Abstract
Previously, we reported that metronidazole MICs are not dependent on the expression levels of nim genes in B. fragilis strains and we compared the proteomes of metronidazole-resistant laboratory B. fragilis strains to those of their susceptible parent strains. Here, we used RT-qPCR to correlate the expression levels of 18 candidate genes in a panel of selected, clinical nim gene-positive and -negative B. fragilis strains to their metronidazole MICs. Metronidazole MICs were correlated with the expression of certain tested genes. Specifically, lactate dehydrogenase expression correlated positively, whereas cytochrome fumarate reductase/succinate dehydrogenase, malate dehydrogenase, phosphoglycerate kinase redox and gat (GCN5-like acetyltransferase), and relA (stringent response) regulatory gene expressions correlated negatively with metronidazole MICs. This result provides evidence for the involvement of carbohydrate catabolic enzymes in metronidazole resistance in B. fragilis. This result was supported by direct substrate utilization tests. However, the exact roles of these genes/proteins should be determined in deletion–complementation tests. Moreover, the exact redox cofactor(s) participating in metronidazole activation need to be identified.
1. Introduction
Species within the former Bacteroides fragilis group (BFG) (which are now classified within the Bacteroides, Parabacteroides, and Phocaeicola genera) are the most frequently isolated opportunistic anaerobic pathogens, which are also important members of the mammalian intestinal microbiota. Of these, B. fragilis is the most pathogenic and accounts for 50% of clinical isolates; however, it is only a minor population among intestinal strains [1]. They are highly resistant to most antimicrobial agents through the use of several antibiotic resistance mechanisms. Metronidazole is a prominent choice to treat infections of B. fragilis because metronidazole is an anti-anaerobic drug that usually elicits low levels of resistance among obligately anaerobic pathogens [2]. However, metronidazole resistance levels among Bacteroides have increased somewhat and increased greatly in developed and developing countries, respectively [3,4]. The best known and most investigated metronidazole resistance mechanism of BFG strains is mediated by the nim genes, a few of which were originally discovered in the 1980s and 1990s [5]. There are now 12 known homologs of nim (i.e., nimA-L) that share 50–80% amino acid homologies, and they are all proposed to act as nitro-reductases [6,7]. They have been localized to both the plasmid and chromosome, and they all require an upstream copy of a Bacteroides-specific insertion sequence (IS) element with promoter sequences to function in metronidazole resistance [5]. Besides nim genes, the other proposed metronidazole resistance mechanisms among BFG strains include increased lactate dehydrogenase, decreased pyruvate-ferredoxin oxidoreductase (PFOR), efflux, reduced iron uptake, and increased DNA repair [5]. However, these latter diverse mechanisms do not operate in all nim-negative-resistant strains and are sometimes found only in laboratory strains after the induction of metronidazole resistance. The nim-mediated mechanism, which is most prevalent among Bacteroides, still has some open questions. However, it is known that the metronidazole MICs of nim-positive strains are sometimes low and unstable, and the expression level of the nim genes correlates poorly with metronidazole MICs, which tend to be flexible. To explain this low correlation, the action of auxiliary factors has been proposed [8].
B. fragilis strains can be classified into two divergent divisions based on genetic differences (e.g., differences in alleles or genetic elements, most importantly the carbapenem resistance gene, cfiA) that can be mapped to specific loci in the genome of this important species [9], and, by our observations, it is also related to the gene expressions of the B. fragilis strains.
The aim of this study was to search for factors, in addition to nim genes, that affect the metronidazole resistance of B. fragilis. We have previously revealed that hemin- and iron-uptake mechanisms are involved in metronidazole resistance, and nim-negative and nim-positive B. fragilis strains behave differently in these regards [10]. Previously, we analyzed the proteomics of nimA-positive and -negative B. fragilis laboratory strains [11]. In addition, here, we analyzed the expression of 18 genes previously identified as resistance candidates in a proteomic study in a collection of nim-positive and -negative clinical B. fragilis strains, and we correlated the expression levels of these genes to measurements of metronidazole MICs to identify possible auxiliary factor(s) involved in metronidazole resistance of B. fragilis. Finally, we studied the effects of C4-dicarboxylic acid supplementation on metronidazole MICs to better understand the hemin dependence of metronidazole resistance.
