Abstract
Background: Acquired 16S rRNA methyltransferases (16S-RMTases) confer high-level resistance to aminoglycosides and are often associated with β-lactam and quinolone resistance determinants. Methods: Using PCR, whole-genome sequencing and conjugation experiments, we conducted a retrospective genomic surveillance study of 16S-RMTase-producing Enterobacterales, collected between 2006 and 2023, to explore transmission dynamics of methyltransferase and associated antibiotic resistance genes. Results: Among the 10,731 consecutive isolates, 150 (1.4%) from 13 species carried armA (92.7%), rmtB (4.7%), and rmtF + rmtB (2.7%) methyltransferase genes. The coexistence of extended-spectrum β-lactamase (blaCTX-M-3/15, blaSHV-12, blaSFO-1), carbapenemase (blaNDM-1/5, blaVIM-1/4/86, blaOXA-48), acquired AmpC (blaCMY-2/4/99, blaDHA-1, blaAAC-1), and plasmid-mediated quinolone resistance (qnrB, qnrS, aac(6′)-Ib-cr) genes within these isolates was also detected. Methyltransferase genes were carried by different plasmids (IncL/M, IncA/C, IncR, IncFIB, and IncFII), suggesting diverse origins and sources of acquisition. armA was co-transferred with blaCTX-M-3/15, blaNDM-1, blaVIM-4/86, blaOXA-48, blaCMY-4, aac(6′)-Ib-cr, qnrB, and qnrS, while rmtF1 was co-transferred with blaSFO-1, highlighting the multidrug-resistant nature of these plasmids. Long-read sequencing of ST6260 K. pneumoniae isolates revealed a novel resistance association, with rmtB1 and blaNDM-5 on the chromosome, blaOXA-232 on a conjugative ColKP3 plasmid, and rmtF1 with blaSFO-1 on self-transmissible IncFIB and IncFII plasmids. Conclusions: The genetic plasticity of plasmids carrying methyltransferase genes suggests their potential to acquire additional resistance genes, turning 16S-RMTase-producing Enterobacterales into a persistent public health threat.
1. Introduction
Aminoglycosides play an important role in the treatment of complicated bacterial infections, often used in synergistic combination with cell-wall-active antibiotics such as β-lactams. They act by specifically binding to the aminoacyl site (A-site) of 16S rRNA in prokaryotic 30S ribosomal subunits, thereby inhibiting protein synthesis with subsequent bacterial death [1]. Their widespread use has contributed to the emergence of aminoglycoside resistance involving target alteration, enzymatic drug modification, decreased permeability, and increased efflux [1,2]. Enzymatic drug inactivation through modification by aminoglycoside acetyltransferases, adenylyltransferases and phosphotransferases is the most common mechanism of aminoglycoside resistance in Gram-negative bacteria. Aminoglycoside-modifying enzymes have a well-defined substrate profile, including either amikacin or gentamicin, but never both, which favors a therapeutic approach. Unlike target modification, only the coexistence of various aminoglycoside-modifying enzymes in association with active efflux can result in high-level, broad-spectrum resistance.
Alteration of ribosomal targets by post-transcriptional methylation is an emerging mechanism, formerly confined to aminoglycoside producers as a mechanism of self-protection to avoid suicide [3]. The 16S rRNA methyltransferases (16S-RMTases) modify specific rRNA nucleotide residues within the A-site that blocks aminoglycosides from binding to their target and results in high-level resistance with a minimum inhibitory concentration (MIC) > 256 mg/L. These enzymes are divided into two classes depending on specific nucleotide residues that they modify. The methylation of the N-7 position of nucleotide G1405 by various 16S-RMTases confers resistance to 4,6-disubstituted 2-deoxystreptamines (kanamycin, amikacin, isepamicin, gentamicin, netilmicin, tobramycin, arbekacin, sisomicin, plazomicin), but not to 4,5-disubstituted (neomycin, ribostamycin, paromomycin) and 4-monosubstituted (apramycin) deoxystreptamines. In contrast, the methylation of the N-1 position of nucleotide A1408 confers resistance to deoxystreptamines of all groups. The methyltransferases of both classes are not active against atypical aminoglycosides (streptomycin, spectinomycin) that lack a deoxystreptamine ring [1,2,3]. The first cases of acquired plasmid-mediated 16S-RMTase in human pathogens were RmtA (ribosomal methyltransferases) in Pseudomonas aeruginosa from Japan [4], and ArmA (aminoglycoside resistance methyltransferase) in Klebsiella pneumoniae from France [5]. Subsequently, seven additional plasmid-borne methyltransferases, encoded by the genes rmtB [6], rmtC [7], rmtD [8], rmtE [9], rmtF [10], rmtG [11], and rmtH [12], have emerged in clinical isolates. They showed high-level resistance to clinically relevant aminoglycosides (i.e., amikacin, gentamicin, tobramycin, and plazomicin), but susceptibility to neomycin and apramycin. These enzymes were confirmed to function as N7-G1405 16S-RMTases [2,3]. In 2007, the first acquired N1-A1408 16S-RMTase was discovered. This enzyme, named NpmA, was identified in an Escherichia coli clinical strain in Japan. NpmA was plasmid-borne and confers resistance to neomycin and apramycin in addition to amikacin, gentamicin, tobramycin, and plazomicin [2,13].
Since their acquisition from clinical isolates of Gram-negative bacteria, 16S-RMTase genes have spread among Enterobacterales, P. aeruginosa, and Acinetobacter baumannii in both clinical and veterinary settings all over the world [14,15]. There are several features of 16S-RMTases that are involved in their dissemination and are of clinical concern. Most of the structural genes have been found in association with mobile genetic elements, such as broad- and narrow-host-range plasmids (e.g., IncL/M, IncA/C and IncF) [14,15] and transposons (e.g., Tn1548 with armA) [16], suggesting that both conjugation and transposition are likely responsible for the acquisition and dissemination of these genes worldwide. Furthermore, 16S-RMTase genes are frequently associated with other antibiotic resistance genes of clinical importance, such as carbapenemase, extended-spectrum β-lactamase (ESBL) and plasmid-mediated quinolone resistance (PMQR) determinants. This could compromise the main classes of antimicrobials used to treat multi-resistant Gram-negative infections [3].
16S-RMTase-producing clinical isolates from Bulgaria were first reported in 2005 [16]. The reported isolates (two Citrobacter freundii, two K. pneumoniae, and one isolate each of E. coli, Salmonella enterica, and Shigella flexneri), collected in 2003, carried armA as part of the Tn1548 transposon along with blaCTX-M-3 on IncL/M plasmids. Following this report, systematic surveillance for 16S-RMTase producers was introduced at the cancer hospital in Sofia. The results from the 2004–2005 study period showed that among 1310 Enterobacterales, 242 P. aeruginosa and 97 A. baumannii, the armA gene was identified in 20 (1.5%) Enterobacterales (7 K. pneumoniae, 3 E. coli, 3 Serratia marcescens, 3 C. freundii, 3 Enterobacter cloacae, and 1 Klebsiella oxytoca). ArmA-mediated aminoglycoside resistance was transferable by conjugation and carried by closely related IncL/M plasmids, which also carried aadA2, dfrA12, sul1, blaTEM-1, and blaCTX-M-3 genes encoding resistance to streptomycin, trimethoprim, sulfonamides, and ß-lactams, respectively. Most of the isolates were genetically different, but shared similar restriction patterns of the armA-encoding plasmids [17]. Based on these findings, the systematic surveillance continued.
In this study, we conducted a retrospective genomic surveillance study of 16S-RMTase-producing Enterobacterales at a Bulgarian hospital from 2006 to 2023, aiming to improve our understanding of the transmission dynamics of methyltransferase genes and their association with other antibiotic resistance determinants.
2. Results
2.1. Identification of 16S rRNA Methyltransferase-Producing Bacterial Isolates
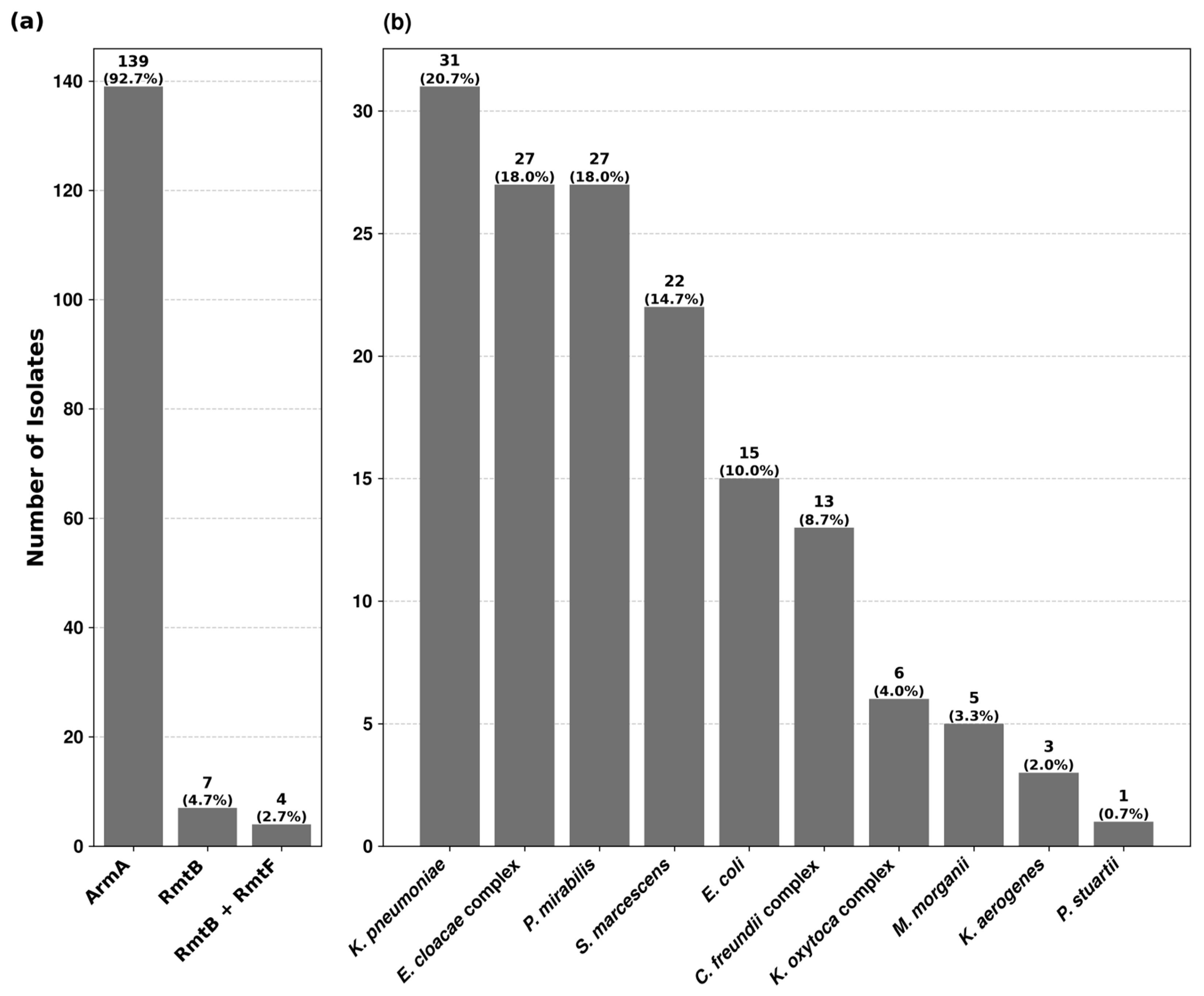
Among the 10,731 isolates, tested for high-level resistance to aminoglycosides, 150 (1.4%) showed concomitant resistance to gentamicin and amikacin with no zone of inhibition around the discs and MICs > 256 mg/L. No isolate was found with high-level resistance to apramycin. PCR analysis revealed the presence of 16S rRNA methyltransferase genes in all 150 isolates (Table S1). 16S-RMTase producers were distributed among thirteen Enterobacterales species. The prevalence rates within each species varied from 0.3% to 16.7%, being the highest in Providencia stuartii (Table S2). Three types of methyltransferase genes were identified, of which armA was the most predominant, followed by rmtB and rmtF. Their distribution among different bacterial isolates is depicted in Figure 1.
