Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand
Abstract
1. Introduction
2. Methodology
2.1. Sample Collection and Antimicrobial Susceptibility Testing
2.2. DNA Extraction and Sequencing
2.3. Bioinformatics and Sequence Analysis
2.4. Multilocus Sequence Typing and Phylogenetic Analysis
3. Results and Discussions
3.1. Clinical Data and Genome Information and Antimicrobial Susceptibility Testing Results of Acinetobacter Non-baumannii Isolates
3.2. Detection of Antimicrobial Resistant Genes (ARGs) from Acinetobacter Non-baumannii
3.3. Detection of Mobile Genetic Elements (MGEs)
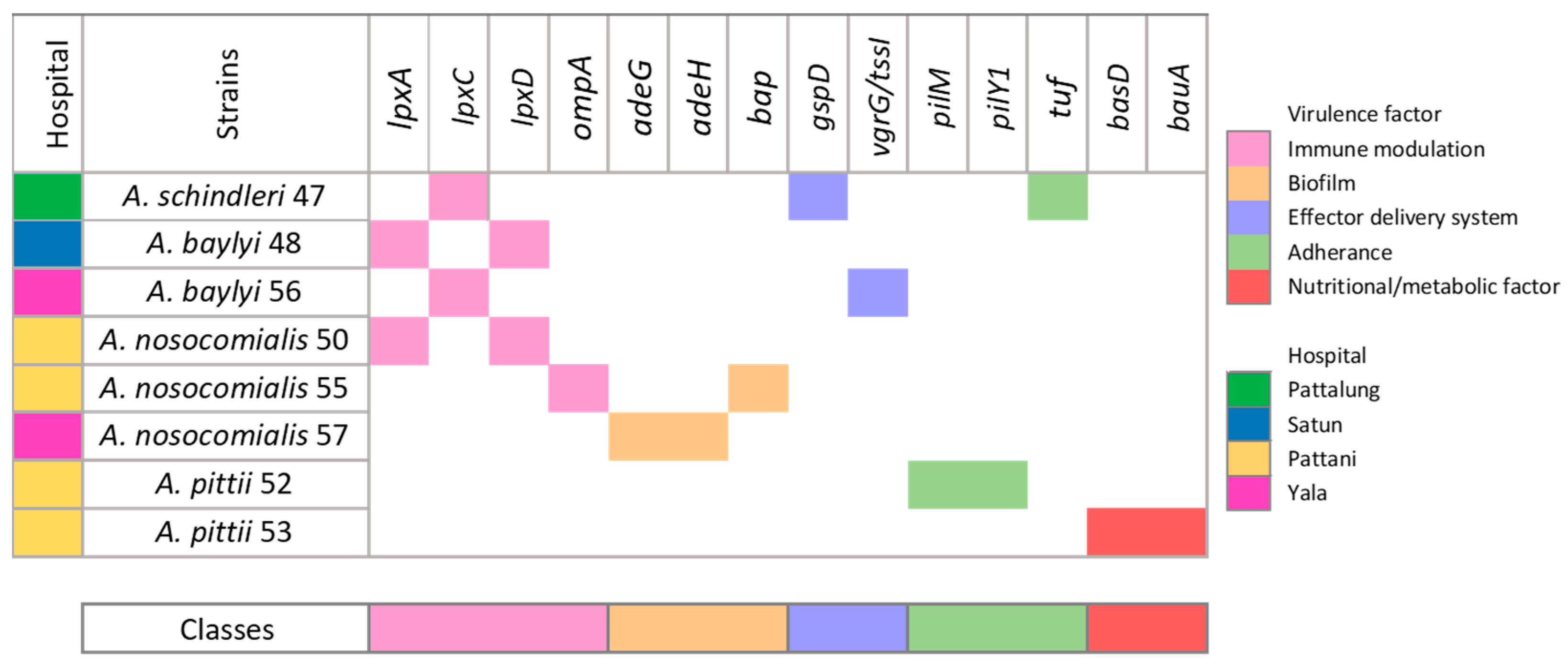
3.4. Evaluation of Virulence-Associated Genes in Eight Acinetobacter Non-baumannii Isolates
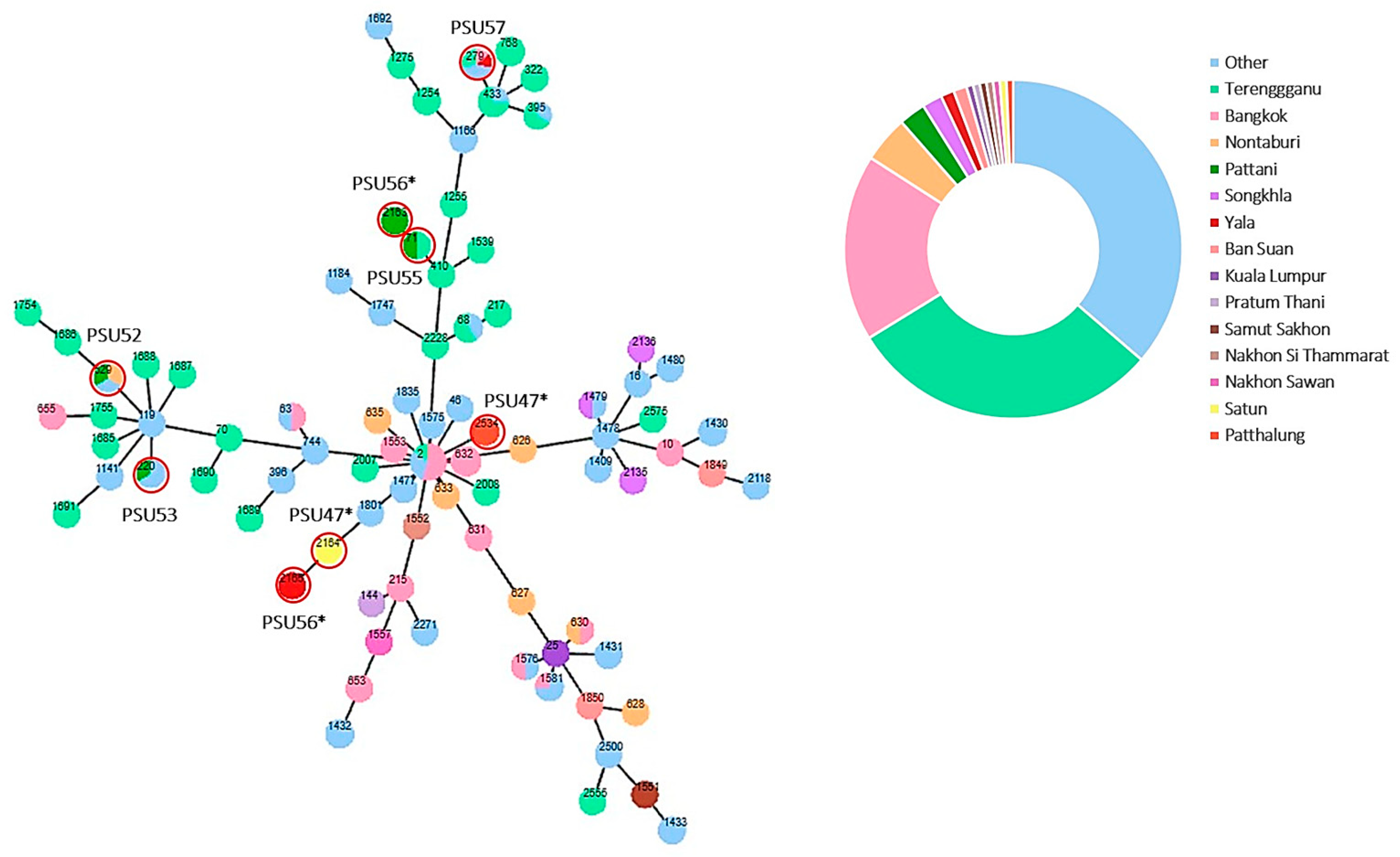
3.5. Multilocus Sequence Typing (MLST) and Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Percival, S.L.; Williams, D.W. Chapter Two—Acinetobacter. In Microbiology of Waterborne Diseases, 2nd ed.; Percival, S.L., Yates, M.V., Williams, D.W., Chalmers, R.M., Gray, N.F., Eds.; Academic Press: London, UK, 2014; pp. 35–48. [Google Scholar]
- Mehrad, B.; Clark, N.M.; Zhanel, G.G.; Lynch, J.P., 3rd. Antimicrobial resistance in hospital-acquired gram-negative bacterial infections. Chest 2015, 147, 1413–1421. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Paulus, T.; Lugenheim, M.; Stefanik, D.; Higgins, P.G.; Edmond, M.B.; Wenzel, R.P.; Seifert, H. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 2012, 64, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Corbella, X.; Pujol, M.; Ayats, J.; Sendra, M.; Ardanuy, C.; Domínguez, M.A.; Liñares, J.; Ariza, J.; Gudiol, F. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1996, 23, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Hsueh, P.R.; Mendes, R.E.; Pfaller, M.A.; Rolston, K.V.; Sader, H.S.; Jones, R.N. The Microbiology of Bloodstream Infection: 20-Year Trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019, 63, e00355-19. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Kou, S.H.; Xie, R.; VanNieuwenhze, M.S.; Qu, J.; Peng, B.; Zheng, J. Non-walled spherical Acinetobacter baumannii is an important type of persister upon β-lactam antibiotic treatment. Emerg. Microbes Infect. 2020, 9, 1149–1159. [Google Scholar] [CrossRef]
- Nguyen, M.; Joshi, S.G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Lin, L.C.; Chang, Y.J.; Liu, C.E.; Soon, M.S. Long-term effectiveness of infection and antibiotic control programs on the transmission of carbapenem-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex in central Taiwan. Med. Mal. Infect. 2015, 45, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Chen, X.; Liu, S.; Zhang, Z.; Li, N.; Dong, C.; Zhang, L.; Wu, H.; Zhao, S. Epidemiological Analysis of Multidrug-Resistant Acinetobacter baumannii Isolates in a Tertiary Hospital over a 12-Year Period in China. Front. Public Health 2021, 9, 707435. [Google Scholar] [CrossRef]
- Dong, J.-F.; Feng, C.-J.; Wang, P.; Li, R.-Q.; Zou, Q.-H. Comparative genomics analysis of Acinetobacter baumannii multi-drug resistant and drug sensitive strains in China. Microb. Pathog. 2022, 165, 105492. [Google Scholar] [CrossRef]
- Krieg, N.R.; Holt, J.G. Bergey’s Manual of Systematic Bacteriology; Yi Hsien Publishing Co.: New Taipei City, Taiwan, 1984. [Google Scholar]