2. Results and Discussion
2.1. Connection between the Metronidazole MICs and nim Gene Expression
Table 1 shows the results of the metronidazole MICs and the expression levels of the nim genes of the eight nim-positive strains. Similarly to the results of our previous studies [8,11], metronidazole MICs and the expression levels of the nim genes were independent of each other (r = 0.185, p = 0.619, r2 = 0.0342).

Table 1.
Strains used, their properties and RT-qPCR experiment results.
However, nim genes are known resistance factors because they transfer the resistance phenotype in conjugation experiments [19,20,21], and they are associated with metronidazole resistance in field studies [4,22]. Therefore, there is a need to account for this lack of correlation even with the same nim gene and IS element pairs. Previously, we proposed the existence of rate-limiting factors that influence the metronidazole resistance of B. fragilis strains [8].
2.2. Examination of the Roles of 18 Genes in Metronidazole Resistance
To investigate this possibility, we measured the expression levels of 18 genes selected according to the results of previous research or from our recent investigations [11] using RT-qPCR to study 15 B. fragilis strains. The cross-correlations between gene expressions and the correlation between gene expression and metronidazole MICs for all 15 B. fragilis strains are shown in Table 2. The cross-correlations between certain genes were very strong (r > 0.7, p < 0.01), indicating their common regulation, although not all genes (except for frdA and frdC, whose expressions correlated well—r = 0.593, p = 0.0192, Table 2) are located on the same operon [23]. Moreover, we detected highly significant correlations between the expression of some genes and metronidazole MICs. In particular, lactate dehydrogenase (ldh) expression correlated positively, whereas cytochrome b fumarate reductase/succinate dehydrogenase (frdC), malate dehydrogenase (mdh), phosphoglycerate kinase (pgk) catabolic and gat (GCN5-related acetyltransferase toxin), and relA (stringent response regulator) regulatory gene expressions correlated negatively with metronidazole MICs. Within the nim-positive and nim-negative strains, we detected cross-correlations between gene expressions; however, we found no significant association between metronidazole MICs and gene expressions, except for mdh and gat, which tended to correlate with metronidazole MICs in the nim-positive and nim-negative groups (Tables S2 and S3), respectively. In addition, the gene cross-correlations of the full set did not overlap with those in the nim-positive and nim-negative groups of strains (cf. Table S2 and Table 3). The lack of statistical confirmation may be due to the low number of strains in each group (eight nim-positive and seven nim-negative strains).

Table 2.
Cross-correlation values between the examined gene expressions and the metronidazole MICs for 15 B. fragilis strains a.

Table 3.
Significances of the one-way variance analyses of the gene expressions and the metronidazole MICs depending on the genetic background in 15 B. fragilis strains.
However, one-way variance analysis (Table 3) demonstrated that frdC, gat, mdh, nanH (sialidase), pgk, and relA gene expression depended on the presence of the nim gene; however, the cfiA gene status did not affect the expression of the studied genes (Table 3). The genes listed above differ from the list in Table 3 because the list above includes and excludes ldh and nanH, respectively. We are currently unable to explain this finding, although the inclusion of nanH indicates a link between metronidazole resistance/nim positivity and virulence.
Although we found no significant association of the examined genes among the nim-negative and nim-positive strains separately, the combined data signalized some good associations in the case of the whole strain set (see above). Therefore, we conclude that no particular enzyme is exclusively correlated with metronidazole resistance in both the nim-negative and -positive strains. However, some of these genes have previously been found to cause metronidazole resistance, e.g., feoAB (described in [24]), acr5 (bmeB, described in [25]) and por (described in [26]). This may be applied to genes not examined here (recA, sod and rhaA), as their role has been demonstrated in metronidazole resistance earlier [27,28,29]. So, on the population level, these genes do not exert a general role; they are important only in individual cases/strains. However, it should be noted that in both nim-positive and nim-negative strains, the possible exceptions of mdh and gat could be significant, respectively (mentioned above).
