Abstract
Background: High-risk Escherichia coli clones, such as sequence type (ST)131 and ST1193, along with multidrug-resistant (MDR) Klebsiella pneumoniae, are globally recognized for their significant role in urinary tract infections (UTIs). This study aimed to provide an overview of the virulence factors, clonal diversity, and antibiotic resistance profiles of extended-spectrum cephalosporin (ESC)-E. coli and K. pneumoniae causing UTIs in humans in the Tebessa region of Algeria. Methods: Forty E. coli and 17 K. pneumoniae isolates exhibiting ESC-resistance were recovered (July 2022–January 2024) from urine samples of patients at three healthcare facilities to be phenotypically and genotypically characterized. Whole genome sequencing (WGS) was performed on the ST1193 clone. Results: Among K. pneumoniae isolates, all except one harbored CTX-M-15, with a single isolate carrying blaCTX-M-194. Additionally, two K. pneumoniae isolates co-harboring blaCTX-M-15 and blaNDM exhibited phenotypic and genotypic hypervirulence traits. Fluoroquinolone resistance (FQR) was detected in 94.1% of K. pneumoniae isolates. The E. coli isolates carried diverse ESC-resistance genes, including CTX-M-15 (87.5%), CTX-M-27 (5%), CTX-M-1, CMY-59, and CMY-166 (2.5% each). Co-carriage of blaESC and blaOXA-48 was identified in three E. coli isolates, while 62.5% exhibited FQR. Phylogenetic analysis revealed that 52.5% of E. coli belonged to phylogroup B2, including the high-risk clonal complex (CC)131 CH40-30 (17 isolates) and ST1193 (one isolate). In silico analysis of the ST1193 genome determined O75:H5-B2 (CH14-64), and the carriage of IncI1-I(Alpha) and IncF [F-:A1:B10] plasmids. Notably, core genome single-nucleotide polymorphism (SNP) analysis demonstrated high similarity between the Algerian ST1193 isolate and a previously annotated genome from a hospital in Northwest Spain. Conclusions: This study highlights the spread and genetic diversity of E. coli CC131 CH40-30 and hypervirulent K. pneumoniae clones in Algeria. It represents the first report of a CTX-M-15-carrying E. coli ST1193 in the region. The findings emphasize the urgent need for antibiotic optimization programs and enhanced surveillance to curb the dissemination of high-risk clones that pose an increasing public health threat in Algeria. A simplified method based on virulence traits for E. coli and K. pneumoniae is proposed here for antimicrobial resistance (AMR) monitoring.
1. Introduction
Urinary tract infections (UTIs) are the second most common infectious disease in humans, after respiratory tract infections, in healthcare-associated centers and the community worldwide [1]. UTIs affect millions of individuals annually, are associated with decreased quality of life, and pose a significant clinical and economic burden [2]. Women are particularly susceptible, with approximately 50% to 60% of women experiencing at least one UTI during their lifetime [3,4]. Data from 1990 to 2021 revealed that UTIs showed an upward trend, with a particularly pronounced disease burden among women, older men, and low–middle sociodemographic index regions [5]. Furthermore, the impact estimation of UTIs in 2021 was 287,200 deaths associated with bacterial antimicrobial resistance (AMR), including 67,467 deaths directly attributable to bacterial AMR, being the fourth leading cause of death associated with AMR worldwide [5,6]. Tropical Latin America and low–middle sociodemographic index regions exhibited the highest impact in terms of prevalence, death rate, and disability-adjusted life-years (DALYs) [5].
A wide range of virulence factors and multidrug-resistant (MDR) pathogenic strains are described in the pathogenicity and resistance of the uropathogenic agents, making it more difficult to manage these complicated infections. Among them, Escherichia coli is the predominant causative agent of UTIs, and Klebsiella pneumoniae is recognized as the second contributor to UTIs. Both E. coli and K. pneumoniae exhibit a variety of virulence factors that enable them to adhere to, invade, and persist in the urinary tract, often resulting in acute or chronic infections [4]. These pathogens also exhibit increasing resistance, especially to cephalosporins, fluoroquinolones, and carbapenems, leading to longer hospital stays, increased healthcare costs, and treatment failures [6]. Globally, 0.26 million deaths (95% uncertainty interval [UI]: 0.18–0.36) were associated with bacterial AMR in UTIs in 2019 [7].