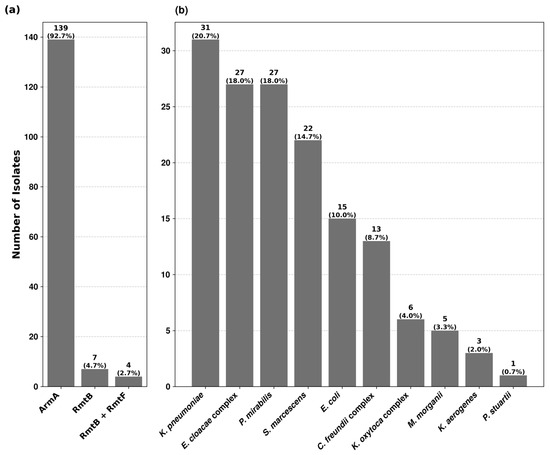
Figure 1.
Prevalence of 16S rRNA methyltransferase genes and their distribution among 16S-RMTase-producing isolates. (a) The first chart shows the prevalence rate of ArmA, RmtB and RmtF methyltransferases among the 150 bacterial isolates. (b) The second chart depicts the distribution of 16S-RMTase-producing isolates across the studied bacterial species.
The most common isolation site was urine (83%, 124/150), followed by wound exudates (13%, 20/150), respiratory samples (3%, 4/150), blood (1/150) and abdominal fluid (1/150). Further information for each bacterial species can be found in Table S1.
2.2. Identification of β-Lactamase and Plasmid-Mediated Quinolone Resistance Genes
ESBL genes were identified in 97% (145/150) of the 16S-RMTase-producing isolates, with blaCTX-M-3 (67%, 97/145) being the most common, followed by blaSHV-12 (19%, 27/145), blaCTX-M-15 (12%, 17/145) and blaSFO-1 (3%, 4/145) (Table S1, Figure 2 and Figure 3).

Figure 2.
Heatmap representation of the acquired antimicrobial resistance genes of (a) Klebsiella pneumoniae; (b) Enterobacter cloacae complex; (c) Serratia marcescens. Phylogenetic trees were constructed based on SNP analysis for (b,c) with PhaME v1.0.4, while for (a), MLVA types were used for phylogenetic tree construction according to Grissa et al. [18] (see Section 4.7). For (a) strains marked with an asterisk (*), resistance data were obtained from PCR and for the remaining strains from WGS. Drug classes are colored consistently, as shown in the legend (right). The only exception from the legend is aac(6′)-Ib-cr5, colored in pale green to indicate dual resistance. Both dendrograms and heatmaps were annotated with iTOL v6.8.1 (https://itol.embl.de/about.cgi, accessed on 20 August 2024). Sequence type (ST) was derived for the species with available MLST schemes. ND, not determined either due to missing alleles or imperfect match. KP, Klebsiella pneumoniae; EA, Enterobacter asburiae; EH, Enterobacter hormaechei; SM, Serratia marcescens.
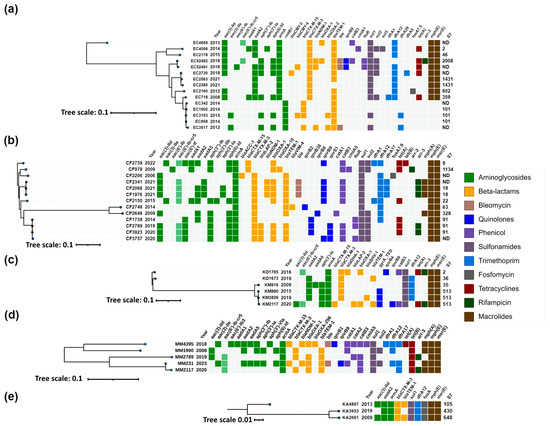
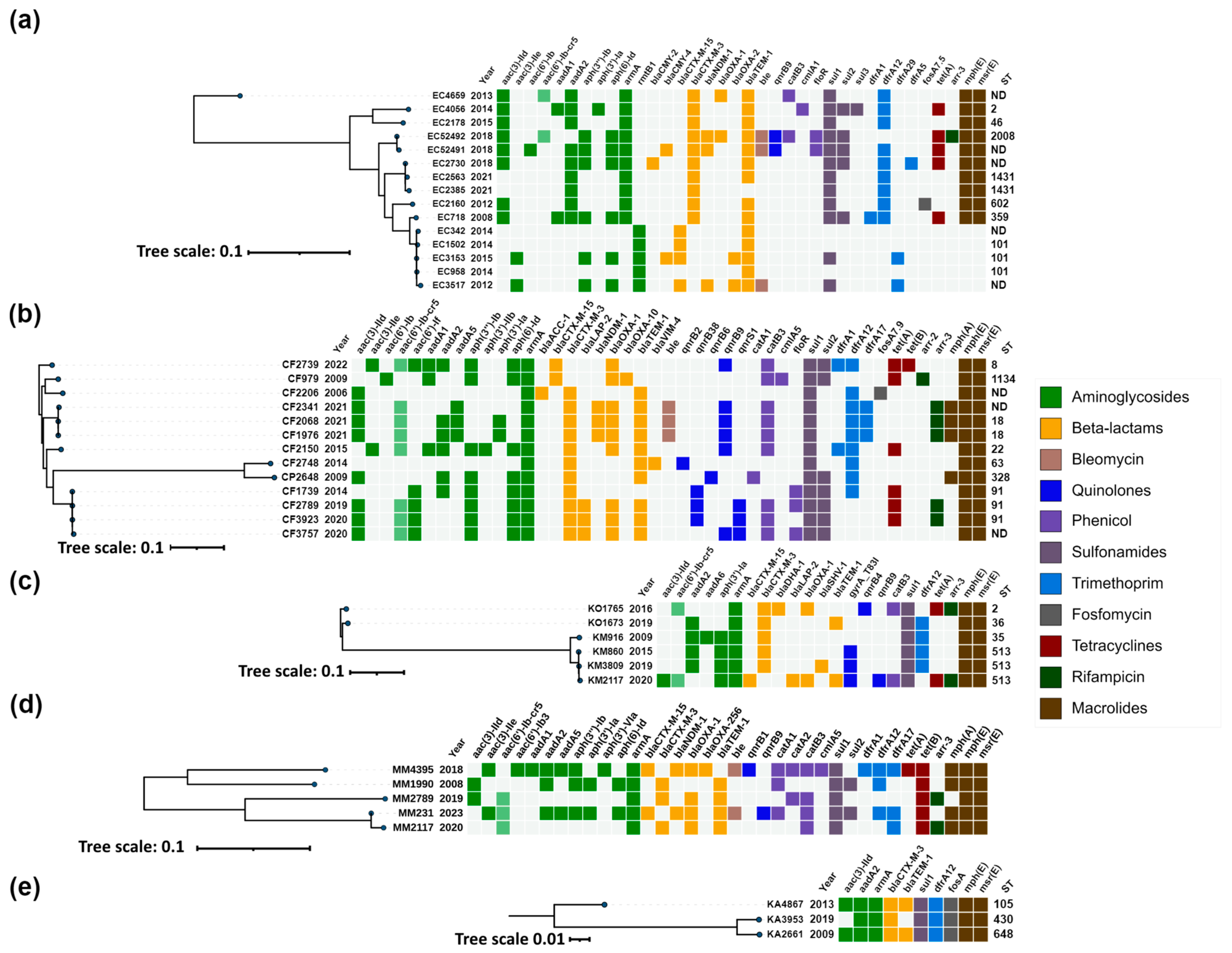
Figure 3.
