Biofilm Development by Mycobacterium avium Complex Clinical Isolates: Effect of Clarithromycin in Ultrastructure
Abstract
1. Introduction
2. Material and Methods
2.1. Strains
2.2. Biofilm Development
2.3. Effect of Clarithromycin on Biofilm Ultrastructure
2.4. Clarithromycin Susceptibility of Planktonic and Sessile Bacteria
2.5. Statistical Analysis
3. Results
3.1. Strains
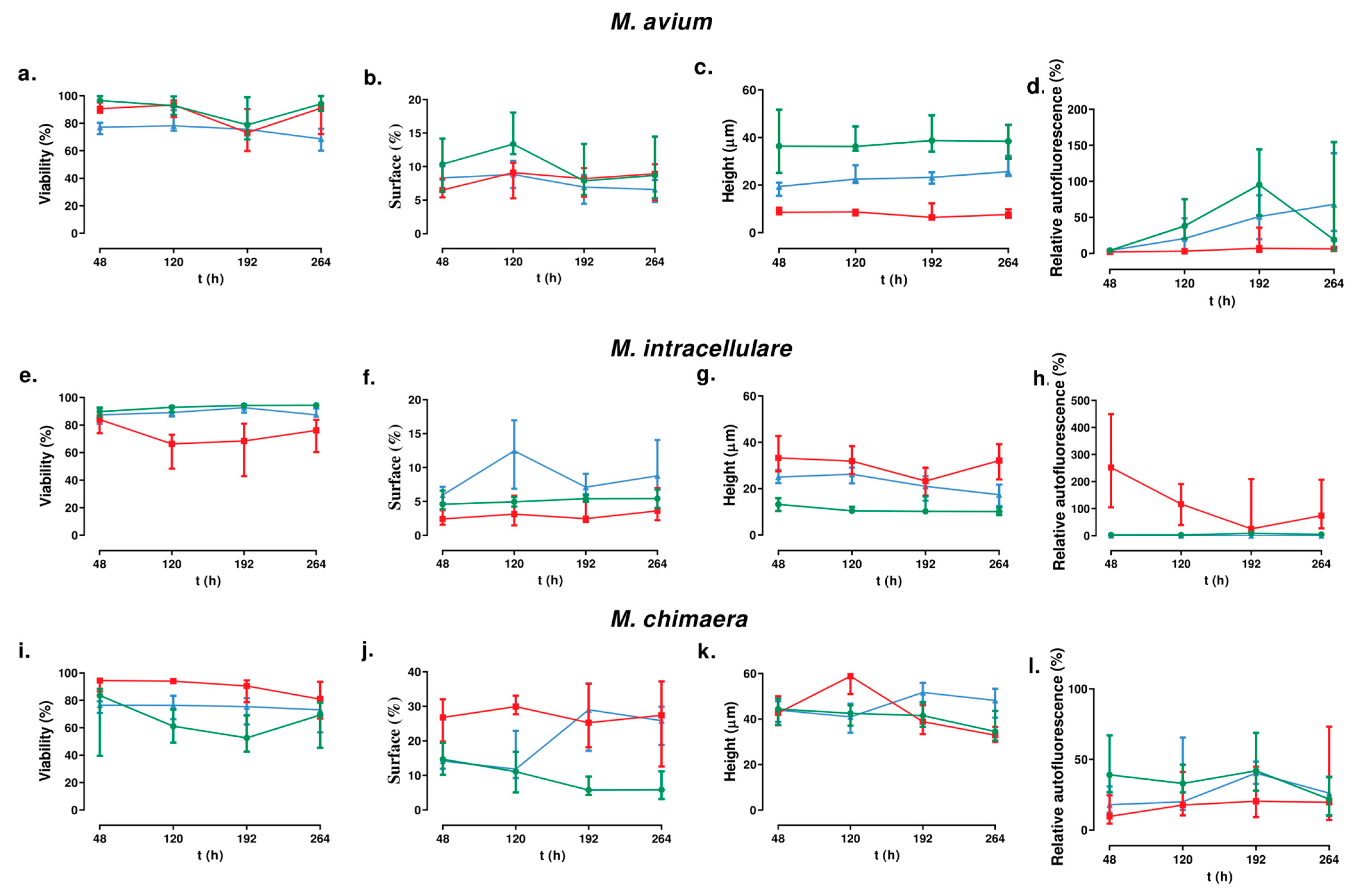
3.2. Biofilm Forming Studies
3.2.1. For M. avium
3.2.2. For M. intracellulare
3.2.3. For M. chimaera
3.3. Antimicrobial Susceptibility of Planktonic and Sessile Bacteria
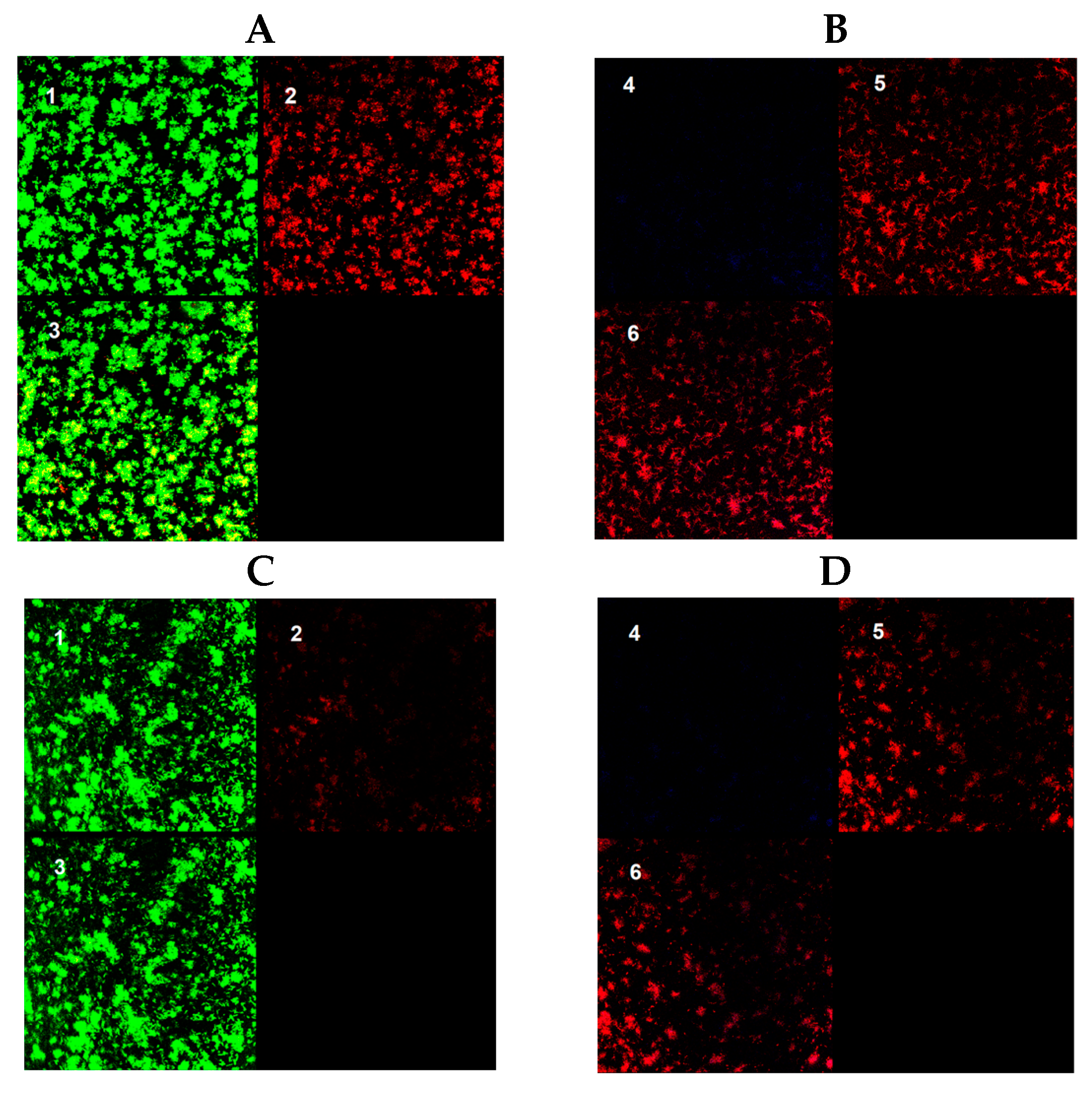
3.4. Effect of Clarithromycin in Biofilm Ultrastructure
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolinsky, E. Nontuberculous Mycobacteria and Associated Diseases. Am. Rev. Respir. Dis. 1979, 119, 107–159. [Google Scholar] [PubMed]
- Daley, C.L. Mycobacterium avium Complex Disease. Microbiol. Spectr. 2017, 5, 663–701. [Google Scholar] [CrossRef] [PubMed]
- Hoefsloot, W.; van Ingen, J.; Andrejak, C.; Angeby, K.; Bauriaud, R.; Bemer, P.; Beylis, N.; Boeree, M.J.; Cacho, J.; Chihota, V.; et al. The Geographic Diversity of Nontuberculous Mycobacteria Isolated from Pulmonary Samples: An NTM-NET Collaborative Study. Eur. Respir. J. 2013, 42, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J.; Andrejak, C.; Böttger, E.C.; Brozek, J.; Griffith, D.E.; Guglielmetti, L.; et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin. Infect. Dis. 2020, 71, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Kasperbauer, S.H.; Daley, C.L. Mycobacterium chimaera Infections Related to the Heater-Cooler Unit Outbreak: A Guide to Diagnosis and Management. Clin. Infect. Dis. 2019, 68, 1244–1250. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.R.; Diekema, D.J.; Edmond, M.B. Mycobacterium chimaera Infections Associated with Contaminated Heater-Cooler Devices for Cardiac Surgery: Outbreak Management. Clin. Infect. Dis. 2017, 65, 669–674. [Google Scholar] [CrossRef]