In addition, the roles of enzymes involved in the central metabolism in B. fragilis should also be considered. The central metabolism varies greatly among bacteria [30], e.g., the central metabolism of Bacteroides differs greatly from that of γ-Proteobacteria, and the latter comprises glycolysis and parts of the tricarboxylic cycle (TCA). However, instead of a complete TCA cycle, Bacteroides have a reductive or reverse TCA (rTCA) branch that is heme dependent, as well as a branch that is heme independent (Figure S1) [31]. Previously, we found that hemin depletion causes metronidazole susceptibility in both nim-negative and nim-positive strains of B. fragilis [10]. Thus, heme may be a rate-limiting factor in the metronidazole resistance of B. fragilis, as proposed above. Our results show that the expression of genes from the glycolytic and rTCA pathways (pgk, frdC, and mdh) correlate negatively with the metronidazole MICs, whereas that of ldh correlates positively. These latter changes can decrease the cellular concentrations of reducing cofactor, which diminishes metronidazole activation, thus inducing resistance.
The Nim enzymes are nitro-reductases that can transfer either six [6] or two electrons to the nitro group of metronidazole, yielding either an amino or a nitroso imidazole, respectively [20]. Recently, in vivo and in vitro experiments demonstrated that a nim group enzyme encoded by Clostridioides difficile strains is a nitro-reductase [7]. In this latter study, it was also confirmed that metronidazole resistance in C. difficile is dependent on hemin [32] through experiments involving the direct addition of metronidazole to assay its modification by recombinant NimB and by transcriptomic analysis. Moreover, genetic (transposon mutagenesis) and biochemical (aromatic nitro-reduction to amine) tests have proven that the nimB gene of some C. difficile strains is responsible for their metronidazole resistance. However, in the in vitro experiments, the metronidazole concentration used, 5 mM, was much higher than that to which the bacteria are usually exposed (the 4 μg/mL breakpoint concentration corresponds to 23.4 μM—a ca. 160-fold difference). It is possible that the hemin dependence of metronidazole resistance is due to the hemin dependence of the NimB protein; however, this does not explain the hemin dependence of nim-negative strains.
2.3. Examination of Addition of C4-dicarboxylic Acids on Metronidazole Resistance
We were also interested in how the addition of intermediates of the rTCA pathway affects metronidazole MICs. We expected that higher oxaloacetate or fumarate concentrations would decrease the redox intermediate concentration (e.g., NADH), thus decreasing metronidazole activation and MICs. In these experiments, we used modified M9 minimal medium supplemented with Tryptone, hemin, vitamin K1, and glucose or C4-dicarboxylic acid. The results are shown in Table S4, with some representative plates shown in Figure S2. Out of six nim-negative or nim-positive strains, four showed no significant difference in metronidazole MICs compared to those obtained on supplemented Columbia agars. However, the MICs of one nim-positive and one nim-negative strains increased in response to glucose, malate, and succinate addition, whereas no changes were observed in response to oxaloacetate or fumarate addition. This latter finding supports our assumption that is noted above (e.g., that the metronidazole resistance is highly dependent on reducing cofactor(s)). Moreover, our findings are consistent with the previous observation on the flexibility of metronidazole MICs and the idea of a rate-limiting step(s) involved in nim action in metronidazole resistance. We propose the following mechanism: the addition of malate and succinate forces the cells to reduce the levels of these compounds at the expense of the pool of reducing cofactors, thus leading to decreased metronidazole activation. We also argue that the C4-dicarboxylic acid uptake rates probably do not affect these processes because one transport protein, the anaerobic C4-dicarboxylic carrier protein, is responsible for their uptake with similar efficiencies in Escherichia coli [33]. The ortholog of this carrier protein is present in the genomes of B. fragilis strains (our unpublished analysis). The observed increase in ldh gene expression is consistent with previous findings, showing the importance of reducing cofactors in metronidazole resistance in anaerobic bacteria [26]. This means that the pyruvate level is the main mediator in this latter process. However, we did not observe a differential expression of por during our experiment. It is possible that withdrawing hydrogen/reducing cofactors from metronidazole activation may be involved in this process. The involvement of frdC (a cytochrome b enzyme) in metronidazole resistance is noteworthy because it can explain, at least partly, the heme dependence of the metronidazole resistance of B. fragilis. Additionally, the negative correlation of the regulatory genes (relA and gat) suggests that a high metabolic state is required for metronidazole to act on cells because these genes have a role in decreasing cellular metabolism.