- Feßler, A.T.; Wang, Y.; Burbick, C.R.; Diaz-Campos, D.; Fajt, V.R.; Lawhon, S.D.; Li, X.-Z.; Lubbers, B.V.; Maddock, K.; Miller, R.A.; et al. Antimicrobial susceptibility testing in veterinary medicine: Performance, interpretation of results, best practices and pitfalls. One Health Adv. 2023, 1, 26. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33th ed.; CLSI Guideline M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Chuang, Y.C.; Cheng, C.Y.; Sheng, W.H.; Sun, H.Y.; Wang, J.T.; Chen, Y.C.; Chang, S.C. Effectiveness of tigecycline-based versus colistin- based therapy for treatment of pneumonia caused by multidrug-resistant Acinetobacter baumannii in a critical setting: A matched cohort analysis. BMC Infect. Dis. 2014, 14, 102. [Google Scholar] [CrossRef]
- Fernández-Mazarrasa, C.; Mazarrasa, O.; Calvo, J.; del Arco, A.; Martínez-Martínez, L. High concentrations of manganese in Mueller-Hinton agar increase MICs of tigecycline determined by Etest. J. Clin. Microbiol. 2009, 47, 827–829. [Google Scholar] [CrossRef]
- Chukamnerd, A.; Jeenkeawpiam, K.; Chusri, S.; Pomwised, R.; Singkhamanan, K.; Surachat, K. BacSeq: A User-Friendly Automated Pipeline for Whole-Genome Sequence Analysis of Bacterial Genomes. Microorganisms 2023, 11, 1769. [Google Scholar] [CrossRef]
- Torsten, S. Abricate, Github 2020. Available online: https://github.com/tseemann/abricate (accessed on 1 January 2020).
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Torsten, S. Mlst Github 2022. Available online: https://github.com/tseemann/mlst (accessed on 1 January 2020).