Heatmap representation of the acquired antimicrobial resistance genes of (a) Escherichia coli; (b) Citrobacter freundii complex; (c) Klebsiella oxytoca complex; (d) Morganella morganii; (e) Klebsiella aerogenes. Phylogenetic trees were constructed based on SNP analysis for (a–e) with PhaME v1.0.4 (see Section 4.7). Drug classes are colored consistently, as shown in the legend (right). The only exception from the legend is aac(6′)-Ib-cr5, colored in pale green to indicate dual resistance. Both dendrograms and heatmaps were annotated with iTOL v6.8.1 (https://itol.embl.de/about.cgi, accessed on 20 August 2024). Sequence type (ST) was derived for the species with available MLST schemes. ND, not determined either due to missing alleles or imperfect match. EC, Escherichia coli; CF, Citrobacter freundii; CP, Citrobacter portucalensis; KO, Klebsiella oxytoca; KM, Klebsiella michiganensis; MM, Morganella morganii; KA, Klebsiella aerogenes.
Carbapenemase genes were identified in 33% (49/150) of the 16S-RMTase-producers. WGS analysis identified blaVIM-1/4/86 allelic variants in 63% (31/49), blaNDM-1/5 in 35% (17/49) and blaOXA-48 in a single E. hormaechei strain.
Acquired AmpC enzymes were identified in 23% (34/150) of the 16S-RMTase-producing isolates, including blaCMY-99 (n = 27), blaCMY-4 (n = 3) and blaDHA-1 (n = 2). blaCMY-2 and blaAAC-1 genes were also detected in one isolate each.
The plasmid-mediated fluoroquinolone resistance gene aac(6′)-Ib-cr was identified in 36% (54/150) of the 16S-RMTase-producing isolates of almost all species, except P. mirabilis and K. aerogenes, while qnrB variants and qnrS1 were found in 17% and 9%, respectively.
2.3. Exploring Genomic Diversity of 16S rRNA Methyltransferase-Producing Isolates
Genomic characteristics, correlation between acquired antimicrobial resistance genes, year of isolation, MLST profiles (where applicable), and phylogenetic relatedness of isolates within each species are illustrated in Figure 2 and Figure 3.
2.3.1. Klebsiella pneumoniae
K. pneumoniae isolates were distributed across six main clusters corresponding to sequence types (STs) ST15, ST147, ST6260, ST659 and ST16, in addition to MLVA type 14, for which STs remained undetermined (Figure 2a).
The ST15 cluster comprised four isolates, three from 2009 and one from 2016. All isolates carried armA, with two (KP4320 and KP305) harboring blaCTX-M-3, while the other two (KP2158 and KP510) contained blaCTX-M-15.
The ST147 cluster included five isolates, collected between 2008 and 2023, all exhibiting armA and blaCTX-M-3. Notably, one isolate, KP740, also harbored the carbapenemase gene blaNDM-1, and the quinolone resistance genes qnrS1 and aac(6′)-Ib-cr5.
K. pneumoniae ST6260 exhibited a concerning array of resistance genes. Long-read sequencing analysis revealed that rmtB1 and blaNDM-5, which were present in all isolates, were chromosomally encoded. Additionally, four isolates carried rmtF genes; two were identified as rmtF1, while the remaining two variants remained undetermined. Manual inspection of the sequences suggested that these variants are most likely also rmtF1. The rmtF genes were hosted on IncFIB(K)-type plasmids, with some of these being multireplicon plasmids that also contained IncFIB(pQil) and IncFII(pKP91) replicons. These plasmids in some isolates also harbored the blaSFO-1 and blaOXA-9 β-lactamase genes. Furthermore, the blaOXA-232 carbapenemase gene was detected in four of these isolates, situated on a small ~6141 bp plasmid of ColKP3 type. rmtF, blaSFO-1, blaOXA-9 and blaOXA-232 were successfully transferred by conjugation, and were confirmed by PCR (Table 1).

Table 1.
Characteristics of transconjugants harboring 16S rRNA methyltransferase and carbapenemase genes.
The ST659 cluster included two isolates from 2007 and 2008, both carrying armA, blaCTX-M-15, blaOXA-1, blaOXA-10 and aac(6′)-Ib-cr5, with one isolate (KP2280) additionally harboring qnrB1.
The MLVA type 14 cluster consisted of three isolates collected in 2008 (two isolates) and 2018 (one isolate), all possessing armA and blaCTX-M-3.
The ST16 cluster, with isolates collected between 2010 and 2019, consistently harbored armA, with most isolates also containing blaCTX-M-3, except for KP1339. Two isolates, KP1339 and KP826, also possessed qnrS1.
The remaining isolates outside these clusters, though unrelated, included KP6024, KP2178, KP414, KP1698, KP1724, and KP2010 and all carried armA, with most harboring blaCTX-M-3 except for KP2010, which carried blaCTX-M-15. KP6024 also carried blaOXA-9.
Transfer of high-level aminoglycoside resistance was achieved from 24/25 K. pneumoniae clinical strains. The armA-positive transconjugants always carried blaCTX-M-3 or blaCTX-M-15 and to a lesser extent, blaOXA-1, blaOXA-10, aac(6′)-Ib-cr, and qnrB1 (Table 2). PCR-based replicon typing (PBRP) showed that armA was mainly carried by IncL/M plasmids. However, in a transconjugant obtained from KP2158, co-carriage of armA and blaCTX-M-15 on an IncR plasmid was detected, while three plasmids harboring armA and blaCTX-M-3, and three others with armA and blaCTX-M-15 could not be typed with the PBRT method.

Table 2.
Characteristics of transconjugants harboring 16S rRNA methyltransferase genes.
2.3.2. Enterobacter cloacae Complex
Most of the isolates were identified as Enterobacter hormaechei, except EA146, which was characterized as Enterobacter asburiae. The presence of armA and blaCTX-M genes was detected across all isolates (Table S1). PCR and sequencing identified CTX-M-3 ESBL in 25/27 isolates. blaOXA-1-like, and quinolone resistance genes aac(6′)-Ib-cr and qnr were detected in 15/27, 16/27 and 13/27 ArmA-producers, respectively. The only carbapenemase, OXA-48-like, was found in the E. hormaechei strain EH3113. BOX-PCR identified eight genotypes (Table S1). Based on PCR results and molecular typing, thirteen isolates were selected for WGS. These included EH3113 (blaOXA-48-like-positive), EH4629 (blaCTX-M-3-negative), ten E. hormaechei isolates with different genetic configurations and genotypes, and the E. asburiae strain EA146.
E. asburiae 146, collected in 2006, carried armA along with blaCTX-M-3, aadA2, dfrA12 and sul1 genes (Figure 2b), similar to the genetic context of armA on the IncL/M pCTX-M3 [19]. Among the E. hormaechei isolates, subjected to WGS analysis, four isolates from the period 2012–2022 formed a cluster corresponding to ST78. The earliest isolate from this cluster (EH4655) carried qnrB4, while subsequent isolates harbored qnrS1. In addition, EH4655 had an acquired blaDHA-1 AmpC gene. All isolates in this cluster also carried blaOXA-1 and aac(6′)-Ib-cr5 along with armA and blaCTXM-3 genes.
Another cluster, covering the years 2016 to 2019 and corresponding to ST200, consisted of five isolates. Within this group, three isolates were identified with blaCTX-M-3, and two with blaCTX-M-15. The presence of the carbapenemase gene, blaOXA-48, was also noted in EH3113. Four isolates from this cluster carried the qnrB1 gene, with one additionally harboring qnrS1.
The remaining three isolates were associated with different sequence types (ST90, ST190, and ST544), and exhibited a similar acquired antimicrobial resistance gene profile with the presence of armA and blaCTX-M-3. Among these, isolates EH5493 and EH1813 were noted for carrying qnrS1 and qnrB1, respectively.
armA-positive transconjugants were obtained from 26/27 clinical isolates. In EH3113, co-transfer of armA with blaOXA-48, blaOXA-1, aadA2, qnrB1 and aac(6′)-Ib-cr on IncL/M plasmids was observed (Table 1). In the remaining transconjugants, armA was consistently located on CTX-M-3 IncL/M plasmids, which were often associated with additional resistance genes such as blaOXA-1 and aac(6′)-Ib-cr (Table 2).
2.3.3. Serratia marcescens
PCR analysis detected the presence of armA and blaCTX-M genes in all isolates (Table S1). In addition, blaVIM or blaNDM were detected in 2 isolates each, blaOXA-1-like and aac(6′)-Ib-cr were identified in 10/22, and qnrB or qnrS in single strains. CTX-M-3 was confirmed in 21/22 isolates. BOX-PCR typing distinguished eight genotypes (Table S1). Based on the PCR and genotyping data, nine isolates were selected for WGS.
S. marcescens isolates, subjected to WGS analysis, were distributed across three sequence types: ST92, ST470, and ST891, with each isolate carrying armA and blaCTX-M genes (Figure 2c). The ST92 group included a single isolate from 2018 that exhibited a diverse array of resistance genes, including armA, blaCTX-M-3, blaNDM-1, and qnrB9. Isolates in the ST470 group, collected between 2010 and 2014, uniformly displayed the presence of armA and blaCTX-M-3. In the ST891 cluster, all isolates harbored armA, with most also carrying blaCTX-M-3; this cluster included SM3560 with CTX-M-15 and two other isolates with VIM-4 carbapenemase. Additionally, a single isolate with an undetermined ST, collected in 2019, was found to harbor armA, blaCTX-M-3, blaNDM-1, and qnrS1.
armA was transferred by conjugation from all donor strains and was carried by IncL/M plasmids along with blaCTX-M-3 or blaCTX-M-15, and in a single case, with blaOXA-1 and aac(6′)-Ib-cr (Table 2). In a transconjugant, obtained from SM5327, co-carriage of armA and blaCTX-M-3 on IncA/C plasmids was observed. Carbapenemase genes were not co-transferred with armA, except for SM4949, whose transconjugant harbored armA with blaCTX-M-3, blaNDM-1, aadA2 and qnrB9 on an IncL/M plasmid (Table 1).