Among E. coli strains, infections of MDR high-risk clones such as ST38, ST131, ST167, ST405, ST410, ST648, and ST1193 are commonly reported [8,9,10]. ST131 is considered the most prominent, while ST1193 appears to be emerging as a significant clone following a trajectory similar to that of the globally dominant ST131 lineage. It exhibits strong virulence potential, efficient human-to-human transmission, and adaptability to both community and hospital environments. Its rise is particularly concerning due to its co-resistance to multiple antibiotic classes and its frequent carriage of key resistance determinants such as blaCTX-M and mutations in gyrA/parC, conferring fluoroquinolone resistance (FQR) [11]. MDR clones are often linked to resistance to FQR, with many also producing cefotaximases (CTX-M), a family of extended-spectrum beta-lactamase (ESBL) enzymes which confer resistance to extended-spectrum cephalosporins (ESC), thereby limiting the effectiveness of this last resort class of antibiotics and significantly reducing available treatment options in clinical settings [9,10,11,12].
On the other hand, ESBL and carbapenemase-encoding K. pneumoniae are globally disseminated and cause infections that are often difficult to treat. According to the 2024 WHO Medically Important Antimicrobials (MIA) List, ESBL-producing and carbapenemase-producing E. coli and K. pneumoniae are highlighted as critical threats due to their resistance to critically important antimicrobials (CIAs), underscoring the need for strengthened antimicrobial stewardship across human, animal, and environmental health sectors [13].
In Algeria and other low- and middle-income countries (LMICs), the prevalence of MDR uropathogens is notably high, exacerbated by the lack of robust antibiotic stewardship programs. The study “Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis”, published in The Lancet [6], reported that the Middle East and North Africa (MENA) region experienced a significant impact from AMR in 2019. Specifically, there were approximately 7687 deaths (95% uncertainty interval: 6663–8969) associated with UTIs in the MENA region during that year.
Recent studies have underscored the growing public health threat posed by MDR uropathogens in Algeria, particularly E. coli and K. pneumoniae. In a study conducted in the Tizi-Ouzou region between 2017 and 2019, E. coli isolates recovered from UTI patients exhibited high levels of resistance to frequently used antibiotics, including ampicillin, trimethoprim-sulfamethoxazole, and fluoroquinolones [14]. In parallel, several investigations have reported the emergence and dissemination of MDR K. pneumoniae in Algeria. Notably, carbapenemase-producing strains such as KPC-2 and OXA-48 have been detected in clinical settings, with clonal dissemination of high-risk lineages like ST258, ST101, and ST512 [15,16]. For instance, KPC-2-producing K. pneumoniae ST512 was isolated from cerebrospinal fluid in 2014 [15], while a 2016–2017 study in Annaba Hospital documented the first cases of KPC-2-producing K. pneumoniae ST258, alongside an outbreak involving ST101 strains harbouring the blaOXA-48 gene in a urology department, suggesting nosocomial transmission [16]. Moreover, isolates collected in 2018 from a military hospital in Oran showed carriage of ESBL-encoding genes such as blaCTX-M and blaTEM, often associated with biofilm formation, which can further complicate treatment and infection control [17]. These data collectively highlight the alarming spread of MDR Enterobacteriaceae and the importance of coordinated national strategies to curb resistance in both community and hospital settings. In Tebessa province, another study demonstrated the correlation between inappropriate antibiotic use and the increasing prevalence of ESBL-producing bacteria [18]. Similar patterns have been observed in other regions, where antibiotics are frequently dispensed without prescriptions or proper medical oversight [19,20]. These challenges are compounded by the limited availability of coordinated national surveillance data, with most reports stemming from academic studies rather than systematic public health monitoring [21].
This study aims to contribute to this growing body of evidence by characterizing the virulence and antibiotic resistance profiles of E. coli and K. pneumoniae isolates causing UTIs in the Tebessa region of Algeria. By exploring a simplified laboratory workflow for AMR monitoring, this work seeks to support the development of cost-effective surveillance approaches. A clearer understanding of local epidemiological trends is essential to guide therapeutic choices, inform infection control measures, and strengthen national AMR surveillance capacity.