- Portell-Buj, E.; López-Gavín, A.; González-Martín, J.; Tudó, G. In Vitro Biofilm Formation in Mycobacterium avium-intracellulare Complex. Arch. Bronconeumol. 2021, 57, 140–141. [Google Scholar] [CrossRef]
- Portell-Buj, E.; González-Criollo, C.; López-Gavín, A.; Fernández-Pittol, M.; Busquets, M.A.; Estelrich, J.; Garrigó, M.; Rubio, M.; Tudó, G.; Gonzalez-Martin, J. Activity of Antibiotics and Potential Antibiofilm Agents against Biofilm-Producing Mycobacterium avium-intracellulare Complex Causing Chronic Pulmonary Infections. Antibiotics 2022, 11, 589. [Google Scholar] [CrossRef]
- Carter, G.; Wu, M.; Drummond, D.C.; Bermudez, L.E. Characterization of Biofilm Formation by Clinical Isolates of Mycobacterium avium. J. Med. Microbiol. 2003, 52, 747–752. [Google Scholar] [CrossRef]
- Carter, G.; Young, L.S.; Bermudez, L.E. A Subinhibitory Concentration of Clarithromycin Inhibits Mycobacterium avium Biofilm Formation. Antimicrob. Agents Chemother. 2004, 48, 4907–4910. [Google Scholar] [CrossRef] [PubMed]
- Keefe, B.F.; Leestemaker-Palmer, A.; Bermudez, L.E. Mycobacterium avium subsp. jominissuis (MAH) Microaggregate Induction of Host Innate Immunity Is Linked to Biofilm Formation. Microb. Pathog. 2021, 157, 104977. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; Hidaka, E.; Katsuyama, T.; Stang, B.; Petrofsky, M.; Bildfell, R.; Bermudez, L.E. The Ability to Form Biofilm Influences Mycobacterium avium Invasion and Translocation of Bronchial Epithelial Cells. Cell Microbiol. 2006, 8, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Egea, M.