2.3.4. Escherichia coli
Two main resistance genes profiles were determined for E.coli isolates, including one with armA and blaCTX-M-3, and one with rmtB and blaCTX-M-15 (Figure 3a). The first profile was presented by two clusters corresponding to ST2008 and ST1431, as well as three unrelated isolates collected between 2013 and 2015, and two additional isolates from 2012 and 2008 with ST602 and ST359, respectively. The isolates from the ST2008 cluster were collected in 2018 and were associated with the presence of blaNDM-1 and qnrB9. Additionally, one of the isolates (EC52491) also carried blaCMY-4, while the other isolate (EC52492) carried blaOXA-1 and aac(6′)-Ib-cr5. The isolates from the ST1431 cluster harbored the classical configuration of armA and blaCTX-M-3, and only one of these isolates (EC2730) from 2018 also contained blaCMY-2. The unrelated isolates collected between 2008 and 2015 from the first profile shared the same configuration of armA and blaCTX-M-3, with one of these isolates (EC4659) from 2013 additionally harboring blaOXA-1 and aac(6′)-Ib-cr5.
The second profile was presented with one cluster corresponding to ST101 and comprised isolates collected between 2012 and 2015. All isolates harbored rmtB1, and all except one (EC958) had blaCTX-M-15 genes. In addition, one isolate (EC3517) harbored the carbapenemase gene blaNDM-1, while another isolate from 2015 (EC3153) carried the blaCMY-4 AmpC gene.
All isolates harboring ArmA and CTX-M-3 yielded transconjugants, which were positive for armA and blaCTX-M-3 on IncL/M plasmids (Table 2). Co-transfer of armA with blaCTX-M-3, blaNDM-1, blaCMY-4, and qnrB9 on IncL/M plasmids was observed in EC52491 (Table 1). In contrast, two transconjugants were obtained from EC52492, including one harboring armA, blaCTX-M-3, blaOXA-1 and aac(6′)-Ib-cr on IncL/M plasmids, and another carrying blaNDM-1 and qnrB9 on IncA/C plasmid.
Conjugal transfer of rmtB1 from E. coli donor strains could not be achieved despite repeated attempts. However, WGS analysis of donors’ plasmid replicons and in silico plasmid analysis showed that in EC3517, EC342 and EC3153, the rmtB1 gene was located on IncFII, IncA/C and IncFII plasmids, respectively. In EC958 and EC1502, the plasmid location of rmtB1 genes was suspected, but could not be proven by the only short-read sequencing data available.
2.3.5. Citrobacter freundii Complex
The isolates of the C. freundii complex, including C. freundii strains and single isolate of Citrobacter portucalensis, were characterized by the consistent presence of armA and blaCTX-M genes (Figure 3b). They exhibited diverse sequence types, with two clusters being clearly distinguished: one corresponding to ST18 and the other to ST91. Outside of these clusters, the isolates displayed unique STs, indicating a lack of relatedness.
The ST18 cluster comprised isolates collected in 2021, all of which were positive for armA, blaCTX-M-3, and also harbored the carbapenemase gene blaNDM-1 along with qnrB9 and aac(6′)-Ib-cr5 quinolone resistance genes. The ST91 cluster also exhibited the ArmA/CTX-M-3 configuration, accompanied by various qnr genes. One isolate from this cluster, CF1739 (2014), carried only qnrB38. Other isolates within this cluster included CF2789 and CF3923, which harbored qnrB38 in combination with qnrS1, and CF3757, which carried both qnrB9 and qnrS1 in addition to aac(6′)-Ib-cr5.
Two isolates, CF2739 (2022) and CF979 (2009), were characterized by the presence of blaCTX-M-15 instead of blaCTX-M-3, with CF2739 also harboring qnrB9 and aac(6′)-Ib-cr5. The earliest isolate in this panel, CF2206 (2006), possessed armA, blaCTX-M-3, and blaACC-1. Other notable isolates included CF2150, CF2748, and CP2648, all of which had the ArmA/CTX-M-3 profile. CF2150 additionally harbored qnrB9 and aac(6′)-Ib-cr5, while CP2648 had qnrB6. CF2748 was distinguished by the production of VIM-4 metallo-β-lactamase and qnrB2.
armA was successfully transferred from all donors, yielding transconjugants with varied genetic contents, but always positive for blaCTX-M-3 or blaCTX-M-15, and blaNDM-1 or blaVIM-4 when present in the donors (Table 1 and Table 2). PBRT confirmed that armA was located on IncL/M plasmids in all transconjugants, except one obtained from CF2739, in which co-carriage of armA and blaCTX-M-15 on the IncFIB plasmid was observed (Table 2).
2.3.6. Klebsiella oxytoca Complex
The isolates of the K. oxytoca complex were predominantly identified as Klebsiella michiganensis, with the exception of two isolates that were classified as K. oxytoca. All isolates within this complex harbored armA and blaCTX-M genes (Figure 3c). The two K. oxytoca isolates were characterized by the ArmA/CTX-M-3 configuration and one of them, KO1765, also carried blaDHA-1, qnrB4 and aac(6′)-Ib-cr5 genes.
The K. michiganensis group included one isolate from 2009, identified as ST35, and three isolates collected between 2015 and 2020, which shared the sequence type ST513 and formed a distinct cluster. Most K. michiganensis isolates exhibited the ArmA/CTX-M-3 profile. An exception was the isolate KM2117, which harbored blaCTX-M-15 instead of blaCTX-M-3, and also carried qnrB9 and aac(6′)-Ib-cr5 quinolone resistance genes.
Thansconjugants carrying armA were obtained from all clinical isolates. The armA gene was consistently located on IncL/M plasmids, which also carried blaCTX-M-3 or blaCTX-M-15, and variably aadA2, blaOXA-1 and aac(6′)-Ib-cr genes (Table 2).
2.3.7. Morganella morganii
M. morganii isolates, though unrelated, consistently harbored armA and blaCTX-M genes (Figure 3d). The majority carried blaCTX-M-3, with the exception of two isolates, MM4395 and MM231, which exhibited blaCTX-M-15. Additionally, MM4395 and MM231 were notable for also having the NDM-1 carbapenemase and qnrB genes, specifically qnrB1 in MM4395 and qnrB9 in MM231, along with aac(6′)-Ib-cr5.
ArmA was transferred in all but one isolate. In transconjugants from MM1990, MM2789 and MM2117, armA was carried by CTX-M-3 IncL/M plasmids with various associated resistance genes (Table 2). In MM4395, a co-transfer of armA with blaNDM-1, blaOXA-1, qnrB1 and aadA2 on IncT plasmids was observed (Table 1). In contrast, no plasmid replicons were detected in MM231, and it failed to transfer any resistance determinants by conjugation, suggesting the chromosomal location of armA and associated resistance genes.
2.3.8. Klebsiella aerogenes
K. aerogenes isolates were distributed across three distinct sequence types—ST105, ST430, and ST648—all of which possessed the armA, blaCTX-M-3, aadA2, dfrA12, and sul1 genes typical for the genetic context of armA on the IncL/M pCTX-M3 plasmid [19], and without other resistance genes (Figure 3e).
Conjugal transfer of ArmA was achieved from all donor strains. armA was always carried by IncL/M plasmids along with blaCTX-M-3 and aadA2 genes (Table 2).
2.3.9. Proteus mirabilis
Molecular typing was performed sequentially on 27 P. mirabilis isolates collected between 2007 and 2021 (Table S1). First, the isolates were identified as belonging to the Pm-1 “Dienes strain” as judged by the Dienes test. PFGE typing of selected isolates from each year showed the presence of identical patterns, suggesting that all isolates belonged to a single clone, which had already been found for ten of the isolates described previously. [20]. PCR assays detected the presence of armA and identical co-harboring genes in all isolates (Table S1). WGS analysis of the representative strain PM1502, collected in 2014, revealed several resistance genes including armA, aac(6′)-Ib, aadA1, aadA2, aph(3″)-Ib, aph(6)-Id (aminoglycoside resistance), blaVIM-1, blaCMY-99, blaSHV-12, blaOXA-9, blaTEM-1 (β-lactam resistance), dfrA1, dfrA12 (trimethoprim resistance), and sul1, sul2 (sulfonamide resistance).
Conjugal transfer of armA from clinical isolates of P. mirabilis could not be achieved and no plasmid replicons were detected in PM1502, suggesting chromosomal location of armA and associated resistance genes. However, this was not investigated further as PM1502 had only short-read sequencing data available.
2.3.10. Providentia stuartii
The only ArmA-producing P. stuartii strain, PS3347, was identified in 2020 (Table S1). This strain was previously described in our study on carbapenem-resistant AmpC producers [21]. PS3347 harbored a diverse array of resistance genes located on an IncA/C conjugative plasmid, including armA, blaVIM-86, blaCMY-4, blaOXA-1, and aac(6′)-lb-cr5 (Table 1).
2.4. Antimicrobial Susceptibility of 16S rRNA Methyltransferase-Producing Isolates
The results of susceptibility testing for each bacterial isolate, along with the resistance genes detected, are shown in Table S4. All isolates were concomitantly highly resistant to amikacin and gentamicin, consistent with the presence of 16S rRNA methyltransferase genes. In addition, almost all isolates were resistant to cephalosporins and most isolates to fluoroquinolones, due to ESBLs with or without carbapenemases and quinolone resistance determinants, respectively. All but two isolates (PS3347 and KP3112) were susceptible to cefiderocol, whereas resistance to ceftazidime–avibactam, imipenem–relebactam and meropenem–vaborbactam was associated with the presence of methalo-β-lactamase genes.
3. Discussion
This retrospective genomic surveillance study, conducted between 2006 and 2023 in a single hospital, is the first systematic evaluation of 16S rRNA methyltransferase-producing Enterobacterales in Bulgaria. The period prevalence was 1.4% (150/10,731), which is consistent with a previous study conducted at the same hospital that identified methyltransferase genes in 1.5% (20/1310) of enterobacterial isolates collected in 2004–2005 [17]. Our results, which show persistence of 16S-RMTase-mediated resistance despite infection control measures [22], suggest that the dissemination of methyltransferase genes was likely due to horizontal transmission rather than clonal spread. The prevalence comparison with analogous studies showed that Bulgaria has a lower prevalence rate than India [23] and Iran [24] where rates of 46.3% (57/123) and 13.0% (40/307) were reported in consecutive isolates, although the small number of isolates may not reflect the true prevalence rate. A similar prevalence rate of 1.1% (20/1770) was observed in Poland [25], whereas in the UK, 0.1% of the 56,172 Enterobacterales isolates collected in 2016 harbored methyltransferase genes [26].
