimicrobial Resistance and β-Lactamase Production among Hospital Dumpsite Isolates. J. Environ. Prot. 2016, 7, 1057–1063. [Google Scholar] [CrossRef]
- Ejikeugwu, C.; Esimone, C.; Iroha, I.; Eze, P.; Ugwu, M.; Adikwu, M. Genotypic and Phenotypic Characterization of MBL Genes in Pseudomonas aeruginosa Isolates from the Non-hospital Environment. J. Pure Appl. Microbiol. 2018, 12, 1877–1885. [Google Scholar] [CrossRef]
- Hurtado, L.; Miranda, C.D.; Rojas, R.; Godoy, F.A.; Añazco, M.A.; Romero, J. Live Feeds Used in the Larval Culture of Red Cusk Eel, Genypterus chilensis, Carry High Levels of Antimicrobial-Resistant Bacteria and Antibiotic-Resistance Genes (ARGs). Animals 2020, 10, 505. [Google Scholar] [CrossRef] [PubMed]
- Rosser, S.; Young, H.-K. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 1999, 44, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic Resistance in the ECOR collection: Integrons an identification of a novel aad gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
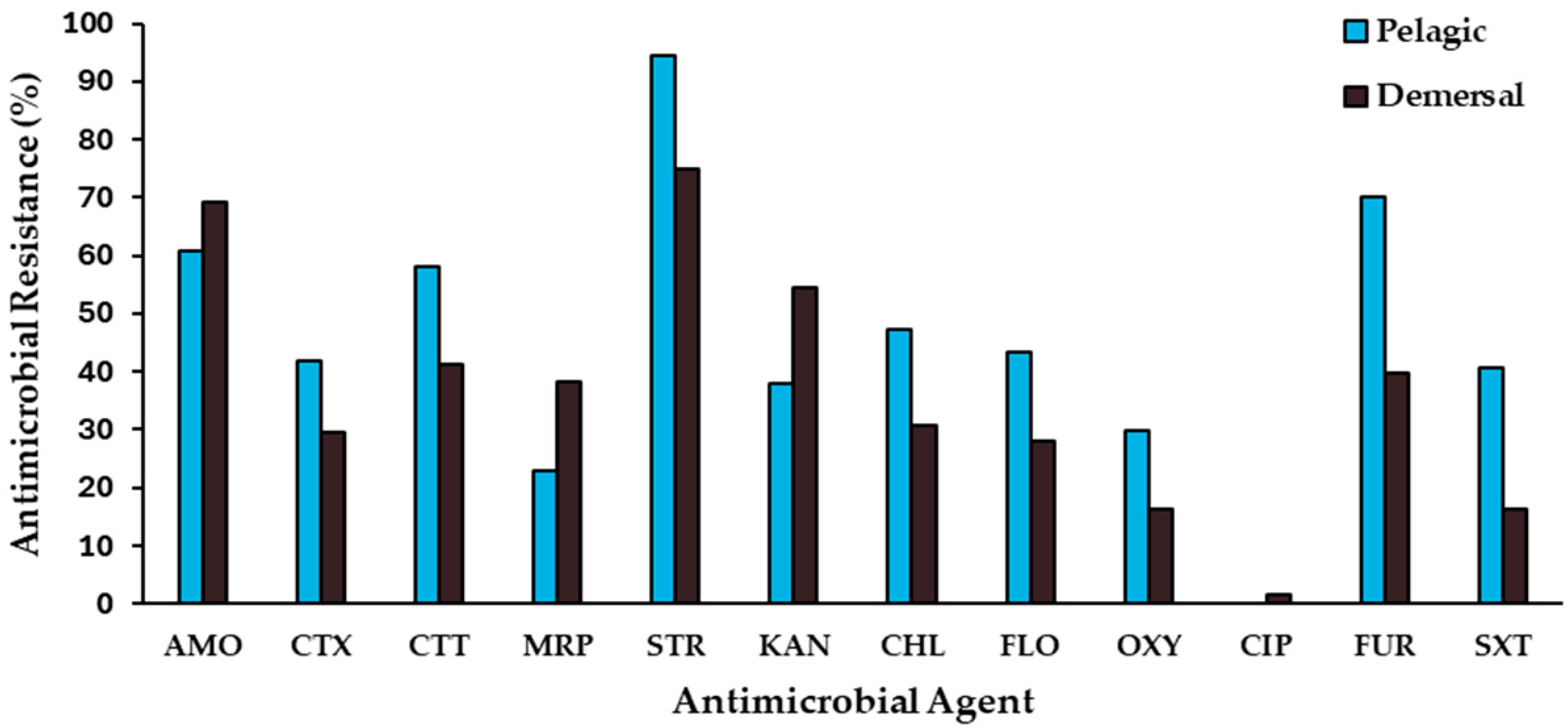
| Habitat | Common Name | Scientific Name | CBCC ± SD (CFU g−1) | Antimicrobial Resistance ± SD (%) | ||||
|---|---|---|---|---|---|---|---|---|
| FLO (30 µg mL−1) | OXY (30 µg mL−1) | AMO (50 µg mL−1) | STR (25 µg mL−1) | CIP (10 µg mL−1) | ||||
| Pelagic | Pacific chub mackerel | Scomber japonicus | 4.93 × 108 ± 4.58 × 108 | 21.41± 8.617 | 0.30 ± 0.07 | 8.69 ± 5.70 | 12.34 ± 5.02 | 0.002 ± 0.001 |
| Yellowtail amberjack | Seriola lalandi | 5.17 × 107 ± 6.13 × 107 | 0.89 ± 1.18 | 0.07 ± 0.03 | 0.95 ± 0.09 | 5.86 ± 6.55 | <0.001 | |
| Jack mackerel | Trachurus murphyi | 4.76 × 107 ± 3.01 × 107 | 0.04 ± 0.06 | 0.02 ± 0.01 | 0.40 ± 0.08 | <0.001 | 5.86 ± 6.55 | |
| Pacific bonito | Sarda chiliensis | 3.29 × 106 ± 2.77 × 105 | 5.11 ± 1.61 | 1.03 ± 0.62 | 5.30 ± 1.43 | 19.55 ± 8.20 | <0.001 | |
| Demersal | Chilean sandperch | Pinguipes chilensis | 3.62 × 109 ± 3.19 × 109 | <0.001 | <0.001 | 0.34 ± 0.07 | 5.28 ± 3.01 | <0.001 |
| Snackerel king-croaker | Menticirrhus ophicephalus | 6.68 × 107 ± 5.16 × 107 | 0.17 ± 0.06 | 0.06 ± 0.05 | 1.36 ± 1.10 | 27.76 ± 7.29 | <0.001 | |