This study is the first to examine the role of multiple proteins/genes on metronidazole resistance in clinical B. fragilis strains. Earlier modeling studies have focused only on laboratory strains of B. fragilis. For example, based on the roles of a limited set of proteins analyzed by two-dimensional protein electrophoresis and northern blotting, Diniz et al. proposed that ldh and por participate in metronidazole activation at certain low levels [26,34]. However, their model was not confirmed by Paunkov et al. [35]. Also, de Freitas et al. analyzed the transcriptome-wide effect of metronidazole on a large number of proteins, and they confirmed that, along with some other proteins, the concentration of activating ferredoxin is important in alleviating metronidazole stress [36]. Based on the results of proteomic studies, Paunkov et al. developed models of how nim and other proteins act in nim-dependent and -independent metronidazole-resistant B. fragilis strains [11].
2.4. Proposal for the Interactions of Redox and Other Proteins in Metronidazole Resistance
Here, we propose that a limited number of genes/proteins are correlated with metronidazole resistance in B. fragilis at the population level. In this study, we highlight the importance of reducing cofactors that are needed for both metronidazole activation and inactivation. The activated metronidazole radical acts by reducing nim and redox cofactor proteins and thiol compounds of the proteome [37]. Thus, the metronidazole resistance mechanism of B. fragilis is complex and nonlinear. This complexity can explain why metronidazole MICs and nim gene expression do not always correlate, especially long after the isolation of strains from clinical specimens. Thus, the process of developing resistance to metronidazole is also complex (Figure 1).
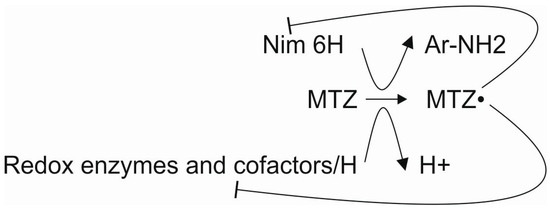
Figure 1.
Interactions of the participants in metronidazole resistance. Lines with the ͱ symbol mean inhibition.
Earlier work suggested that ferredoxin is responsible for reducing metronidazole [26]; however, we were unable to find a role for PFOR (negative association), as we observed increased PFOR and PFOR activities in laboratory metronidazole-resistant B. fragilis strains [35]. Thus, more work is needed to determine significant associations between gene expression and metronidazole MICs in “field” strains nim-negative and -positive B. fragilis. In particular, more strains need to be analyzed to prove the roles of those genes. In addition, the roles of the genes that had positive or negative correlations with metronidazole resistance should be confirmed by deletion–complementation analysis. In particular, frdC is a good candidate for these experiments because it also contains heme. Identifying with more certainty which redox cofactor activates metronidazole also remains a future task. The starting point for this study was a proteomic analysis of metronidazole resistant laboratory strains [11], but only some of them proved to be effective in metronidazole resistance on a population level; therefore, we believe that the genes that had a role in our study (ldh, frdC, mdh, pgk, gat and relA) are significant/valid contributors in this important kind of antibiotic resistance mechanism.
