Examining Antimicrobial Resistance in Escherichia coli: A Case Study in Central Virginia’s Environment
Abstract
1. Introduction
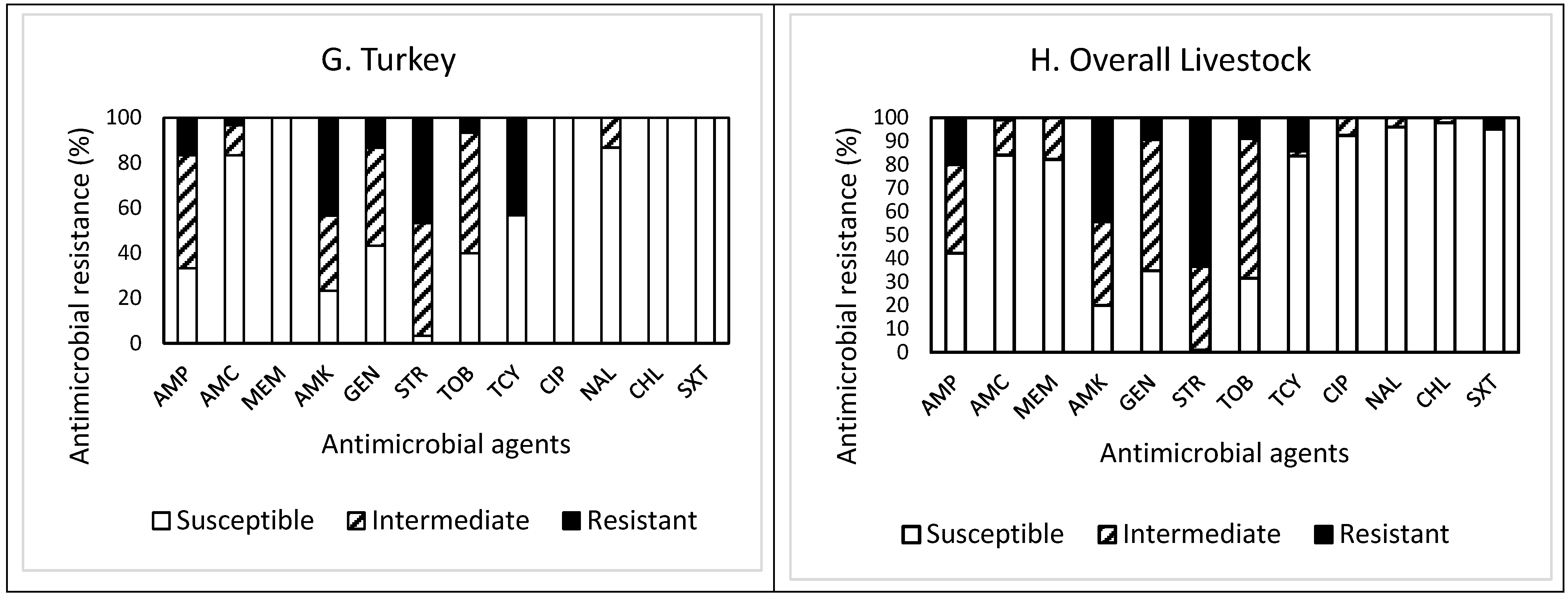
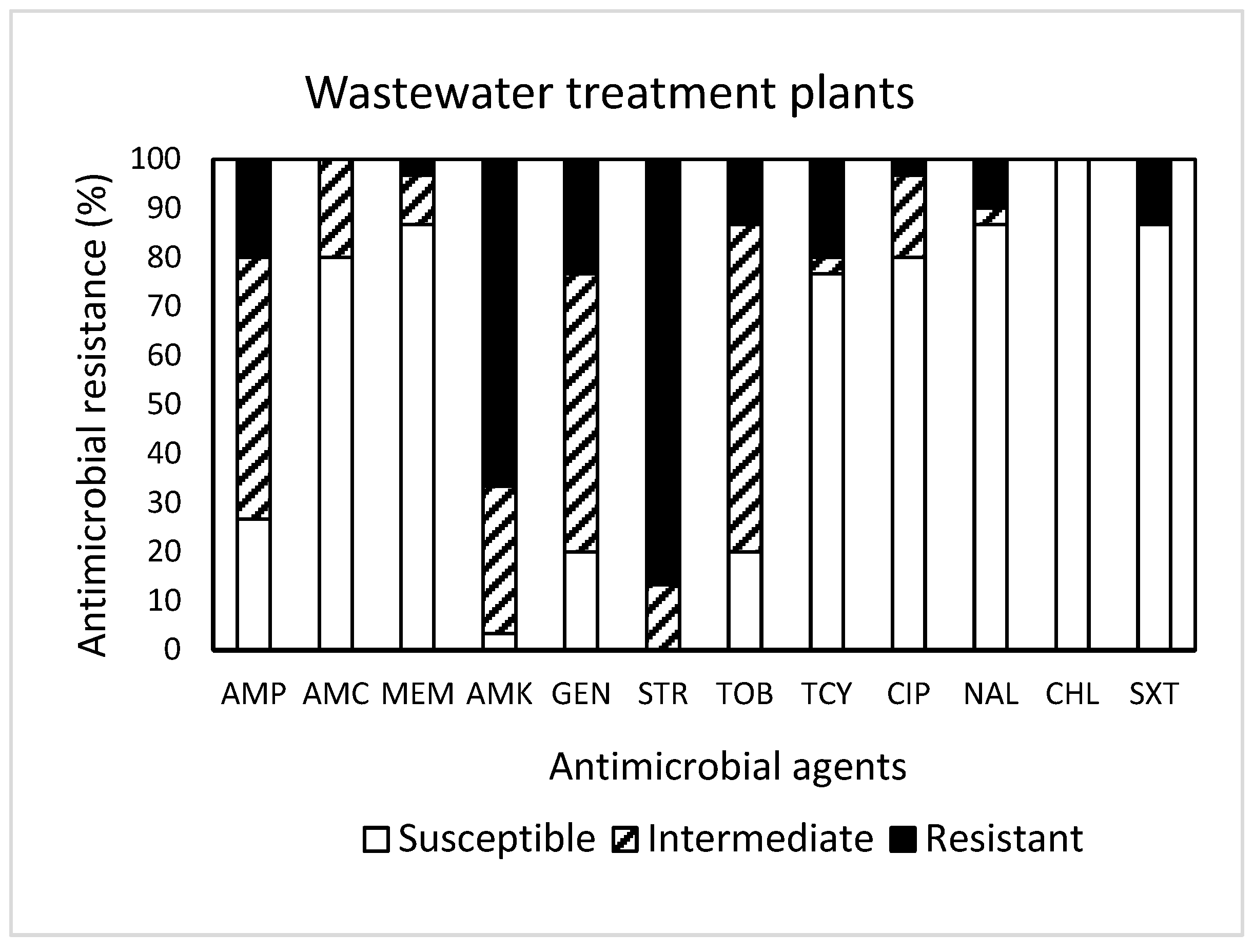
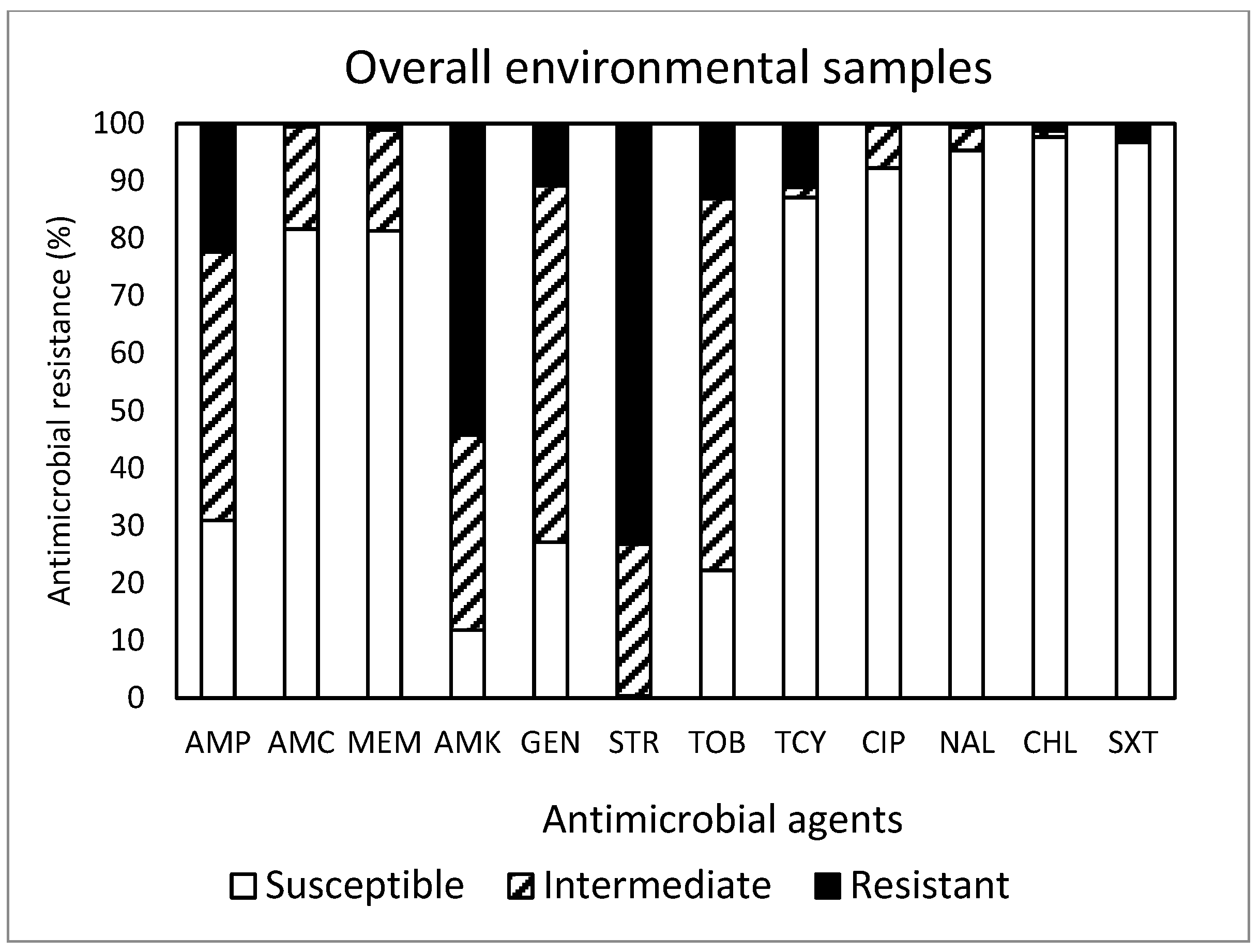
2. Results and Discussion
3. Materials and Methods
3.1. Sample Collection
3.2. E. coli Isolation
3.3. AMR Test
3.4. Data Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wüst, F. Antibiotic resistance of E. coli in sewage ad sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, A. Prevalence and antibiotic resistance pattern of Campylobacter species in foods of animal origin. Vet. World 2014, 7, 681–684. [Google Scholar]
- Sáenz, Y.; Zarazaga, M.; Lantero, M.; Gastanares, M.; Baquero, F.; Torres, C. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997–1998. Antimicrob. Agents Chemother. 2000, 44, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Kulis, J.; Thomson, B.; Chapman, T.; Mawhinney, D. Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 2006, 366, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Geneva Environment Network. Antimicrobial Resistance and the Environment-Update. 2023. Available online: https://www.genevaenvironmentnetwork.org/resources/updates/antimicrobial-resistance-and-the-environment (accessed on 20 December 2023).
- Fletcher, S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ. Health Prev. Med. 2015, 20, 243–252. [Google Scholar] [CrossRef]
- Cherny, S.S.; Chowers, M.; Obolski, U. Patterns of Antibiotic Cross-Resistance by Bacterial Sample Source: A Retrospective Cohort Study. medRxiv 2022. Available online: https://www.medrxiv.org/content/10.1101/2022.03.31.22273223v2 (accessed on 21 December 2023).