| Pacific sandperch | Prolatilus jurgularis | 1.75 × 108 ± 2.74 × 107 | <0.001 | <0.001 | 0.80 ± 0.75 | 13.15 ± 15.58 | <0.001 | |
| Common hake | Merluccius gayi | 8.43 × 107 ± 4.46 × 107 | 0.26 ± 0.05 | 0.01 ± 0.01 | 0.36 ± 0.11 | 0.62 ± 0.13 | <0.001 | |
| Habitat | Common Name | Scientific Name | CBCC ± SD (CFU g−1) | Antimicrobial Resistance ± SD (%) | ||||
|---|---|---|---|---|---|---|---|---|
| FLO (30 µg mL−1) | OXY (30 µg mL−1) | AMO (50 µg mL−1) | STR (25 µg mL−1) | CIP (10 µg mL−1) | ||||
| Pelagic | Pacific chub mackerel | Scomber japonicus | 4.50 × 108 ± 3.90 × 108 | 23.78 ± 9.12 | 0.25 ± 0.03 | 9.34 ± 4.45 | 10.71 ± 4.90 | 0.001 ± 0.001 |
| Yellowtail amberjack | Seriola lalandi | 3.38 × 107 ± 4.63 × 107 | 1.48 ± 1.23 | 0.27 ± 0.34 | 4.76 ± 5.03 | 12.73 ± 16.36 | <0.001 | |
| Jack mackerel | Trachurus murphyi | 1.69 × 107 ± 1.77 × 107 | 0.10 ± 0.13 | 0.08 ± 0.10 | 0.56 ± 0.47 | 14.83 ± 3.97 | <0.001 | |
| Pacific bonito | Sarda chiliensis | 1.72 × 106 ± 1.12 × 105 | 6.27 ± 0.65 | 1.54 ± 0.25 | 5.77 ± 0.58 | 18.87 ± 3.32 | <0.001 | |
| Demersal | Chilean sandperch | Pinguipes chilensis | 1.42 × 109 ± 1.49 × 109 | <0.001 | <0.001 | 0.68 ± 0.87 | 2,81 ± 0,85 | <0.001 |
| Snackerel king-croaker | Menticirrhus ophicephalus | 3.61 × 107 ± 3.56 × 107 | 0.22 ± 0.04 | 0.03 ± 0.02 | 0.42 ± 0.01 | 25.37 ± 11.42 | <0.001 | |
| Pacific sandperch | Prolatilus jurgularis | 1.30 × 107 ± 8.75 × 106 | <0.001 | <0.001 | 9.41 ± 11.54 | 8.59 ± 2.17 | <0.001 | |
| Common hake | Merluccius gayi | 2.87 × 106 ± 3.29 × 106 | 6.90 ± 0.03 | 0.04 ± 0.04 | 8.37 ± 6.09 | 20.67 ± 7.77 | <0.001 | |
| Genus | Pelagic | Demersal | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. lalandi | S. japonicus | S. chiliensis | T. murphyi | Total | M. gayi | M. ophicephalus | P. chilensis | P. jugularis | Total | ||
| Acinetobacter | 1 | 1 | 1 | 1 | 2 | ||||||
| Aliivibrio | 4 | 4 | 8 | 8 | |||||||
| Brevibacterium | 1 | 1 | 1 | ||||||||
| Brochothrix | 1 | 4 | 1 | 6 | 6 | ||||||
| Myroides | 1 | 1 | 1 | ||||||||
| Moellerella | 1 | 1 | 1 | 1 | 2 | ||||||
| Photobacterium | 1 | 1 | 1 | 3 | 4 | 5 | |||||
| Proteus | 1 | 1 | 1 | ||||||||