3. Materials and Methods
3.1. Bacterial Strains and Cultivation
Sixteen B. fragilis test strains (Table 1 and Table S1) with known genetic backgrounds were stored in 20% glycerol stocks at −80 °C and cultivated on supplemented Columbia blood agar medium (SCA, Columbia base, supplemented with 2.5% defibrinated sheep blood, 0.5% laked sheep blood, 0.3 mg/mL L-cysteine, 1 µg/mL vitamin K1) or in supplemented brain–heart infusion broth (BHIS, brain–heart infusion broth supplemented with 2.5% yeast extract, 10 µg/mL hemin and 1 µg/mL vitamin K1) under anaerobic conditions (85% N2, 10% H2, 5% CO2, Concept 400 anaerobic cabinet (Ruskinn, Bridgend, UK)) at 37 °C. The strains include both nim-positive and -negative B. fragilis strains whose cfiA gene statuses are known (Table 1). Parallelly with metronidazole MIC determinations, we used the same SCA plates for inoculations of 5 mL of BHIS for RNA isolation and cell suspensions to determine MICs. To test the effect of C4-dicarboxylic acids on metronidazole resistance, we used a semi-defined M9-based agar medium (48 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, 1.6 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 1% casein peptone Type I (Neogene), 0.625% yeast extract, 10 mM glucose or 15 mM C4-dicarboxylic acid (oxaloacetate/D(-) malate/fumarate/succinate), 10 µg/mL hemin, 1 µg/mL vitamin K1) to perform MIC measurements.
3.2. Metronidazole MIC Measurements
Metronidazole MICs were measured using a gradient method (Etest, bioMérieux, Hungary Ltd., Budapest, Hungary). First, McFarland density suspensions were made in a phosphate-buffered saline solution (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4 and 1.8 mM KH2PO4, pH 7.4), with which we inoculated the surface of SCA plates by cotton swabs, and applied the Etest strips, and after anaerobic cultivation at 37 °C for 48 h, we read the plates.
3.3. RT-qPCR
We extracted total RNA from 5 mL BHIS cultures for RT-qPCR experiments using the HighPure RNA Isolation Kit (Roche). The quantity and quality of RNA were assessed using the Qubit 4 fluorometer and the Qubit RNA BR and RNA Integrity kits (Thermo Fisher Scientific, Waltham, MA, USA). Of 32 candidate genes identified in previous proteomic studies [11], we chose 18 and designed primer pairs using the Primer3 Plus software (www.primer3plus.com). During primer design, we took into account the possibility that the cfiA-positive and -negative strains may differ in their respective sequences. Therefore, the consensus nucleotide sequences of the selected genes were obtained from the complete genomic sequences of the cfiA-negative and -positive strains B. fragilis 638R (GenBank Acc. No. NC_016776) and B. fragilis 3130 (GenBank Acc. No. LJVI01), respectively, to design primer pairs. We used the gap, rrn, and rpoD genes as endogenous controls. The nucleotide sequences of the primers used are shown in Table S1. The 10 μL RT-qPCR reactions contained 5.6 μL kit components (Verso 1-step SYBR RT-PCR mastermix, Thermo Fisher Scientific), 0.2 μL each primer (35 μM), 3 μL H2O, and 1 μL total RNA. The reactions were incubated in an RT-PCR instrument (QuantStudio 3, Thermo Fisher Scientific) in 100 μL 96-well plates using the following conditions: 35 cycles consisting of 55 °C 20 min, 95 °C 15 min; 95 °C 15 s, 55 °C 30 s, 72 °C 30 s. The melting curves were recorded using 3 technical replicates. We detected the expression of the nim genes in 8 nim-positive B. fragilis strains by amplifying nim PCR products using the same conditions as those described above, except the 35 PCR cycles consisted of two steps (55 °C 20 min, 95 °C 15 min; 95 °C 15 s, 60 °C 1 min; melting curve) because three nim gene types were included.
3.4. Data Analysis
We used the amplification threshold values (CT) from RT-qPCR experiments to calculate the ratios of gene expression by the ΔΔCT method. The calculations were performed by the Relative Quantitation App on the Thermo Fisher Scientific webpage (www.thermofisher.com). One-way variance (ANOVA), Spearman’s rank, and cross-correlation values were calculated using SigmaPlot 12 software (Sigmaplot, Erkrath, Germany).