We characterized three types of 16S rRNA methyltransferases genes. Among them, armA was the predominant type (92.7%), whereas rmtB1 (4.7%) and rmtF1 + rmtB1 (2.7%) were only infrequently identified. armA was widely distributed in Enterobacterales, comprising thirteen species, while other methyltransferase genes were more species-specific, such as rmtF, which was found only in K. pneumoniae, and rmtB in E. coli and K. pneumoniae. During the study period, further spread of ArmA involving isolates of P. mirabilis, M. morganii, P. stuartii and K. aerogenes was observed. Furthermore, isolates with rmtF and rmtB genes were identified in our hospital, with rmtF being reported for the first time in Bulgaria. Our results confirm the researchers’ findings that ArmA and RmtB are the two most frequently reported 16S RMTases, and at the same time reveal the specificity of their distribution within the hospital [3,14].
16S rRNA methyltransferases were often reported to coexist with β-lactamase genes. A high percentage of 16S-RMTase-producing Enterobacterales, in our study, harbored ESBL genes (97%), with blaCTX-M-3 being the most common, which is in accordance with our previous studies [16,17]. In addition, during the study period, associations of blaCTX-M-15 with armA or rmtB in many species, and blaSFO-1 with rmtF and rmtB in K. pneumoniae, were identified using WGS analysis. Although associations of blaCTX-M-15 with armA and rmtB have been reported previously [27,28], as well as one case of E. coli possessing blaSFO-1 with rmtB [29], the coexistence of blaSFO-1 with rmtF, as far as we are aware, has not been reported to date. One-third of the 16S RMTase-producing Enterobacterales were also found to carry carbapenemase genes, with blaVIM and blaNDM being dominant. The carbapenemases NDM-5 and OXA-232 were found only in isolates of K. pneumoniae ST6260. VIM-1 was detected exclusively in isolates belonging to the P. mirabilis clone, while NDM-1 was widely distributed among different species. Half of these isolates belonged to the Enterobacter spp., S. marcescens, C. freundii, Providencia spp., and M. morganii (ESCPM) group and were previously described in our study on carbapenemase-producing enterobacteria with natural AmpC enzymes [21]. These results are in agreement with globally reported findings showing that 16S RMTase genes are commonly associated with carbapenemase genes [23,30,31]. Other antibiotic resistance genes frequently associated with 16S-RMTase genes in this study were the plasmid-mediated quinolone resistance determinants aac(6′)-Ib-cr (36%) and qnr (26%), whose co-existence with carbapenemase, ESBL, armA and rmtB has already been reported [27]. These associations lead to co-selection of antibiotic resistance genes by numerous antimicrobials and compromise the clinical use of the three main groups of bactericidal antibiotics for the treatment of life-threatening bacterial infections.
In this study, 16S rRNA methyltransferases were found to be associated with broad-host-range plasmids of diverse incompatibility groups, reflecting multiple sources of acquisition through horizontal gene transfer. In total, 116 isolates (77%) were found to carry the methyltransferase gene on a plasmid. Of these, one hundred and nine isolates of thirteen bacterial species harbored armA on at least five plasmid types, with IncL/M being the dominant plasmid type found. The majority of IncL/M plasmids carrying armA also carry blaCTX-M-3, similar to the IncL/M plasmid pCTX-M3 in C. freundii [19] and pIP1204 in K. pneumoniae [5], on which the armA gene was initially identified. While neither carbapenemase nor plasmid-mediated quinolone resistance genes were detected in our previous studies [16,17], in this study, various carbapenemase (blaNDM-1, blaVIM-4, blaVIM-86, blaOXA-48), PMQR (aac(6′)-Ib-cr, qnrS, qnrB), and other β-lactamase (blaCMY-4, blaOXA-1, blaOXA-10) genes were found on IncL/M plasmids co-transferred with armA. The blaCTX-M-15 gene instead of blaCTX-M-3 was also detected in armA-carrying IncL/M plasmids from some isolates. Furthermore, armA was found on IncA/C, IncR, IncFIB and IncT self-transmissible plasmids co-transferred with either blaCTX-M-15/blaCTX-M-3 or blaVIM-86/blaNDM-1 and other resistance genes.
armA was frequently co-located with blaNDM-1 or other carbapenemase genes (especially blaVIM-86, blaVIM-4 and blaOXA-48) on the same plasmids, with some of them additionally carrying genes for fluoroquinolone resistance, such as aac(6′)-Ib-cr and qnrB. The acquisition of these multidrug-resistant plasmids would have led to simultaneous resistance to most β-lactams, including carbapenems, as well as aminoglycosides and fluoroquinolones, the three major groups of antimicrobial agents active against Gram-negative bacteria.
A few high-risk clones were found to be associated with the ArmA gene, including K. pneumoniae ST147, C. freundii ST18 and a P. mirabilis clone endemic to Bulgaria, all of which have been reported to carry carbapenemase genes, such as blaNDM-1 in K. pneumoniae ST147 [32] and C. freundii ST18 [33], and blaVIM-1 in the P. mirabilis clone [34]. Besides these clones, an abundance of minor STs in ArmA-producing Enterobacterales was found in this study. On the basis of the clonal diversity observed by MLST and MLVA, it can be assumed that the persistence of methyltransferase genes in our hospital is driven by plasmid transmission rather than clonal spread.
rmtB was initially identified on a plasmid in association with blaTEM-1 in a clinical isolate of S. marcescens from Japan [6]. In this study, rmtB1 was identified in five E. coli strains of the high-risk ST101 clone, which emerged as sporadic cases during a limited period (2012–2015). rmtB was found on non-conjugative IncFII and IncA/C plasmids in 3/5 isolates, which could explain the lack of further dissemination. The only blaNDM-1-positive strain, EC3517, appeared to be similar to the reported outbreak strain in another Bulgarian hospital in 2012 as both ST101 strains harbored blaCTX-M-15 in addition to blaNDM-1 and rmtB. However, while we identified rmtB on a non-conjugative IncFII plasmid, the transconjugants obtained from the outbreak strains were found to carry rmtB with blaNDM-1 on an untypable plasmid [35].
The most important finding of this study was the identification of K. pneumoniae ST6260 in our hospital, which was found to emerge in 2022 (Table S1). Long-read sequencing revealed a novel resistance association of RmtF1 and RmtB1 methyltransferases, NDM-5 and OXA-232 carbapenemases, and SFO-1 ESBL. The rmtB1 and blaNDM-5 were chromosomally located in all isolates. The rmtF1 was hosted on conjugative IncFIB(K) plasmids, with some of these being multireplicon plasmids that also contained IncFIB(pQil) and IncFII(pKP91) replicons. The blaOXA-232 carbapenemase gene was situated on a small ~6141 bp plasmid of ColKP3 type that could be transferred by conjugation. The blaSFO-1 ESBL gene was plasmid-born and co-transferred with rmtF1. In contrast to RmtB1 and NDM-5, plasmid-encoded RmtF1, OXA-232 and SFO-1 were variously present in some of the six investigated isolates. Based on the patient metadata (Table S1), which indicates that the first isolate of K. pneumoniae ST6260, KP3746, originated from an outpatient and the reported nosocomial spread of K. pneumoniae ST6260 in another Bulgarian hospital [36], it can be assumed that K. pneumoniae ST6260 is already spreading between hospitals in Bulgaria. Therefore, a national genome-wide survey is currently underway to determine the extent of the emerging clonal spread of K. pneumoniae ST6260 and to prevent its further expansion.
4. Materials and Methods
4.1. Bacterial Isolates, Identification and Susceptibility Testing
From January 2006 to October 2023, a total of 10,731 consecutive, nonduplicate enterobacterial isolates were collected from patients’ specimens in a 252-bed oncology hospital in Sofia, Bulgaria. Clinical specimens originated from infected or colonized patients admitted to hospital wards or attending outpatient departments. Bacterial isolates were initially identified by using GNI cards on the VITEK®2 system (bioMérieux, Marcy l’Étoile, France). The selected high-level aminoglycoside-resistant isolates were re-identified by MALDI-TOF Biotyper (Bruker Daltonics GmbH, Bremen, Germany) with MALDI Reference 2022 Library v.4.0.
Antimicrobial susceptibility testing was carried out by broth microdilution using the MicroScan NM-EN52 panel (Beckman Coulter, Inc., Brea, CA, USA) by following the manufacturer’s protocol. Susceptibility to cefiderocol, ceftazidime–avibactam, imipenem–relebactam, and meropenem–vaborbactam was determined by the disk diffusion method with MASTDISCS® (Mast group, Merseyside, UK). Susceptibility testing results were interpreted in accordance with EUCAST clinical breakpoints v13.0. E. coli ATCC 25922 was used for quality control.
4.2. Screening of Aminoglycoside-Resistant Isolates and 16S rRNA Methytransferase Gene Detection
All isolates were screened for high-level resistance to amikacin, gentamicin and apramycin by the Kirby–Bauer disc diffusion method. Isolates that exhibited no zone of inhibition around 15 µg apramycin discs (Oxoid, Basingstoke, UK) or 30 µg amikacin and 10 µg gentamicin discs (BD, Sparks, MD, USA) were further confirmed by gradient strip MIC (bioMérieux, Marcy l’Étoile, France) [2]. When they showed an MIC > 256 mg/L to amikacin and gentamicin, indicating N7-G1405 methyltransferase production, or MIC > 256 mg/L to apramycin, indicating the N1-A1408 enzyme, a multiplex PCR to detect armA, rmtA-F and npmA [37,38,39] was performed as described in Table S5.
4.3. Detection of β-Lactamase and Plasmid-Mediated Quinolone Resistance Genes
16S rRNA methyltransferases-positive isolates were further examined for associated β-lactamase and plasmid-mediated quinolone resistance genes. Class A, class B, and class D carbapenemase genes were detected with primer pairs from prior publications [40,41,42,43,44] used in previously established multiplex PCR [45], with the PCR conditions detailed in Supplementary Table S6. blaCTX-M genes were detected as described previously [46]. For further differentiation of the allelic variants, the amplicons were purified using the Agencourt AMPure XP beads (Beckman Coulter, Fullerton, CA, USA) and sequenced with the GeXP Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA). blaOXA-1/2/9/10-like genes [47] were screened with a protocol listed in Supplementary Table S7. Plasmid-mediated AmpC β-lactamase genes were detected as previously published [48]. PMQR determinants (qnrA, qnrB, qnrC, qnrD, qnrS, qepA) [49,50,51,52] were detected as described in Supplementary Table S8. Identification of the aac(6′)-Ib-cr acetyltransferase gene was performed as previously reported [53].