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016, 44, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Chusri, S.; Chongsuvivatwong, V.; Rivera, J.I.; Silpapojakul, K.; Singkhamanan, K.; McNeil, E.; Doi, Y. Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob. Agents Chemother. 2014, 58, 4172–4179. [Google Scholar] [CrossRef]
- Yang, L.; Dong, N.; Xu, C.; Ye, L.; Chen, S. Emergence of ST63 Pandrug-Resistant Acinetobacter pittii Isolated From an AECOPD Patient in China. Front. Cell. Infect. Microbiol. 2021, 11, 739211. [Google Scholar] [CrossRef]
- Nemec, A.; De Baere, T.; Tjernberg, I.; Vaneechoutte, M.; van der Reijden, T.J.; Dijkshoorn, L. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2001, 51, 1891–1899. [Google Scholar] [CrossRef]
- Dortet, L.; Legrand, P.; Soussy, C.J.; Cattoir, V. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 2006, 44, 4471–4478. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, H.; Ijaz, M.; Ahmed, A.; Javed, M.U.; Shah, S.F.A.; Anwaar, F. Discrepancies between phenotypic and genotypic identification methods of antibiotic resistant genes harboring Staphylococcus aureus. Microb. Pathog. 2023, 184, 106342. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 2011, 63, 1061–1067. [Google Scholar] [CrossRef]
- Bala, A.; Uhlin, B.E.; Karah, N. Insights into the genetic contexts of sulfonamide resistance among early clinical isolates of Acinetobacter baumannii. Infect. Genet. Evol. 2023, 112, 105444. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Mendes, R.E.; Gales, A.C. Global Epidemiology and Mechanisms of Resistance of Acinetobacter baumannii-calcoaceticus Complex. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2023, 76 (Suppl. S2), S166–S178. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, C.; Wu, J.; Jiang, R.; Mi, Z.; Huang, Z. A novel aminoglycoside-modifying enzyme gene aac(6′)-Ib in a pandrug-resistant Acinetobacter baumannii strain. J. Hosp. Infect. 2009, 73, 184–185. [Google Scholar] [CrossRef]
- Bergogne-Bérézin, E.; Towner, K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef]
- Novotna, G.; Janata, J. A New Evolutionary Variant of the Streptogramin A Resistance Protein, Vga(A)LC, from Staphylococcus haemolyticus with Shifted Substrate Specificity towards Lincosamides. Antimicrob. Agents Chemother. 2006, 50, 4070–4076. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, M.; Thamaraiselvan, S.; Bulent, B.; Cecilia Stalsby, L.; Ashok, J.T.; Nades, P.; Nachimuthu, R. Transfer of Antibiotic Resistance Genes from Gram-Positive Bacterium to Gram-negative Bacterium. bioRxiv 2020. [Google Scholar] [CrossRef]
- Touchon, M.; Cury, J.; Yoon, E.-J.; Krizova, L.; Cerqueira, G.C.; Murphy, C.; Feldgarden, M.; Wortman, J.; Clermont, D.; Lambert, T.; et al. The Genomic Diversification of the Whole Acinetobacter Genus: Origins, Mechanisms, and Consequences. Genome Biol. Evol. 2014, 6, 2866–2882. [Google Scholar] [CrossRef] [PubMed]
- Palmen, R.; Vosman, B.; Buijsman, P.; Breek, C.K.D.; Hellingwerf, K.J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. Microbiology 1993, 139, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Courvalin, P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 1994, 38, 1447–1451. [Google Scholar] [CrossRef]
- Brisson-Noël, A.; Arthur, M.; Courvalin, P. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J. Bacteriol. 1988, 170, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Bertram, J.; Strätz, M.; Dürre, P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J. Bacteriol. 1991, 173, 443–448. [Google Scholar] [CrossRef]
- Kanj, Z. Acinetobacter Infection: Treatment and Prevention. Available online: https://www.uptodate.com/contents/acinetobacter-infection-treatment-and-prevention/print#H1768507054 (accessed on 8 November 2023).