- Nyirabahizi, E.; Tyson, G.H.; Dessai, U.; Zhao, S.; Kabera, C.; Crarey, E.; Womack, N.; Crews, M.K.; Stain, E.; Tate, H. Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 2020, 115, 107280. [Google Scholar] [CrossRef]
- Van den Bogaard, A.; Stobberingh, E. Epidemiology of resistance to antibiotics: Links between animals and humans. Int. J. Antimicrob. Agents 2000, 14, 327–335. [Google Scholar] [CrossRef]
- Cornaglia, G.; Hryniewicz, W.; Jarlier, V.; Kahlmeter, G.; Mittermayer, H.; Stratchounski, L.; Baquero, F.; ESCMID Study Group for Antimicrobial Resistance Surveillance. European recommendations for antimicrobial resistance surveillance. Clin. Microbiol. Infect. 2004, 10, 349–383. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T. The global epidemic nature of antimicrobial resistance and the need to monitor and manage it locally. Clin. Infect. Dis. 1997, 24 (Suppl. S1), S2–S8. [Google Scholar] [CrossRef]
- M100–S25; Performance Standards for Antimicrobial Disk Susceptibility Tests. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2015.
- United States Department of Agriculture. Antimicrobial Resistance Action Plan. 2014. Available online: https://www.usda.gov/sites/default/files/documents/usda-antimicrobial-resistance-action-plan.pdf (accessed on 21 December 2023).
- Hammerum, A.W.; Heuer, O.E.; Emborg, H.-D.; Bagger-Skjøt, L.; Jensen, V.F.; Rogues, A.-M.; Skov, R.L.; Agersø, Y.; Brandt, C.T.; Seyfarth, A.M.; et al. Danish integrated antimicrobial resistance monitoring and research program. Emerg. Infect. Dis. 2007, 13, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.; Aidara-Kane, A.; Dailey, J. Agriculture and Food Production as Drivers of the Global Emergence and Dissemination of Antimicrobial Resistance. 2017. Available online: http://resistancecontrol.info/2017/agriculture-and-food-production-as-drivers-of-the-global-emergence-and-dissemination-of-antimicrobial-resistance/ (accessed on 19 December 2023).
- Centers for Disease Control and Prevention. Outpatient Antibiotic Prescriptions—United States, 2015. 2017. Available online: https://www.cdc.gov/antibiotic-use/data/report-2015.html (accessed on 19 December 2023).
- Virginia Department of Health. 2018 Virginia State and Regional Cumulative Antibiogram. 2020. Available online: https://www.vdh.virginia.gov/haiar/ar/ (accessed on 19 December 2023).
- Burch, T.R.; Stokdyk, J.P.; Firnstahl, A.D.; Kieke, A.A., Jr.; Cook, R.M.; Opelt, S.A.; Spencer, S.K.; Durso, L.M.; Borchardt, M.A. Microbial source tracking and land use associations for antibiotic resistance genes in private wells influenced by human and livestock fecal sources. J. Environ. Qual. 2023, 52, 270–286. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Storteboom, H.; Arabi, M.; Davis, J.G.; Crimi, B.; Pruden, A. Identification of antibiotic-resistance-gene molecular signatures. suitable as tracers of pristine River, urban, and agricultural sources. Environ. Sci. Technol. 2010, 44, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Olivares-Rieumont, S.; Knapp, C.W.; Lima, L.; Werner, D.; Bowen, E. Antibiotic resistance gene abundances associated with waste discharges to the Almendares river near Havana, Cuba. Environ. Sci. Technol. 2011, 45, 418–424. [Google Scholar] [CrossRef]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, J.; Hynds, P.; Pot, M.; Adley, C.C.; Ryan, M.P. Evaluation of levels of antibiotic resistance in groundwater-derived E. coli isolates in the Midwest of Ireland and elucidation of potential predictors of resistance. Hydrogeol. J. 2017, 25, 939–951. [Google Scholar] [CrossRef]
- Poeta, P.; Radhouani, H.; Igrejas, G.; Gonçalves, A.; Carvalho, C.; Rodrigues, J.; Vinué, L.; Somalo, S.; Torres, C. Seagulls of the Berlengas natural reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl. Environ. Microbiol. 2008, 74, 7439–7441. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Igrejas, G.; Pinto, L.; Gonalves, A.; Coelho, C.; Rodrigues, J.; Poeta, P. Molecular characterization of antibiotic resistance in enterococci recovered from seagulls (Larus cachinnans) representing an environmental health problem. J. Environ. Monit. 2011, 13, 2227–2233. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; Gonzalez-Acuna, D.; Klaassen, M.; Eden, J.-S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; Holmes, E.C. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019, 17, 31. [Google Scholar] [CrossRef]