4.4. Conjugation Experiments and PCR-Based Replicon Typing
Transferability of 16S rRNA methyltransferase and carbapenemase genes was examined by mating on filters with the sodium azide-resistant E. coli J53 recipient strain as previously described [54]. Transconjugants were selected on Luria–Bertani agar containing sodium azide (100 mg/L) supplemented with either amikacin (50 mg/L) and gentamicin (50 mg/L) or 0.5 mg/L meropenem. Transconjugants were confirmed by susceptibility testing and PCR assays for methytransferase and co-transferred resistance genes. In addition, a singleplex PCR for the detection of aadA2 was performed using previously published primers [55] with a protocol described in Supplementary Table S9.
Incompatibility typing of the plasmids carrying 16S rRNA methyltransferases genes was performed by PCR-based replicon typing (PBRT), involving a combination of multiplex and singleplex PCR panels targeting 21 plasmid replicons as detailed in a previous study [56].
All PCR assays were carried on a Gentier E96 Real-time PCR Sysetem (Xi’an Science and Technology Co., Ltd., Xi’an, China). All amplicons were visualized on a QIAxcel Advanced high-resolution capillary electrophoresis system (Qiagen, Hilden, Germany).
4.5. Typing of the 16S rRNA Methytransferase-Producing Isolates
16S rRNA methytransferase-positive isolates of K. pneumoniae, P. mirabilis, E. cloacae complex and S. marcescens, which were selectively sequenced, were subjected to molecular typing to delineate the clonality of the different isolates. K. pneumoniae isolates were typed using multi-locus variable number of tandem repeats analysis (MLVA8+) [57]. P. mirabilis isolates were typed using the Dienes test [58,59] and the unique strains were further characterized by pulsed field gel electrophoresis (PFGE) using the restriction enzymes ApaI [20]. The E. cloacae complex and S. marcescens isolates were typed by using the BOX-PCR fingerprinting technique, as detailed in a previous study [60].
4.6. Whole-Genome Sequencing
Genomic DNA extraction for PCR and whole-genome sequencing was performed using the PureLink™ Genomic DNA Mini Kit (Thermo Fisher Scientific, Missouri, TX, USA) adhering to the manufacturer’s guidelines, with all homogenization steps carried out by pipetting.
In total, 58% (87/150) of the 16S-RMTase-producing Enterobacterales were subjected to whole-genome sequencing. This included all isolates of E. coli (n = 15), the C. freundii complex (n = 13), the K. oxytoca complex (n = 6), M. morganii (n = 5), K. aerogenes (n = 3), and P. stuartii (n = 1). Additionally, selected isolates of K. pneumoniae (21/31), E. cloacae complex (13/27), S. marcescens (9/27), and one representative strain of the P. mirabilis clone were included. Short-read sequencing was performed using an Illumina DNA Prep kit (Illumina, San Diego, CA, USA) on a MiSeq V3 (2 × 300 bp) or NextSeq 550 with V2.5 (2 × 150 bp) mid-output flow cell (Illumina, San Diego, CA, USA). Additionally, DNA samples from KP146, KP448, KP3112, KP3161 and KP3648 were sequenced on a MinION Mk1C using the Rapid Barcoding Kit 96 (SQK-RBK110.96) and FLO-MIN106D (R9.4.1) (Oxford Nanopore Technologies, Oxford, UK) with minor adjustments. The final library pool size selection was carried out using 0.4× SPRI magnetic particles to exclude fragments smaller than 1.5 kb [61].
4.7. Bioinformatic Analysis
The quality of raw sequencing reads was evaluated using FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc, accessed on 20 August 2024). Quality trimming and filtering for short and long reads were conducted with fastp v0.23.2 [62] and filtlong v0.2.1 (https://github.com/rrwick/Filtlong, accessed on 20 August 2024). Short-read assemblies were generated with Unicycler v0.4.8 [63]. Long-read assemblies were created with Trycycler v0.5.3 [64] and polished with MEDAKA v1.7.3 (ONT, https://github.com/nanoporetech/medaka, accessed on 20 August 2024). In silico identification was performed with rMLST [65]. Assemblies were annotated with Bakta v 1.7 [66] (database v5.0-full). Antimicrobial resistance determinants and phenotype prediction were carried out using AMRFinderPlus v3.11.4 [67] and ResFinder v4.3.1 [68] (database v2022-05-24). Plasmid analysis was conducted using PlasmidFinder v2.1 with the database v2023-01-18 [69]. MLST profiles were inferred from the assembled genomes using mlst v2.23.0 (Seemann T, mlst Github, https://github.com/tseemann/mlst, accessed on 20 August 2024).
Phylogenetic trees for all individual species, except for K. pneumoniae, were constructed using PhaME v1.0.4 [70], considering only SNPs within the coding regions of the core genome. For K. pneumoniae, as not all isolates were subjected to WGS, we used multi-locus variable number of tandem repeats analysis (MLVA) for phylogenetic purposes. It was performed according to the MLVA8+ method as previously described [57]. MLVA types were then used for phylogenetic tree construction according to Grissa et al. [18]. All resulting phylogenetic trees were linked to antimicrobial resistance patterns and visualized using iTOL v6.8.1 [71].
5. Conclusions
We conducted a retrospective genomic study of 150 consecutive 16S rRNA methyltransferase-producing Enterobacterales, collected between 2006 and 2023, to clarify the transmission dynamics of methytransferase and associated antibiotic resistance genes using PCR, WGS and conjugation experiments. We found that the armA, rmtB and rmtF methyltransferase genes were carried by different plasmid Inc types (IncL/M, IncA/C, IncR, IncT, IncFIB, and IncFII), suggesting diverse origins and sources of acquisition due to the large number and variety of host organisms. We showed that persistence of the armA gene was associated with the spread of IncL/M-type conjugative plasmids, whose broad-host range included thirteen of the species presented in this study. It was observed that armA-carrying IncL/M plasmids also harbor blaCTX-M-3 or blaCTX-M-15. Furthermore, carbapenemase (blaNDM-1, blaVIM-4, blaVIM-86, blaOXA-48), PMQR (aac(6′)-Ib-cr, qnrS, qnrB), and other β-lactamase (blaCMY-4, blaOXA-1, blaOXA-10) genes were also found on these plasmids. Long-read sequencing of ST6260 K. pneumoniae isolates revealed a novel resistance association of RmtF1 and RmtB1 methyltransferases, NDM-5 and OXA-232 carbapenemases, and SFO-1 ESBL. The rmtB1 and blaNDM-5 were found on the chromosome; blaOXA-232 was carried by conjugative plasmids of the ColKP3 type, while rmtF1 was hosted on self-transmissible IncFIB and IncFII plasmids, and was co-transferred with the blaSFO-1 ESBL gene. The genetic plasticity and adaptability of plasmids containing methyltransferase genes suggest that, by acquiring more potent resistance genes, they have the potential to minimize the continuing threat to public health from 16S rRNA methyltransferase-producing Enterobacterales.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics13100950/s1: Table S1: Properties of 16S rRNA methytransferase-producing Enterobacterales isolates collected from 2006 to 2023; Table S2: Prevalence of 16S rRNA methyltransferase producers among different Enterobacterales species; Table S3: Characteristics of transconjugants harboring 16S rRNA methyltransferase genes; Table S4: Antimicrobial susceptibility of 16S rRNA methytransferase-producing Enterobacterales isolates; Table S5: 16S rRNA methyltransferase (16S-RMTase) mPCR; Table S6: Carbapenemase mPCR; Table S7: OXA mPCR; Table S8: Plasmid-mediated quinolone resistance (PMQR) mPCR; Table S9: aadA2 PCR.
Author Contributions
Conceptualization, S.S., I.S. and I.C.; methodology I.S., S.S., I.N.I. and I.C.; software and validation I.S., I.N.I., D.D., Y.H. and S.S.; formal analysis, S.S. and I.S.; investigation, S.S., I.S. and S.G.; resources S.S., E.D. and I.N.I.; writing—original draft preparation, S.S. and I.S.; writing—review and editing, I.S., S.G., I.N.I., D.D., Y.H., E.D., I.C. and S.S.; visualization, I.S., D.D. and S.S.; funding acquisition, S.S. and I.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Regional Development Fund through the Operational Program Science and Education for Smart Growth 2014–2020 under Grant number BG05M2OP001-1.002-0001-C04 “Fundamental Translational and Clinical Research in Infection and Immunity”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated during this study are included in this article and its Supplementary Materials. Relevant links and references to additional sources are provided in the text. Draft genome sequences have been uploaded to the National Center for Biotechnology Information (NCBI) database and are accessible under BioProject IDs PRJNA1077246 and PRJNA1132711. The complete genome sequences of the ST6260 K. pneumoniae isolates (KP146, KP448, KP3112, KP3161, and KP3648) are available under BioProject ID PRJEB70839.
Conflicts of Interest
The authors declare no conflicts of interests.