| Providencia | 1 | 1 | 1 | ||||||||
| Pseudoalteromonas | 2 | 2 | 2 | ||||||||
| Pseudomonas | 5 | 7 | 16 | 9 | 37 | 11 | 7 | 18 | 55 | ||
| Psychrobacter | 4 | 1 | 5 | 2 | 2 | 4 | 9 | ||||
| Shewanella | 4 | 1 | 2 | 6 | 13 | 4 | 2 | 6 | 19 | ||
| Vibrio | 5 | 2 | 7 | 2 | 6 | 9 | 6 | 23 | 30 | ||
| Total | 21 | 11 | 24 | 18 | 74 | 16 | 28 | 9 | 15 | 68 | 142 |
| Habitat | Fish Species | No. Isolates | MDR | ARI | Enzyme Production | Plasmid Content | Intl1 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | MBL | 0 | 1 | 2 | 3 | ||||||
| Pelagic | Seriola lalandi | 21 | 14 | 0.4 | 4 | 0 | 5 | 9 | 6 | 1 | 0 |
| Scomber japonicus | 11 | 8 | 0.42 | 1 | 0 | 7 | 3 | 1 | 0 | 0 | |
| Sarda chiliensis | 24 | 22 | 0.51 | 6 | 2 | 14 | 7 | 2 | 1 | 0 | |
| Trachurus murphyi | 18 | 16 | 0.48 | 1 | 1 | 8 | 7 | 2 | 1 | 1 | |
| Demersal | Merluccius gayi | 16 | 13 | 0.52 | 7 | 0 | 5 | 9 | 2 | 0 | 0 |
| Menticirrhus ophicephalus | 28 | 14 | 0.36 | 4 | 0 | 12 | 10 | 5 | 1 | 1 | |
| Pinguipes chilensis | 9 | 6 | 0.26 | 0 | 0 | 7 | 2 | 0 | 0 | 0 | |
| Prolatilus jurgularis | 15 | 8 | 0.29 | 0 | 0 | 8 | 7 | 0 | 0 | 0 | |
| Total | 142 | 101 | 0.37 | 23 | 3 | 66 | 54 | 18 | 4 | 2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda, C.D.; Concha, C.; Hurtado, L.; Urtubia, R.; Rojas, R.; Romero, J. Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile. Antibiotics 2024, 13, 332. https://doi.org/10.3390/antibiotics13040332
Miranda CD, Concha C, Hurtado L, Urtubia R, Rojas R, Romero J. Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile. Antibiotics. 2024; 13(4):332. https://doi.org/10.3390/antibiotics13040332
Chicago/Turabian StyleMiranda, Claudio D., Christopher Concha, Luz Hurtado, Rocío Urtubia, Rodrigo Rojas, and Jaime Romero. 2024. "Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile" Antibiotics 13, no. 4: 332. https://doi.org/10.3390/antibiotics13040332
APA StyleMiranda, C. D., Concha, C., Hurtado, L., Urtubia, R., Rojas, R., & Romero, J. (2024). Occurrence of Antimicrobial-Resistant Bacteria in Intestinal Contents of Wild Marine Fish in Chile. Antibiotics, 13(4), 332. https://doi.org/10.3390/antibiotics13040332