- Doyle, M.; Loneragan, H.; Scott, M.; Singer, S. Antimicrobial resistance: Challenges and perspectives. Compr. Rev. Food Sci. Food Saf. 2013, 12, 234–248. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Li, J.; Cao, J.; Zhu, Y.; Chen, Q.; Shen, F.; Wu, Y.; Xu, S.; Fan, H.; Da, G.; Huang, R.; et al. Global survey of antibiotic resistance genes in air. Environ. Sci. Technol. 2018, 52, 10975–10984. [Google Scholar] [CrossRef]
- Andrade, L.; Kelly, M.; Hynds, P.; Weatherill, J.; Majury, A.; O’Dwyer, J. Groundwater resources as a global reservoir for antimicrobial-resistant bacteria. Water Res. 2020, 170, 115360. [Google Scholar] [CrossRef]
- Zainab, S.M.; Junaid, M.; Xu, N.; Malik, R.N. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020, 187, 116455. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, D.; Xie, J.; Zhang, Y.; Wan, Y.; Zhang, Y.; Shi, X. Impacts of farmland application of antibiotic-contaminated manures on the occurrence of antibiotic residues and antibiotic resistance genes in soil: A meta-analysis study. Chemosphere 2020, 300, 134529. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Initiatives for Addressing Antimicrobial Resistance in the Environment. 2018. Available online: https://wellcome.org/sites/default/files/antimicrobial-resistance-environment-report.pdf (accessed on 19 December 2023).
- Coleman, B.L.; Louie, M.; Salvadori, M.I.; McEwen, S.A.; Neumann, N.; Sibley, K.; Irwin, R.J.; Jamieson, F.B.; Daignault, D.; Majury, A.; et al. Contamination of Canadian private drinking water sources with antimicrobial resistant Escherichia coli. Water Res. 2013, 47, 3026–3036. [Google Scholar] [CrossRef]
- Li, X.; Atwill, E.R.; Antaki, E.; Applegate, O.; Bergamaschi, B.; Bond, R.F.; Chase, J.; Ransom, K.M.; Samuels, W.; Watanabe, N.; et al. Fecal indicator and pathogenic bacteria and their antibiotic resistance in alluvial groundwater of an irrigated agricultural region with dairies. J. Environ. Qual. 2015, 44, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-L.; Li, H.; Zhou, X.-Y.; Zhao, Y.; Su, J.-Q.; Zhang, X.; Huang, F.-Y. An underappreciated hotspot of antibiotic resistance: The groundwater near the municipal solid waste landfill. Sci. Total Environ. 2017, 609, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, E.; Chiriac, C.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.-L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.Z.; Zou, H.Y.; Wu, D.L.; Chen, S.; He, L.Y.; Zhang, M.; Bai, H.; Ying, G.G. Swine farming elevated the proliferation of Acinetobacter with the prevalence of antibiotic resistance genes in the groundwater. Environ. Int. 2020, 136, 105484. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Method 1603. Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane Thermotolerant Escherichia coli Agar (Modified mTEC). EPA 821-R-02-023. 2002. Available online: https://www.epa.gov/sites/default/files/2015-08/documents/method_1103-1_2010.pdf (accessed on 22 December 2023).
- Kim, C.; Fatani, A.; Almuqati, R.; Rahemi, A.; Abujamous, A.; Wynn, C.; Nartea, T.; Ndegwa, E.; Rutto, L.; Dhakal, R. Prevalence and antimicrobial resistance of foodborne pathogens in value-added commodities procured from farmers’ markets in Central Virginia. J. Food Saf. 2021, 41, e12931. [Google Scholar] [CrossRef]
- Magiorakos, A.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration (FDA). FDA Releases 2012 and 2013 NARMS Integrated Annual Report; Finds Some Improvement in Antibiotic Resistance Trends in Recent Years. 2015. Available online: http://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm457825.htm (accessed on 19 December 2023).