References
- Magnet, S.; Blanchard, J.S. Molecular Insights into Aminoglycoside Action and Resistance. Chem. Rev. 2005, 105, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.I.; Doi, Y.; Arakawa, Y. Aminoglycoside Resistance: Updates with a Focus on Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2020, 34, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Wachino, J.-I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
- Yokoyama, K.; Doi, Y.; Yamane, K.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Yagi, T.; Kato, H.; Arakawa, Y. Acquisition of 16S RRNA Methylase Gene in Pseudomonas aeruginosa. Lancet 2003, 362, 1888–1893. [Google Scholar] [CrossRef] [PubMed]
- Galimand, M.; Courvalin, P.; Lambert, T. Plasmid-Mediated High-Level Resistance to Aminoglycosides in Enterobacteriaceae Due to 16S RRNA Methylation. Antimicrob. Agents Chemother. 2003, 47, 2565–2571. [Google Scholar] [CrossRef]
- Doi, Y.; Yokoyama, K.; Yamane, K.; Wachino, J.I.; Shibata, N.; Yagi, T.; Shibayama, K.; Kato, H.; Arakawa, Y. Plasmid-Mediated 16S RRNA Methylase in Serratia marcescens Conferring High-Level Resistance to Aminoglycosides. Antimicrob. Agents Chemother. 2004, 48, 491–496. [Google Scholar] [CrossRef]
- Wachino, J.I.; Yamane, K.; Shibayama, K.; Kurokawa, H.; Shibata, N.; Suzuki, S.; Doi, Y.; Kimura, K.; Ike, Y.; Arakawa, Y. Novel Plasmid-Mediated 16S RRNA Methylase, RmtC, Found in a Proteus mirabilis Isolate Demonstrating Extraordinary High-Level Resistance against Various Aminoglycosides. Antimicrob. Agents Chemother. 2006, 50, 178–184. [Google Scholar] [CrossRef]
- Doi, Y.; De Oliveira Garcia, D.; Adams, J.; Paterson, D.L. Coproduction of Novel 16S RRNA Methylase RmtD and Metallo-β-Lactamase SPM-1 in a Panresistant Pseudomonas aeruginosa Isolate from Brazil. Antimicrob. Agents Chemother. 2007, 51, 852–856. [Google Scholar] [CrossRef]
- Lee, C.S.; Hu, F.; Rivera, J.I.; Doi, Y. Escherichia coli Sequence Type 354 Coproducing CMY-2 Cephalosporinase and RmtE 16S RRNA Methyltransferase. Antimicrob. Agents Chemother. 2014, 58, 4246–4247. [Google Scholar] [CrossRef]
- Galimand, M.; Courvalin, P.; Lambert, T. RmtF, a New Member of the Aminoglycoside Resistance 16S RRNA N7 G1405 Methyltransferase Family. Antimicrob. Agents Chemother. 2012, 56, 3960–3962. [Google Scholar] [CrossRef]
- Bueno, M.F.C.; Francisco, G.R.; O’Hara, J.A.; De Oliveira Garcia, D.; Doi, Y. Coproduction of 16S RRNA Methyltransferase RmtD or RmtG with KPC-2 and CTX-M Group Extended-Spectrum β-Lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013, 57, 2397–2400. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, J.A.; McGann, P.; Snesrud, E.C.; Clifford, R.J.; Waterman, P.E.; Lesho, E.P.; Doi, Y. Novel 16S RRNA Methyltransferase RmtH Produced by Klebsiella pneumoniae Associated with War-Related Trauma. Antimicrob. Agents Chemother. 2013, 57, 2413–2416. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.I.; Shibayama, K.; Kurokawa, H.; Kimura, K.; Yamane, K.; Suzuki, S.; Shibata, N.; Ike, Y.; Arakawa, Y. Novel Plasmid-Mediated 16S RRNA M1A1408 Methyltransferase, NpmA, Found in a Clinically Isolated Escherichia coli Strain Resistant to Structurally Diverse Aminoglycosides. Antimicrob. Agents Chemother. 2007, 51, 4401–4409. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hu, F. Research Updates of Plasmid-Mediated Aminoglycoside Resistance 16S RRNA Methyltransferase. Antibiotics 2022, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.I.; Arakawa, Y. Exogenously Acquired 16S RRNA Methyltransferases Found in Aminoglycoside-Resistant Pathogenic Gram-Negative Bacteria: An Update. Drug Resist. Updat. 2012, 15, 133–148. [Google Scholar] [CrossRef]
- Galimand, M.; Sabtcheva, S.; Courvalin, P.; Lambert, T. Worldwide Disseminated armA Aminoglycoside Resistance Methylase Gene Is Borne by Composite Transposon Tn1548. Antimicrob. Agents Chemother. 2005, 49, 2949–2953. [Google Scholar] [CrossRef]
- Sabtcheva, S.; Saga, T.; Kantardjiev, T.; Ivanova, M.; Ishii, Y.; Kaku, M. Nosocomial Spread of armA-Mediated High-Level Aminoglycoside Resistance in Enterobacteriaceae Isolates Producing CTX-M-3 β-Lactamase in a Cancer Hospital in Bulgaria. J. Chemother. 2008, 20, 593–599. [Google Scholar] [CrossRef]
- Grissa, I.; Bouchon, P.; Pourcel, C.; Vergnaud, G. On-Line Resources for Bacterial Micro-Evolution Studies Using MLVA or CRISPR Typing. Biochimie 2008, 90, 660–668. [Google Scholar] [CrossRef]
- Gołȩbiewski, M.; Kern-Zdanowicz, I.; Zienkiewicz, M.; Adamczyk, M.; Zyliǹska, J.; Baraniak, A.; Gniadkowski, M.; Bardowski, J.; Cegłowski, P. Complete Nucleotide Sequence of the PCTX-M3 Plasmid and Its Involvement in Spread of the Extended-Spectrum β-Lactamase Gene BlaCTX-M-3. Antimicrob. Agents Chemother. 2007, 51, 3789–3795. [Google Scholar] [CrossRef]
- Ivanov, I.; Sabtcheva, S.; Dobreva, E.; Todorova, B.; Velinov, T.Z.; Borissova, V.; Petrova, I.; Ivancheva, K.; Asseva, G.; Padeshki, P.; et al. Prevalence of Carbapenemase Genes among 16S RRNA Methyltransferase-Producing Enterobacteriaceae Isolated from Cancer Patients. Probl. Infect. Parasit. Dis. 2014, 42, 10–13. [Google Scholar]
- Sabtcheva, S.; Stoikov, I.; Ivanov, I.N.; Donchev, D.; Lesseva, M.; Georgieva, S.; Teneva, D.; Dobreva, E.; Christova, I. Genomic Characterization of Carbapenemase-Producing Enterobacter hormaechei, Serratia marcescens, Citrobacter freundii, Providencia stuartii, and Morganella morganii Clinical Isolates from Bulgaria. Antibiotics 2024, 13, 455. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Cataldo, M.A.; Dancer, S.J.; De Angelis, G.; Falcone, M.; Frank, U.; Kahlmeter, G.; Pan, A.; Petrosillo, N.; Rodríguez-Baño, J.; et al. ESCMID Guidelines for the Management of the Infection Control Measures to Reduce Transmission of Multidrug-Resistant Gram-Negative Bacteria in Hospitalized Patients. Clin. Microbiol. Infect. 2014, 20, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Wangkheimayum, J.; Paul, D.; Dhar, D.; Nepram, R.; Chetri, S.; Bhowmik, D.; Chakravarty, A.; Bhattacharjee, A. Occurrence of Acquired 16S RRNA Methyltransferase-Mediated Aminoglycoside Resistance in Clinical Isolates of Enterobacteriaceae within a Tertiary Referral Hospital of Northeast India. Antimicrob. Agents Chemother. 2017, 61, e01037-16. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh Sefidan, F.; Mohammadzadeh-Asl, Y.; Ghotaslou, R. High-Level Resistance to Aminoglycosides Due to 16S RRNA Methylation in Enterobacteriaceae Isolates. Microb. Drug Resist. 2019, 25, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Piekarska, K.; Zacharczuk, K.; Wolkowicz, T.; Rzeczkowska, M.; Bareja, E.; Olak, M.; Gierczynski, R. Distribution of 16S RRNA Methylases Among Different Species of Aminoglycoside-Resistant Enterobacteriaceae in a Tertiary Care Hospital in Poland. Adv. Clin. Exp. Med. 2016, 25, 539–544. [Google Scholar] [CrossRef]
- Taylor, E.; Bal, A.M.; Balakrishnan, I.; Brown, N.M.; Burns, P.; Clark, M.; Diggle, M.; Donaldson, H.; Eltringham, I.; Folb, J.; et al. A Prospective Surveillance Study to Determine the Prevalence of 16S RRNA Methyltransferase-Producing Gram-Negative Bacteria in the UK. J. Antimicrob. Chemother. 2021, 76, 2428–2436. [Google Scholar] [CrossRef]
- Wei, D.D.; Wan, L.G.; Yu, Y.; Xu, Q.F.; Deng, Q.; Cao, X.W.; Liu, Y. Characterization of Extended-Spectrum β-Lactamase, Carbapenemase, and Plasmid Quinolone Determinants in Klebsiella pneumoniae Isolates Carrying Distinct Types of 16S RRNA Methylase Genes, and Their Association with Mobile Genetic Elements. Microb. Drug Resist. 2015, 21, 186–193. [Google Scholar] [CrossRef]
- Ayad, A.; Drissi, M.; de Curraize, C.; Dupont, C.; Hartmann, A.; Solanas, S.; Siebor, E.; Amoureux, L.; Neuwirth, C. Occurence of armA and RmtB Aminoglycoside Resistance 16S RRNA Methylases in Extended-Spectrum β-Lactamases Producing Escherichia coli in Algerian Hospitals. Front. Microbiol. 2016, 7, 1409. [Google Scholar] [CrossRef]
- Zhao, J.Y.; Zhu, Y.Q.; Li, Y.N.; Mu, X.D.; You, L.P.; Xu, C.; Qin, P.; Ma, J.L. Coexistence of SFO-1 and NDM-1 β-Lactamase Genes and Fosfomycin Resistance Gene FosA3 in an Escherichia coli Clinical Isolate. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef]
- Fournier, C.; Poirel, L.; Despont, S.; Kessler, J.; Nordmann, P. Increasing Trends of Association of 16S RRNA Methylases and Carbapenemases in Enterobacterales Clinical Isolates from Switzerland, 2017–2020. Microorganisms 2022, 10, 615. [Google Scholar] [CrossRef]