- Villalón, P.; Valdezate, S.; Medina-Pascual, M.J.; Carrasco, G.; Vindel, A.; Saez-Nieto, J.A. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenem-hydrolysing oxacillinase and metallo-β-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J. Antimicrob. Chemother. 2013, 68, 550–553. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Tiku, V. Acinetobacter baumannii: Virulence Strategies and Host Defense Mechanisms. DNA Cell Biol. 2022, 41, 43–48. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef] [PubMed]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Imperi, F.; Carattoli, A.; Visca, P. Deciphering the Multifactorial Nature of Acinetobacter baumannii Pathogenicity. PLoS ONE 2011, 6, e22674. [Google Scholar] [CrossRef]



| Strains | Isolation Sources | Location | Estimated Genome Size (bp) * | N50 Value |
|---|---|---|---|---|
| A. schindleri PSU47 | Endotracheal tube | Phatthalung | 3,347,770 | 93,667 |
| A. baylyi PSU48 | Nasopharynx | Satun | 3,601,820 | 79,944 |
| A. nosocomialis PSU50 | Rectum | Pattani | 4,015,432 | 122,902 |
| A. pittii PSU52 | Rectum | Pattani | 3,986,945 | 110,976 |
| A. pittii PSU53 | Rectum | Pattani | 3,917,437 | 75,803 |
| A. nosocomialis PSU55 | Endotracheal tube | Pattani | 4,047,440 | 93,952 |
| A. baylyi PSU56 | Nasopharynx | Yala | 3,820,703 | 216,777 |
| A. nosocomialis PSU57 | Rectum | Yala | 4,091,066 | 103,279 |
| Isolates | Mobile Genetic Elements (MGEs) | ARGs Found on MGEs | ||
|---|---|---|---|---|
| Plasmid | Insertion Sequences (n **) | Composite Transposons | ||
| A. schindleri PSU47 | ISOur1, IS1007, ISAca1, ISAcsp5, IS18, ISAba11 | cn_2205_ISOur1, cn_2205_IS1007 | ||
| A. baylyi PSU48 | IS17(3), ISAba27, ISAba34, | |||
| ISAcsp5 * | ||||
| A. nosocomialis PSU50 | ISAba26, ISAba13, IS17, ISAha3, ISAba27, ISAba33 (2), ISAba1, ISOur1, IS18, ISAba14 | cn_26436_ISAba33, cn_8866_IS18 | ||
| ISCfr1 * | ||||
| A. pittii PSU52 | rep5d * | vga(A)LC | ||
| ISAba45, ISAba26, ISAba50, ISAba13, IS17, ISAba33, IS26 | cn_4415_ISAba33 | |||
| A. pittii PSU53 | ISAba1, ISAba33, ISAba26, ISPst2, ISAha3, ISAba31 | |||
| A. nosocomialis PSU55 | cn_3572_IS1008 * | sul2, aac(3)-Iid, sul1, blaIMP-14 | ||
| ISCfr1 * | ||||
| IS1008, ISAba14, ISAba1, ISAba33, ISAba10(2), IS17, ISAha3, ISAba27, ISAba25 | ||||
| A. baylyi PSU56 | IS17 * | |||
| IS18 * | ||||
| IS17(2), ISAba27, ISAba14 | ||||
| A. nosocomialis PSU57 | IS18, ISAba125, ISAba27(2), ISAha3, ISAba33, ISAba21, IS1007 | cn_8866_IS18, cn_1062_IS1007, cn_2153_IS1007 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaikhan, T.; Chukamnerd, A.; Singkhamanan, K.; Nokchan, N.; Chintakovid, N.; Chusri, S.; Pomwised, R.; Wonglapsuwan, M.; Surachat, K. Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand. Antibiotics 2024, 13, 149. https://doi.org/10.3390/antibiotics13020149
Yaikhan T, Chukamnerd A, Singkhamanan K, Nokchan N, Chintakovid N, Chusri S, Pomwised R, Wonglapsuwan M, Surachat K. Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand. Antibiotics. 2024; 13(2):149. https://doi.org/10.3390/antibiotics13020149
Chicago/Turabian StyleYaikhan, Thunchanok, Arnon Chukamnerd, Kamonnut Singkhamanan, Natakorn Nokchan, Nutwadee Chintakovid, Sarunyou Chusri, Rattanaruji Pomwised, Monwadee Wonglapsuwan, and Komwit Surachat. 2024. "Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand" Antibiotics 13, no. 2: 149. https://doi.org/10.3390/antibiotics13020149
APA StyleYaikhan, T., Chukamnerd, A., Singkhamanan, K., Nokchan, N., Chintakovid, N., Chusri, S., Pomwised, R., Wonglapsuwan, M., & Surachat, K. (2024). Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand. Antibiotics, 13(2), 149. https://doi.org/10.3390/antibiotics13020149