4. Conclusions
In this study, we assessed the connection between metronidazole MICs and the expression of 18 genes in a wide selection of B. fragilis clinical strains. The expression of metabolic genes ldh, frdC, mdh, and pgk correlated with metronidazole resistance independently of the presence of nim genes. This finding emphasizes that redox intermediates may be crucial in both metronidazole activation and enzymatic inactivation. However, the exact identities of the enzymes and intermediates involved in both processes need to be confirmed experimentally. Roles for some regulatory proteins (gat, relA) were also found and not all (genes)/proteins could be examined here as they were differentially expressed at the protein level. Thus, the list of examined genes should also be increased.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13030207/s1 Table S1. RT-qPCR target genes and primer sequences. Table S2. Cross-correlation values of gene expressions metronidazole resistance for eight nim-positive B. fragilis strains. Table S3. Cross-correlation values of gene expressions metronidazole resistance for seven nim-negative B. fragilis strains. Figure S1. The tricarboxylic acid pathways of B. fragilis. Figure S2. Examples of the Etest results on modified M9 medium. Table S4. Effects of C4 dicarboxylic acid supplementation on metronidazole MICs of selected nim-positive and negative B. fragilis strains.
Author Contributions
Conceptualization: D.L. and J.S. Methodology: B.M., A.P., M.K. and J.S. Formal Analysis: D.L., J.S., E.N. and K.B. Data Curation: J.S. Writing—Original Draft Preparation: J.S. and E.N. Writing—Review and Editing: B.M., A.P., M.K., K.B., E.N., D.L. and J.S. Funding Acquisition: D.L., J.S. and K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a bilateral grant of the National Research, Development and Innovation Office in Hungary (NKFIH), and the Austrian Science Fund (FWF) in Austria (grant numbers ANN_130760 and I 4234, respectively), and obtained partial support from Interreg V-A Romania-Hungary Programme (project eMS code: ROHU339 HEALTH-PREGN-ROHU).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets of this study are available from the corresponding author upon reasonable request.
Acknowledgments
Bakhtiyar Mahmood is a Gedeon Richter Talent Fund student.
Conflicts of Interest
The authors declare no conflicts of interests.
References
- Wexler, H.M. Bacteroides: The good, the bad and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Dingsdag, S.A.; Hunter, N. Metronidazole: An update on metabolism, structure-cytotoxicity and resistance mechanisms. J. Antimicrob. Chemother. 2018, 73, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.U.E.; Nord, C.E. on behalf of the ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe 20 years of experience. Clin. Microbiol. Infect. 2011, 17, 371–379. [Google Scholar] [CrossRef]
- Sethi, S.; Shukla, R.; Bala, K.; Gautam, V.; Angrup, A.; Ray, P. Emerging metronidazole resistance in Bacteroides spp. and its association with the nim gene: A study from North India. J. Glob. Antimicrob. Resist. 2019, 16, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Alauzet, C.; Lozniewski, A.; Marchandin, H. Metronidazole resistance and nim genes in anaerobes: A review. Anaerobe 2019, 55, 40–53. [Google Scholar] [CrossRef]
- Carlier, J.P.; Sellier, N.; Rager, M.N.; Reysset, G. Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic strains of Bacteroides fragilis. Antimicrob. Agents Chemother. 1997, 41, 1495–1499. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Dureja, C.; Youngblom, M.A.; Topf, M.A.; Shen, W.J.; Gonzales-Luna, A.J.; Deshpande, A.; Hevener, K.E.; Freeman, J.; Wilcox, M.H.; et al. Decoding a cryptic mechanism of metronidazole resistance among globally disseminated fluoroquinolone-resistant Clostridioides difficile. Nat. Commun. 2023, 14, 4130. [Google Scholar] [CrossRef]