| Sample | Nature of AMR b | Quantity of Antimicrobial Agents c | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Source (n a) | Location (n) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | MDR d (≥3) | |
| Farm animal /livestock | Cattle (45) | 3 | R | 46.7 | 17.8 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| R + I | 15.6 | 4.4 | 22.2 | 28.9 | 20.0 | 6.7 | 2.2 | 0.0 | 0.0 | NA e | |||
| Chicken (45) | 3 | R | 37.8 | 33.3 | 6.7 | 4.4 | 2.2 | 0 | 0 | 0.0 | 0.0 | 2.2 | |
| R + I | 17.8 | 15.6 | 17.8 | 6.7 | 13.3 | 17.8 | 6.7 | 0.0 | 4.4 | NA | |||
| Goat (30) | 3 | R | 13.3 | 26.7 | 26.7 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| R + I | 3.3 | 13.3 | 10.0 | 10.0 | 26.7 | 16.7 | 0.0 | 6.7 | 0.0 | NA | |||
| Horse (15) | 1 | R | 20.0 | 13.3 | 26.7 | 40.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| R + I | 0.0 | 0.0 | 6.7 | 13.3 | 40.0 | 33.3 | 0.0 | 6.7 | 0.0 | NA | |||
| Pig (30) | 2 | R | 6.7 | 53.3 | 23.3 | 10.0 | 0.0 | 3.3 | 0.0 | 0.0 | 0.0 | 10.0 | |
| R + I | 0.0 | 3.3 | 0.0 | 23.3 | 30.0 | 16.7 | 20.0 | 6.7 | 0.0 | NA | |||
| Sheep (30) | 1 | R | 20.0 | 40.0 | 26.7 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 | |
| R + I | 0.0 | 0.0 | 6.7 | 30.0 | 40.0 | 13.3 | 10.0 | 0.0 | 0.0 | NA | |||
| Turkey (30) | 1 | R | 26.7 | 30.0 | 10.0 | 10.0 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 6.7 | |
| R + I | 6.7 | 10.0 | 13.3 | 23.3 | 16.7 | 23.3 | 6.7 | 0.0 | 0.0 | NA | |||
| Total (225) | 14 | R | 27.1 | 31.1 | 16.0 | 7.6 | 0.9 | 0.4 | 0.0 | 0.0 | 0.0 | 2.7 | |
| R + I | 8.0 | 7.6 | 12.4 | 19.6 | 24.4 | 16.4 | 8.4 | 2.2 | 0.9 | NA | |||
| Wild avian | Goose (45) | 3 | R | 33.3 | 28.9 | 17.8 | 4.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| R + I | 0.0 | 4.4 | 6.7 | 26.7 | 42.2 | 15.6 | 2.2 | 2.2 | 0.0 | NA | |||
| Seagull (30) | 2 | R | 10.0 | 53.3 | 23.3 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.3 | |
| R + I | 0.0 | 0.0 | 3.3 | 10.0 | 50.0 | 16.7 | 20.0 | 0.0 | 0.0 | NA | |||
| Total (75) | 5 | R | 24.0 | 38.7 | 20.0 | 5.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | |
| R + I | 0.0 | 2.7 | 5.3 | 20 | 45.3 | 16.0 | 9.3 | 1.3 | 0.0 | NA | |||
| Land use | Crop land (30) | 2 | R | 33.3 | 20 | 26.7 | 13.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| R + I | 0.0 | 6.7 | 10.0 | 26.7 | 30.0 | 16.7 | 10.0 | 0.0 | 0.0 | NA | |||
| Forest land (30) | 2 | R | 10.0 | 40.0 | 26.7 | 13.3 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| R + I | 0.0 | 0.0 | 3.3 | 20.0 | 33.3 | 33.3 | 6.7 | 3.3 | 0.0 | NA | |||
| Pasture land (30) | 2 | R | 16.7 | 43.3 | 30.0 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| R + I | 0.0 | 0.0 | 0.0 | 13.3 | 33.3 | 33.3 | 10.0 | 6.7 | 0.0 | NA | |||