-C.; García-Pedrazuela, M.; Mahillo, I.; Esteban, J. Effect of Ciprofloxacin in the Ultrastructure and Development of Biofilms Formed by Rapidly Growing Mycobacteria. BMC Microbiol. 2015, 15, 18. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Egea, M.-C.; Garcia-Pedrazuela, M.; Mahillo, I.; Garcia, M.J.; Esteban, J. Autofluorescence as a Tool for Structural Analysis of Biofilms Formed by Nonpigmented Rapidly Growing Mycobacteria. Appl. Environ. Microbiol. 2013, 79, 1065–1067. [Google Scholar] [CrossRef]
- Fish, D.N.; Gotfried, M.H.; Danziger, L.H.; Rodvold, K.A. Penetration of Clarithromycin into Lung Tissues from Patients Undergoing Lung Resection. Antimicrob. Agents Chemother. 1994, 38, 876–878. [Google Scholar] [CrossRef]
- Woods, G.L.; Brown-Elliott, B.A.; Conville, P.S.; Desmond, E.P.; Hall, G.S.; Lin, G.; Pfyffer, G.E.; Ridderhof, J.C.; Siddiqi, S.H.; Wallace, R.J.; et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes, 2nd ed.; CLSI Standards: Guidelines for Health Care Excellence; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011; ISBN 978-1-56238-746-4. [Google Scholar]
- Hernandes, C.; Coppede, J.d.S.; Bertoni, B.W.; França, S.d.C.; Pereira, A.M.S. Flash Microbiocide: A Rapid and Economic Method for Determination of MBC and MFC. Am. J. Plant Sci. 2013, 4, 850. [Google Scholar] [CrossRef]
- Tortoli, E. Microbiological Features and Clinical Relevance of New Species of the Genus Mycobacterium. Clin. Microbiol. Rev. 2014, 27, 727–752. [Google Scholar] [CrossRef]
- Tortoli, E.; Rindi, L.; Garcia, M.J.; Chiaradonna, P.; Dei, R.; Garzelli, C.; Kroppenstedt, R.M.; Lari, N.; Mattei, R.; Mariottini, A.; et al. Proposal to Elevate the Genetic Variant MAC-A, Included in the Mycobacterium avium Complex, to Species Rank as Mycobacterium chimaera Sp. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1277–1285. [Google Scholar] [CrossRef]