- Mahmood, B.; Juhász, H.; Leitsch, D.; Sóki, J. The effects of identical nim gene-insertion sequence combinations on the expression of the nim genes and metronidazole resistance in Bacteroides fragilis strains. Anaerobe 2023, 81, 102739. [Google Scholar] [CrossRef]
- García, N.; Gutiérrez, G.; Lorenzo, M.; Vadillo, S.; Píriz, S.; Quesada, A. Gene Context and DNA Rearrangements in the Carbapenemase Locus of Division II Strains of Bacteroides fragilis. Antimicrob. Agents Chemother. 2009, 53, 2677–2678. [Google Scholar] [CrossRef][Green Version]
- Paunkov, A.; Gutenbrunner, K.; Sóki, J.; Leitsch, D. Haemin deprivation renders Bacteroides fragilis hypersusceptible to metronidazole and cancels high-level metronidazole resistance. J. Antimicrob. Chemother. 2022, 77, 1027–1031. [Google Scholar] [CrossRef]
- Paunkov, A.; Hummel, K.; Strasser, D.; Sóki, J.; Leitsch, D. Proteomic analysis of metronidazole resistance in the human facultative pathogen Bacteroides fragilis. Front. Microbiol. 2023, 14, 1158086. [Google Scholar] [CrossRef]
- Löfmark, S.; Fang, H.; Hedberg, M.; Edlund, C. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob. Agents Chemother. 2005, 49, 1253–1256. [Google Scholar] [CrossRef]
- Sóki, J.; Fodor, E.; Hecht, D.W.; Edwards, R.; Rotimi, V.O.; Kerekes, I.; Urbán, E.; Nagy, E. Molecular characterization of imipenem-resistant, cfiA-positive Bacteroides fragilis isolates from the USA, Hungary and Kuwait. J. Med. Microbiol. 2004, 53, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Baaity, Z.; Jamal, W.; Rotimi, V.O.; Burián, K.; Leitsch, D.; Somogyvári, F.; Nagy, E.; Sóki, J. Molecular characterization of metronidazole resistant Bacteroides strains from Kuwait. Anaerobe 2021, 69, 102357. [Google Scholar] [CrossRef] [PubMed]
- Wareham, D.W.; Wilks, M.; Ahmed, D.; Brazier, J.S.; Miller, M. Anaerobic sepsis due to multidrug-resistant Bacteroides fragilis: Microbiological cure and clinical response with linezolid therapy. Clin. Infect. Dis. 2005, 40, e67–e68. [Google Scholar] [CrossRef] [PubMed]
- Podglajen, I.; Breuil, J.; Casin, I.; Collatz, E. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J. Bacteriol. 1995, 177, 5270–5275. [Google Scholar] [CrossRef] [PubMed]
- Sárvári, K.P.; Sóki, J.; Kristóf, K.; Juhász, E.; Miszti, C.; Latkóczy, K.; Melegh, S.Z.; Urbán, E. A multicentre survey of the antibiotic susceptibility of clinical Bacteroides species from Hungary. Infect. Dis. 2018, 50, 372–380. [Google Scholar] [CrossRef]
- Privitera, G.; Sebald, M.; Fayolle, F. Common regulatory mechanism of expression and conjugative ability of a tetracycline resistance plasmid in Bacteroides fragilis. Nature 1979, 278, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Sóki, J.; Gal, M.; Brazier, J.S.; Rotimi, V.O.; Urbán, E.; Nagy, E.; Duerden, B.I. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 2006, 57, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Reysset, G. Genetics of 5-nitroimidazole resistance in Bacteroides species. Anaerobe 1996, 2, 59–69. [Google Scholar] [CrossRef]
- Husain, F.; Veeranagouda, Y.; Hsi, J.; Meggersee, R.; Abratt, V.; Wexler, H.M. Two multidrug-resistant clinical isolates of Bacteroides fragilis carry a novel metronidazole resistance nim gene (nimJ). Antimicrob. Agents Chemother. 2013, 57, 3767–3774. [Google Scholar] [CrossRef]