| Urban land (60) | 2 | R | 10.0 | 36.7 | 36.7 | 13.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.3 | |
| R + I | 0.0 | 0.0 | 0.0 | 23.3 | 43.3 | 23.3 | 10 | 1.7 | 0.0 | NA | |||
| Total (120) | 8 | R | 17.5 | 35.0 | 30.0 | 11.7 | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.8 | |
| R + I | 0.0 | 1.7 | 4.2 | 20.8 | 35.0 | 26.7 | 9.2 | 2.5 | 0.0 | NA | |||
| Wastewater treatment plant (30) | 2 | R | 16.7 | 33.3 | 23.3 | 6.7 | 3.3 | 6.7 | 3.3 | 0.0 | 0.0 | 20.0 | |
| R + I | 3.3 | 0.0 | 3.3 | 30.0 | 20.0 | 23.3 | 10.0 | 0.0 | 10.0 | NA | |||
| Overall (450) | 29 | R | 23.3 | 33.6 | 20.9 | 8.2 | 0.9 | 0.7 | 0.2 | 0.0 | 0.0 | 3.1 | |
| R + I | 4.2 | 4.7 | 8.4 | 20.7 | 30.4 | 19.6 | 8.9 | 2.0 | 1.1 | NA | |||
| Sample | Number of Sampling Location | Occurrence of Antimicrobial Resistance d | |||
|---|---|---|---|---|---|
| Type (n a) | Source (n) | ||||
| Farm animal/livestock (225) | Cattle (45) | 3 | 3.10 ± 1.37 C * | 2.32 ± 1.05 f * | 2.32 ± 1.05 D ** |
| Chicken (45) | 3 | 2.71 ± 1.57 ef | 2.71 ± 1.57 CD | ||
| Goat (30) # | 3 | 3.27 ± 1.49 bcde | 3.27 ± 1.49 BC | ||
| Horse (15) # | 1 | 4.07 ± 1.02 a | 4.07 ± 1.02 A | ||
| Pig (30) | 2 | 3.93 ± 1.09 ab | 3.93 ± 1.09 AB | ||
| Sheep (30) ǂ | 1 | 3.42 ± 0.86 abcd | 3.42 ± 0.86 ABC | ||
| Turkey (30) ǂ | 1 | 3.02 ± 0.40 de | 3.02 ± 1.40 C | ||
| Wild avian (75) | Goose (45) | 3 | 3.41 ± 0.91 BC | 3.18 ± 0.91 cde | 3.18 ± 0.91 B |
| Seagull (30) | 2 | 3.77 ± 0.80 abc | 3.77 ± 0.80 A | ||
| Water (120) b∆ | Crop land (30) | 2 | 3.72 ± 0.98 AB | 3.38 ± 1.13 abcd | 3.38 ± 1.13 A |
| Forest land (30) | 2 | 3.85 ± 1.06 abc | 3.85 ± 1.06 A | ||
| Pasture land (30) | 2 | 3.82 ± 0.90 abc | 3.82 ± 0.90 A | ||
| Urban land (30) | 2 | 3.83 ± 0.76 abc | 3.83 ± 0.76 A | ||
| Waste (30) c∆ | Treatment plant (30) | 2 | 3.97 ± 1.66 A | 3.97 ± 1.66 a | |
| Sample | GPS Coordinates in Degrees, Minutes, and Seconds | ||
|---|---|---|---|
| Type | Source (n a) | Location (n) | |
| Farm animal /livestock | Cattle (45) | 3 | 36°54′6.084″ N, 76°44′51.54″ W |
| 37°9′53.2008″ N, 77°24′38.2176″ W | |||
| 37°16′6.8988″ N, 77°27′57.744″ W | |||
| Chicken (45) | 3 | 37°13′56.3484″ N, 77°26′37.0284″ W | |
| 37°4′59.664″ N, 76°43′26.076″ W | |||
| 37°15′5.76″ N, 77°18′1.224″ W | |||
| Goat (30) | 3 | 37°13′51.7692″ N, 77°26′52.4472″ W | |
| 37°4′59.664″ N, 76°43′26.076 W | |||
| 37°15′5.76″ N, 77°18′1.224″ W | |||
| Horse (15) | 1 | 37°11′26.7252″ N, 77°25′27.2496″ W | |
| Pig (30) | 2 | 37°37′34.932″ N, 76°53′35.412″ W | |
| 37°4′59.664″ N, 76°43′26.076″ W | |||
| Sheep (30) | 1 | 37°13′58.2384″ N, 77°25′53.922″ W | |
| Turkey (30) | 1 | 37°37′34.932″ N, 76°53′35.412″ W | |
| Wild avian | Goose (45) | 3 | 37°14′25.44″ N, 77°25′14.88″ W |
| 37°16′6.8988″ N, 77°27′57.744″ W | |||
| 37°13′43.8672″ N, 77°26′15.414″ W | |||