- Meier, E.; Pennington, K.; Gallo de Moraes, A.; Escalante, P. Characteristics of Mycobacterium avium Complex (MAC) Pulmonary Disease in Previously Treated Lung Cancer Patients. Respir. Med. Case Rep. 2017, 22, 70–73. [Google Scholar] [CrossRef]
- Fennelly, K.P.; Ojano-Dirain, C.; Yang, Q.; Liu, L.; Lu, L.; Progulske-Fox, A.; Wang, G.P.; Antonelli, P.; Schultz, G. Biofilm Formation by Mycobacterium abscessus in a Lung Cavity. Am. J. Respir. Crit. Care Med. 2016, 193, 692–693. [Google Scholar] [CrossRef]
- Walker, J.; Moore, G.; Collins, S.; Parks, S.; Garvey, M.I.; Lamagni, T.; Smith, G.; Dawkin, L.; Goldenberg, S.; Chand, M. Microbiological Problems and Biofilms Associated with Mycobacterium chimaera in Heater-Cooler Units Used for Cardiopulmonary Bypass. J. Hosp. Infect. 2017, 96, 209–220. [Google Scholar] [CrossRef]
- Falkinham, J.O. Mycobacterium avium Complex: Adherence as a Way of Life. AIMS Microbiol. 2018, 4, 428–438. [Google Scholar] [CrossRef]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2017, 8, 2651. [Google Scholar] [CrossRef] [PubMed]
- Pradal, I.; Esteban, J.; Mediero, A.; García-Coca, M.; Aguilera-Correa, J.J. Contact Effect of a Methylobacterium Sp. Extract on Biofilm of a Mycobacterium Chimaera Strain Isolated from a 3T Heater-Cooler System. Antibiotics 2020, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.B.; Agdestein, A.; Olsen, I.; Nilsen, S.F.; Holstad, G.; Djønne, B. Biofilm Formation by Mycobacterium Avium Isolates Originating from Humans, Swine and Birds. BMC Microbiology 2009, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Deng, W.; Liu, M.; Xie, J. Mycobacterium Biofilms: Factors Involved in Development, Dispersal, and Therapeutic Strategies against Biofilm-Relevant Pathogens. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 269–279. [Google Scholar] [CrossRef] [PubMed]