- Taylor, E.; Sriskandan, S.; Woodford, N.; Hopkins, K.L. High Prevalence of 16S RRNA Methyltransferases among Carbapenemase-Producing Enterobacteriaceae in the UK and Ireland. Int. J. Antimicrob. Agents 2018, 52, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Pajand, O.; Rahimi, H.; Badmasti, F.; Gholami, F.; Alipour, T.; Darabi, N.; Aarestrup, F.M.; Leekitcharoenphon, P. Various Arrangements of Mobile Genetic Elements among CC147 Subpopulations of Klebsiella pneumoniae Harboring blaNDM-1: A Comparative Genomic Analysis of Carbapenem Resistant Strains. J. Biomed. Sci. 2023, 30, 73. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Hansen, F.; Nielsen, H.L.; Jakobsen, L.; Stegger, M.; Andersen, P.S.; Jensen, P.; Nielsen, T.K.; Hansen, L.H.; Hasman, H.; et al. Use of WGS Data for Investigation of a Long-Term NDM-1-Producing Citrobacter freundii Outbreak and Secondary in vivo Spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 2016, 71, 3117–3124. [Google Scholar] [CrossRef]
- Markovska, R.; Schneider, I.; Keuleyan, E.; Ivanova, D.; Lesseva, M.; Stoeva, T.; Sredkova, M.; Bauernfeind, A.; Mitov, I. Dissemination of a Multidrug-Resistant VIM-1- and CMY-99-Producing Proteus mirabilis Clone in Bulgaria. Microb. Drug Resist. 2017, 23, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Savov, E.; Nazli, A.; Trifonova, A.; Todorova, I.; Gergova, I.; Nordmann, P. Outbreak Caused by NDM-1- and RmtB-Producing Escherichia coli in Bulgaria. Antimicrob. Agents Chemother. 2014, 58, 2472–2474. [Google Scholar] [CrossRef] [PubMed]
- Markovska, R.; Stankova, P.; Popivanov, G.; Gergova, I.; Mihova, K.; Mutafchiyski, V.; Boyanova, L. Emergence of BlaNDM-5 and BlaOXA-232 Positive Colistin- and Carbapenem-Resistant Klebsiella pneumoniae in a Bulgarian Hospital. Antibiotics 2024, 13, 677. [Google Scholar] [CrossRef]
- Berçot, B.; Poirel, L.; Nordmann, P. Updated Multiplex Polymerase Chain Reaction for Detection of 16S RRNA Methylases: High Prevalence among NDM-1 Producers. Diagn. Microbiol. Infect. Dis. 2011, 71, 442–445. [Google Scholar] [CrossRef]
- Davis, M.A.; Baker, K.N.K.; Orfe, L.H.; Shah, D.H.; Besser, T.E.; Call, D.R. Discovery of a Gene Conferring Multiple-Aminoglycoside Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2666–2669. [Google Scholar] [CrossRef]
- Hidalgo, L.; Hopkins, K.L.; Gutierrez, B.; Ovejero, C.M.; Shukla, S.; Douthwaite, S.; Prasad, K.N.; Woodford, N.; Gonzalez-Zorn, B. Association of the Novel Aminoglycoside Resistance Determinant RmtF with NDM Carbapenemase in Enterobacteriaceae Isolated in India and the UK. J. Antimicrob. Chemother. 2013, 68, 1543–1550. [Google Scholar] [CrossRef]
- Gröbner, S.; Linke, D.; Schütz, W.; Fladerer, C.; Madlung, J.; Autenrieth, I.B.; Witte, W.; Pfeifer, Y. Emergence of Carbapenem-Non-Susceptible Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Isolates at the University Hospital of Tübingen, Germany. J. Med. Microbiol. 2009, 58, 912–922. [Google Scholar] [CrossRef]
- Cole, J.M.; Schuetz, A.N.; Hill, C.E.; Nolte, F.S. Development and Evaluation of a Real-Time PCR Assay for Detection of Klebsiella pneumoniae Carbapenemase Genes. J. Clin. Microbiol. 2009, 47, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Naas, T.; Nordmann, P. Pyrosequencing as a Rapid Tool for Identification of GES-Type Extended-Spectrum β-Lactamases. J. Clin. Microbiol. 2006, 44, 3008–3011. [Google Scholar] [CrossRef]
- Goudarzi, H.; Mirsamadi, E.S.; Ghalavand, Z.; Hakemi Vala, M.; Mirjalali, H.; Hashemi, A. Rapid Detection and Molecular Survey of BlaVIM, BlaIMP and BlaNDM Genes among Clinical Isolates of Acinetobacter baumannii Using New Multiplex Real-Time PCR and Melting Curve Analysis. BMC Microbiol. 2019, 19, 122. [Google Scholar] [CrossRef]
- Mendes, R.E.; Kiyota, K.A.; Monteiro, J.; Castanheira, M.; Andrade, S.S.; Gales, A.C.; Pignatari, A.C.C.; Tufik, S. Rapid Detection and Identification of Metallo-β-Lactamase-Encoding Genes by Multiplex Real-Time PCR Assay and Melt Curve Analysis. J. Clin. Microbiol. 2007, 45, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Stoikov, I.; Ivanov, I.N.; Donchev, D.; Teneva, D.; Dobreva, E.; Hristova, R.; Sabtcheva, S. Genomic Characterization of IMP-Producing Pseudomonas aeruginosa in Bulgaria Reveals the Emergence of IMP-100, a Novel Plasmid-Mediated Variant Coexisting with a Chromosomal VIM-4. Microorganisms 2023, 11, 2270. [Google Scholar] [CrossRef]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for Rapid Detection of Genes Encoding CTX-M Extended-Spectrum β-Lactamases. J. Antimicrob. Chemother. 2005, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Mlynarcik, P.; Chalachanova, A.; Vagnerovă, I.; Holy, O.; Zatloukalova, S.; Kolar, M. PCR Detection of Oxacillinases in Bacteria. Microb. Drug Resist. 2020, 26, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC β-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Wang, M.; Guo, Q.; Xu, X.; Wang, X.; Ye, X.; Wu, S.; Hooper, D.C.; Wang, M. New Plasmid-Mediated Quinolone Resistance Gene, qnrC, Found in a Clinical Isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 2009, 53, 1892–1897. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Hasman, H.; Xia, S.; Aarestrup, F.M. qnrD, a Novel Gene Conferring Transferable Quinolone Resistance in Salmonella enterica Serovar Kentucky and Bovismorbificans Strains of Human Origin. Antimicrob. Agents Chemother. 2009, 53, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.I.; Suzuki, S.; Arakawa, Y. Plasmid-Mediated qepA Gene among Escherichia coli Clinical Isolates from Japan. Antimicrob. Agents Chemother. 2008, 52, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.; Sabtcheva, S.; Saga, T.; Ishii, Y.; Kaku, M.; Kantardjiev, T. A Rapid and Versatile Assay for Screening of Aac(6’)-Ib-Cr in Multidrug-Resistant Enterocbacteriaceae. Probl. Inf. Parasit. Dis. 2018, 46, 5–8. [Google Scholar]
- Livermore, D.M.; Jones, C.S. Characterization of NPS-1, a Novel Plasmid-Mediated Beta-Lactamase, from Two Pseudomonas aeruginosa Isolates. Antimicrob. Agents Chemother. 1986, 29, 99–103. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, Y.; Li, Y.; Zhang, K.; Liu, L.; Wang, H.; Tian, J.; Ying, H.; Shi, L.; Yu, T. Characterization and Horizontal Transfer of qacH-Associated Class 1 Integrons in Escherichia coli Isolated from Retail Meats. Int. J. Food Microbiol. 2017, 258, 12–17. [Google Scholar] [CrossRef]
- Ruekit, S.; Wangchuk, S.; Dorji, T.; Tshering, K.P.; Pootong, P.; Nobthai, P.; Serichantalergs, O.; Poramathikul, K.; Bodhidatta, L.; Mason, C.J. Molecular Characterization and PCR-Based Replicon Typing of Multidrug Resistant Shigella Sonnei Isolates from an Outbreak in Thimphu, Bhutan. BMC Res. Notes 2014, 7, 95. [Google Scholar] [CrossRef]
- Donchev, D.; Ivanov, I.N.; Stoikov, I.; Sabtcheva, S.; Kalchev, Y.; Murdjeva, M.; Dobreva, E.; Hristova, R. Improvement and Validation of a Multi-Locus Variable Number of Tandem Repeats Analysis (MLVA8+) for Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. Microorganisms 2023, 11, 444. [Google Scholar] [CrossRef]
- Skirrow, M.B. The Dienes (Mutual Inhibition) Test in the Investigation of Proteus Infections. J. Med. Microbiol. 1969, 2, 471–477. [Google Scholar] [CrossRef][Green Version]
- Pfaller, M.A.; Mujeeb, I.; Hollis, R.J.; Jones, R.N.; Doern, G.V. Evaluation of the Discriminatory Powers of the Dienes Test and Ribotyping as Typing Methods for Proteus mirabilis. J. Clin. Microbiol. 2000, 38, 1077–1080. [Google Scholar] [CrossRef]
- Adler, A.; Hussein, O.; Ben-david, D.; Masarwa, S.; Navon-venezia, S.; Schwaber, M.J.; Carmeli, Y.; on behalf of the Post-Acute-Care Hospital Carbapenem-Resistant Enterobacteriaceae Working Group. Persistence of Klebsiella pneumoniae ST258 as the Predominant Clone of Carbapenemase-Producing Enterobacteriaceae in Post-Acute-Care Hospitals in Israel, 2008–2013. J. Antimicrob. Chemother. 2015, 70, 89–92. [Google Scholar] [CrossRef]
- Alvarez-Arevalo, M.; Sterndorff, E.B.; Faurdal, D.; Jørgensen, T.S.; Mourched, A.-S.; Vuksanovic, O.; Saha, S.; Weber, T. Extraction and Oxford Nanopore Sequencing of Genomic DNA from Filamentous Actinobacteria. STAR Protoc. 2023, 4, 101955. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus Long-Read Assemblies for Bacterial Genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Shakya, M.; Ahmed, S.A.; Davenport, K.W.; Flynn, M.C.; Lo, C.-C.C.; Chain, P.S.G.G. Standardized Phylogenetic and Molecular Evolutionary Analysis Applied to Species across the Microbial Tree of Life. Sci. Rep. 2020, 10, 1723. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).