| Seagull (30) | 2 | 37°9′53.2836″ N, 77°22′3.7272″ W | |
| 37°13′43.8672″ N, 77°26′15.414″ W | |||
| Land use | Crop land (30) | 2 | 37°13′28.6″ N, 77°26′46.7″W |
| 37°14′04.2″ N, 77°26′26.4″W | |||
| Forest land (30) | 2 | 37°24′59.472″ N, 78°38′10.6872″ W | |
| 37°15′28.1592″ N, 78°29′10.2768″ W | |||
| Pasture land (30) | 2 | 37°13′07.0″ N, 77°22′29.1″W | |
| 37°20′2.68″ N, 77°13′48.35″W | |||
| Urban land (60) | 2 | 37°13′9.2424″ N, 77°24′59.508″ W | |
| 37°13′12.5436″ N, 77°24′54.8568″ W | |||
| Wastewater treatment plant (30) | 2 | 37°14′19.968″ N, 77°23′35.52″ W | |
| 37°17′42″ N, 77°15′32.4″ W | |||
| Antimicrobial Category | Antimicrobial Agent and Its Abbreviation | Concentration (µg/Disk) | Zone Diameter (mm) | ||
|---|---|---|---|---|---|
| S | I | R | |||
| Penicillins | Ampicillin (AMP) | 10 | >17 | 14–16 | <13 |
| β-lactamase inhibitor combinations | Amoxicillin–clavulanic acid (AMC) | 30 | >18 | 14–17 | <13 |
| Carbapenems | Meropenem (MEM) | 10 | >23 | 20–22 | <19 |
| Aminoglycosides | Amikacin (AMK) | 30 | >17 | 15–16 | <14 |
| Gentamicin (GEN) | 10 | >15 | 13–14 | <12 | |
| Streptomycin (STR) | 10 | >15 | 12–14 | <11 | |
| Tobramycin (TOB) | 10 | >15 | 13–14 | <12 | |
| Tetracyclines | Tetracycline (TCY) | 30 | >15 | 12–14 | <11 |
| Fluoroquinolones | Ciprofloxacin (CIP) | 5 | >21 | 16–20 | <15 |
| Quinolones | Nalidixic acid (NAL) | 30 | >19 | 14–18 | <13 |
| Phenicols | Chloramphenicol (CHL) | 30 | >18 | 13–17 | <12 |
| Folate pathway inhibitors | Trimethoprim–sulfamethoxazole (SXT) | 25 | >16 | 11–15 | <10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.; Riley, A.; Sriharan, S.; Nartea, T.; Ndegwa, E.; Dhakal, R.; Zheng, G.; Baffaut, C. Examining Antimicrobial Resistance in Escherichia coli: A Case Study in Central Virginia’s Environment. Antibiotics 2024, 13, 223. https://doi.org/10.3390/antibiotics13030223
Kim C, Riley A, Sriharan S, Nartea T, Ndegwa E, Dhakal R, Zheng G, Baffaut C. Examining Antimicrobial Resistance in Escherichia coli: A Case Study in Central Virginia’s Environment. Antibiotics. 2024; 13(3):223. https://doi.org/10.3390/antibiotics13030223
Chicago/Turabian StyleKim, Chyer, Allissa Riley, Shobha Sriharan, Theresa Nartea, Eunice Ndegwa, Ramesh Dhakal, Guolu Zheng, and Claire Baffaut. 2024. "Examining Antimicrobial Resistance in Escherichia coli: A Case Study in Central Virginia’s Environment" Antibiotics 13, no. 3: 223. https://doi.org/10.3390/antibiotics13030223
APA StyleKim, C., Riley, A., Sriharan, S., Nartea, T., Ndegwa, E., Dhakal, R., Zheng, G., & Baffaut, C. (2024). Examining Antimicrobial Resistance in Escherichia coli: A Case Study in Central Virginia’s Environment. Antibiotics, 13(3), 223. https://doi.org/10.3390/antibiotics13030223