2. Results
The present study comprised 57 non-duplicate isolates, 40 E. coli and 17 K. pneumoniae, recovered from urine samples at three healthcare facilities in northeast Algeria from July 2022 to January 2024 (Tables S1 and S2).
2.1. Escherichia coli Collection
A total of 40 non-duplicate E. coli isolates were recovered from patients in three healthcare facilities in the Tebessa region, Algeria, including 9 isolates from male and 31 from female patients. All isolates tested positive for the uidA target-specific species gene [22]. Phenotypic characterization on MacConkey Lactose agar revealed that four isolates (10%) were non-lactose fermenters (NLF) (Table S1).
2.1.1. Antimicrobial Susceptibility Testing (AST) and Genotypic Characterization of bla Genes
Antimicrobial susceptibility testing (AST) revealed that all E. coli isolates were resistant to ampicillin, cefuroxime, cefotaxime, and ceftazidime. As shown in Figure 1, most of the isolates (≥50%) exhibited resistance to aztreonam, amoxicillin-clavulanic acid, nalidixic acid, sulfamethoxazole, ciprofloxacin, doxycycline, and tobramycin. Notably, four isolates (10%) were resistant to ertapenem. All isolates were classified as MDR, exhibiting resistance to at least one drug in three or more antimicrobial categories [23], and 62.5% demonstrated FQR (Figure 1).
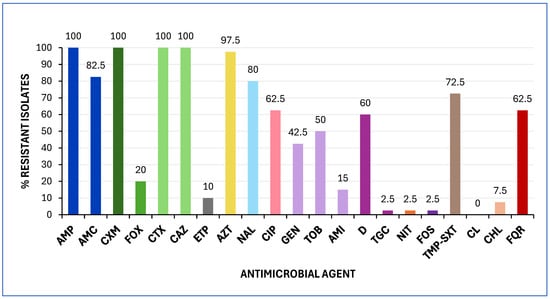
Figure 1.
Prevalence of antimicrobial resistance among the 40 E. coli isolates analysed in this study. Minimum inhibitory concentrations (MICs) were interpreted according to EUCAST 2025 and CLSI 2024 clinical breakpoints. Abbreviations: AMP, ampicillin; AMC, amoxicillin/clavulanic acid; CXM, cefuroxime; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; AZT, aztreonam; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin; AMI, amikacin; D, doxycycline; TGC, tigecycline; NIT, nitrofurantoin; FOS, fosfomycin; TMP-SXT, trimethoprim-sulfamethoxazole; CL, colistin; CHL, chloramphenicol; FQR, fluoroquinolone resistance.
The isolates were subsequently analysed by PCR to detect the presence of blaESBL (SHV, CTX-M), blaCMY, and blaCARBA (VIM, IMP, OXA, NDM, KPC) genes, followed by Sanger sequencing. The collection exhibited different ESC enzymes, including CTX-M-15 (87.5%), CTX-M-27 (5%), as well as CTX-M-1, CMY-59, and CMY-166 (2.5% each). The blaOXA-48 gene was detected in three E. coli isolates resistant to ertapenem, which also harboured CTX-M-27 (two isolates) and CMY-59 (one isolate). None of the isolates tested positive for mcr (1 to 5) genes or showed polymyxin resistance (Table S1).
2.1.2. Virulence Traits
The screening of virulence factors associated with enhanced urinary tract colonization indicated that 18 out of the 40 isolates (45%) met the criteria for UPEC status, defined by the presence of at least three of the specific virulence genes (chuA, fyuA, vat, and yfcV) [24]. Individually, chuA, fyuA, vat, and yfcV were detected in 77.5%, 80%, 5%, and 52.5% of the isolates, respectively.
Furthermore, 72.5% of the isolates were classified as ExPEC based on the presence of at least two of the five key virulence markers: papAH, sfa/focDE, afa/draBC, kpsMII, and iutA [25]. The individual prevalence of these genes among the isolates was 40%, 0%, 30%, 57.5%, and 77.5%, respectively (Table S1).