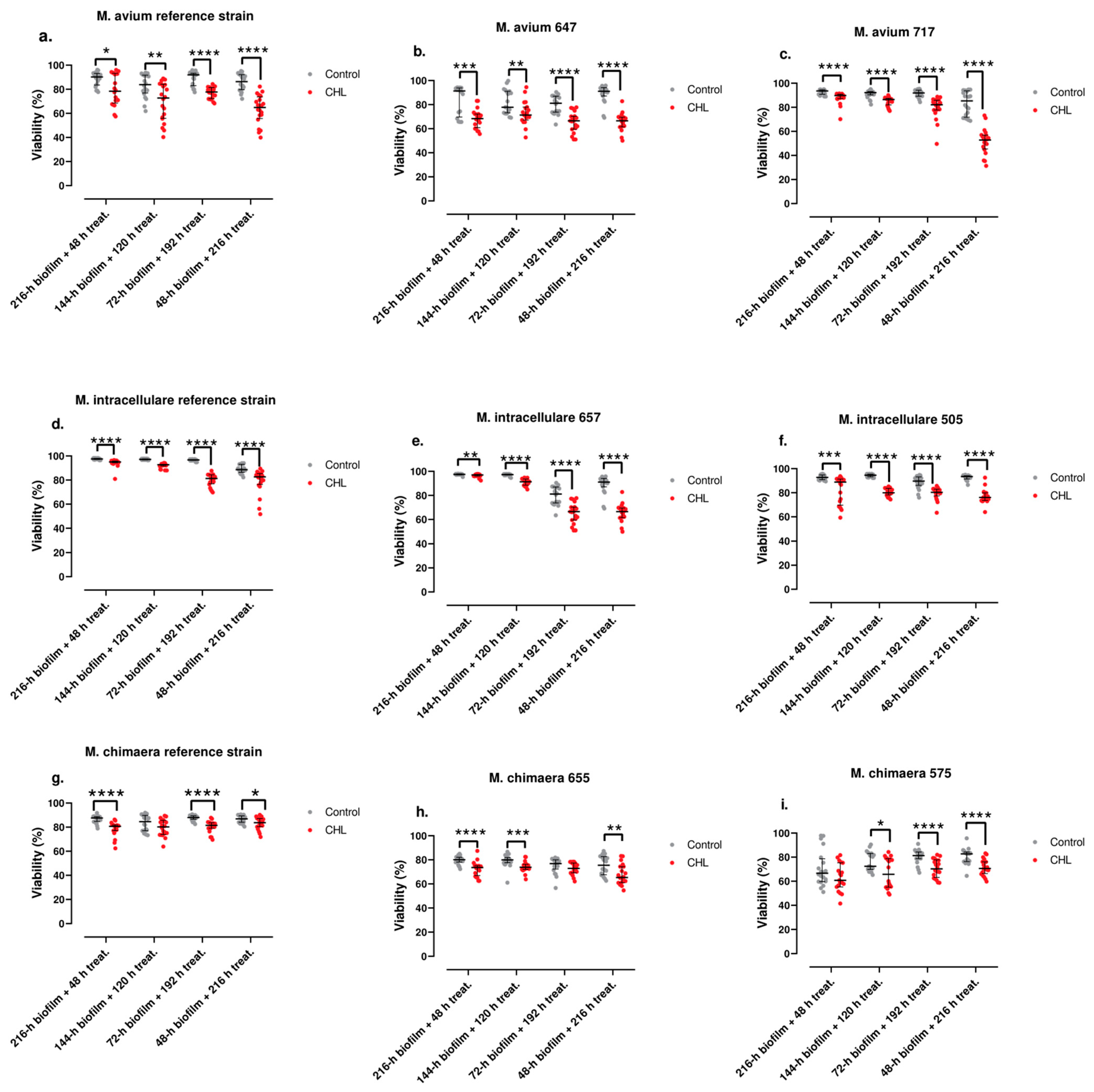
| Strain | MIC (µg/mL) | MBC (µg/mL) | MBIC (µg/mL) |
|---|---|---|---|
| M. avium CECT 670 | 4 | <256 | 16 |
| M. avium 717 | 8 | 8 | 8 |
| M. avium 647 | ≤1 | ≤1 | ≤1 |
| M. intracellulare 669 | ≤1 | 32 | ≤1 |
| M. intracellulare 505 | ≤1 | ≤1 | ≤1 |
| M. intracellulare 657 | ≤1 | 32 | ≤1 |
| M. chimaera DSM 756 | ≤1 | 32 | 16 |
| M. chimaera 575 | ≤1 | 8 | ≤1 |
| M. chimaera 655 | 8 | 16 | ≤1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akir, A.; Senhaji-Kacha, A.; Muñoz-Egea, M.C.; Esteban, J.; Aguilera-Correa, J.J. Biofilm Development by Mycobacterium avium Complex Clinical Isolates: Effect of Clarithromycin in Ultrastructure. Antibiotics 2024, 13, 263. https://doi.org/10.3390/antibiotics13030263
Akir A, Senhaji-Kacha A, Muñoz-Egea MC, Esteban J, Aguilera-Correa JJ. Biofilm Development by Mycobacterium avium Complex Clinical Isolates: Effect of Clarithromycin in Ultrastructure. Antibiotics. 2024; 13(3):263. https://doi.org/10.3390/antibiotics13030263
Chicago/Turabian StyleAkir, Arij, Abrar Senhaji-Kacha, Maria Carmen Muñoz-Egea, Jaime Esteban, and John Jairo Aguilera-Correa. 2024. "Biofilm Development by Mycobacterium avium Complex Clinical Isolates: Effect of Clarithromycin in Ultrastructure" Antibiotics 13, no. 3: 263. https://doi.org/10.3390/antibiotics13030263
APA StyleAkir, A., Senhaji-Kacha, A., Muñoz-Egea, M. C., Esteban, J., & Aguilera-Correa, J. J. (2024). Biofilm Development by Mycobacterium avium Complex Clinical Isolates: Effect of Clarithromycin in Ultrastructure. Antibiotics, 13(3), 263. https://doi.org/10.3390/antibiotics13030263