- Vishwanath, S.; Shenoy, P.A.; Chawla, K. Antimicrobial Resistance Profile and Nim Gene Detection among Bacteroides fragilis Group Isolates in a University Hospital in South India. J. Glob. Infect. Dis. 2019, 11, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Baughn, A.D.; Malamy, M.H. The essential role of fumarate reductase in haem-dependent growth stimulation of Bacteroides fragilis. Microbiology (Reading) 2003, 149, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Veeranagouda, Y.; Husain, F.; Boente, R.; Moore, J.; Smith, C.J.; Rocha, E.R.; Patrick, S.; Wexler, H.M. Deficiency of the ferrous iron transporter FeoAB is linked with metronidazole resistance in Bacteroides fragilis. J. Antimicrob. Chemother. 2014, 69, 2634–2643. [Google Scholar] [CrossRef] [PubMed]
- Pumbwe, L.; Chang, A.; Smith, R.L.; Wexler, H.M. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb. Drug Resist. 2007, 13, 96–101. [Google Scholar] [CrossRef]
- Diniz, C.G.; Farias, L.M.; Carvalho, M.A.; Rocha, E.R.; Smith, C.J. Differential gene expression in a Bacteroides fragilis metronidazole-resistant mutant. J. Antimicrob. Chemother. 2004, 54, 100–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steffens, L.S.; Nicholson, S.; Paul, L.V.; Nord, C.E.; Patrick, S.; Abratt, V.R. Bacteroides fragilis RecA protein overexpression causes resistance to metronidazole. Res. Microbiol. 2010, 161, 346–354. [Google Scholar] [CrossRef]
- Samarawickrema, N.A.; Brown, D.M.; Upcroft, J.A.; Thammapalerd, N.; Upcroft, P. Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 1997, 40, 833–840. [Google Scholar] [CrossRef]
- Patel, E.H.; Paul, L.V.; Casanueva, A.I.; Patrick, S.; Abratt, V.R. Overexpression of the rhamnose catabolism regulatory protein, RhaR: A novel mechanism for metronidazole resistance in Bacteroides thetaiotaomicron. J. Antimicrob. Chemother. 2009, 64, 267–273. [Google Scholar] [CrossRef]
- Huynen, M.A.; Dandekar, T.; Bork, P. Variation and evolution of the citric-acid cycle: A genomic perspective. Trends Microbiol. 1999, 7, 281–291. [Google Scholar] [CrossRef]
- Baughn, A.D.; Malamy, M.H. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: Implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 2002, 99, 4662–4667. [Google Scholar] [CrossRef] [PubMed]
- Boekhoud, I.M.; Sidorov, I.; Nooij, S.; Harmanus, C.; Bos-Sanders, I.; Viprey, V.; Spittal, W.; Clark, E.; Davies, K.; Freeman, J.; et al. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J. Antimicrob. Chemother. 2021, 76, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Janausch, I.G.; Zientz, E.; Tran, Q.H.; Kröger, A.; Unden, G. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 2002, 1553, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, S.; Suzuki, T.; Yamamoto, M.; Nakamura, M. Lactate dehydrogenase activity as a cause of metronidazole resistance in Bacteroides fragilis NCTC 11295. J. Antimicrob. Chemother. 1991, 28, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Paunkov, A.; Sóki, J.; Leitsch, D. Modulation of Iron Import and Metronidazole Resistance in Bacteroides fragilis Harboring a nimA Gene. Front. Microbiol. 2022, 13, 898453. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, M.C.; Resende, J.A.; Ferreira-Machado, A.B.; Saji, G.D.; de Vasconcelos, A.T.; da Silva, V.L.; Nicolás, M.F.; Diniz, C.G. Exploratory Investigation of Bacteroides fragilis Transcriptional Response during In vitro Exposure to Subinhibitory Concentration of Metronidazole. Front. Microbiol. 2016, 7, 1465. [Google Scholar] [CrossRef]
- Leitsch, D.; Kolarich, D.; Wilson, I.B.; Altmann, F.; Duchêne, M. Nitroimidazole action in Entamoeba histolytica: A central role for thioredoxin reductase. PLoS Biol. 2007, 5, e211. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).