2.1.3. Clonal Groups
Using the PCR method described by Clermont et al. [26,27], five E. coli phylogroups were identified among the 40 isolates. Phylogroups B2 and D were the most prevalent, accounting for 52.5% and 22.5% of the isolates, respectively. The remaining isolates belonged to phylogroups B1 (12.5%), A (7.5%), and F (5%) (Table 1).
Presumptive identification of the pandemic clonal complex (CC)131 was performed via PCR screening for the rfbO25b and fliCH4 gene markers [28,29] with 17 out of 40 isolates (42.5%) testing positive for both. Additionally, the fliCH5 gene, typically associated with clones such as ST1193 or CC131 of clade A [9,30] was determined in one NLF isolate (2.5%) (Table S1).
Clonotyping identified 14 distinct fumC-fimH combinations. Notably, 42.5% of the isolates exhibited clonotype CH40-30, which is typically associated with CC131 [9,31,32]. The second most prevalent clonotype, CH26-5, was detected in 15% of the isolates. Notably, UPEC status was associated with phylogroup B2 isolates, whereas ExPEC status was linked to both B2 and D isolates (Table 1).

Table 1.
Clonotypes determined among the 40 E. coli isolates.
Table 1.
Clonotypes determined among the 40 E. coli isolates.
| Phylogroup | Clonotype (CH) a | No Isolates | UPEC Status b | ExPEC Status c | FQR |
|---|---|---|---|---|---|
| B2 | CH40-30 | 17 | 15 | 16 | 17 |
| CH14-64 | 1 | 1 | 1 | 1 | |
| CH108-Neg | 2 | 2 | 2 | 1 | |
| CH1012-Neg | 1 | 0 | 0 | 1 | |
| D | CH26-5 | 6 | 0 | 6 | 1 |
| CH26-Neg | 3 | 0 | 3 | 0 | |
| B1 | CH65-27 | 2 | 0 | 0 | 0 |
| CH27-54 | 1 | 0 | 0 | 0 | |
| CH65-32 | 1 | 0 | 0 | 0 | |
| CH7-604 | 1 | 0 | 0 | 0 | |
| A | CH11-54 | 1 | 0 | 1 | 1 |
| CH11-Neg | 1 | 0 | 0 | 1 | |
| CH7-94 | 1 | 0 | 0 | 0 | |
| F | CH88-Neg | 2 | 0 | 0 | 2 |
a Eight distinct fimH alleles were determined within the 40 E. coli. (Neg): Nine isolates showed no amplification of the 489-bp internal sequence of the fimH gene targeted for clonotyping [31] despite all 40 E. coli isolates testing positive for fimH using primers and conditions described by Johnson and Stell [33]. b UPEC status. + positive for ≥3 of the following genes: chuA, fyuA, vat, and yfcV [24]. c ExPEC status. + positive for ≥2 of these five markers: papAH, sfa/focDE, afa/draBC, kpsMII, and iutA [25]. FQR, fluoroquinolone resistance.
Characterization of Clonal Complex (CC) 131 Isolates
The most prevalent clonal group O25b:H4-B2 (CH40-30) CC131, identified in 17 isolates, exhibited FQR (100%), carriage of blaCTX-M-15 (100%), and virulence profiles accomplishing ExPEC (16 of 17, 94.1%) and UPEC (15 of 17, 88.2%) status (Table 1). Remarkably, one CC131 isolate displayed non-susceptibility to ertapenem, though the underlying genetic mechanism could not be determined. High levels of resistance were observed against tobramycin (16 isolates, 94.1%), amoxicillin/clavulanic acid (15 isolates, 88.2%), sulfamethoxazole (13 isolates, 76.5%), gentamycin (13 isolates, 76.5%), and doxycycline (10 isolates, 58.8%).
Using the virotyping scheme proposed by Dahbi et al. [30], virotype F was identified in seven isolates (41.2%), while virotype E was detected in three isolates (17.6%). Among the extraintestinal virulence markers included in this scheme, the specific pilus tip adhesin molecule type II associated with pyelonephritis (papGII) was present in all isolates, the secreted autotransporter toxin (sat) was detected in 14 isolates (82.3%), and the K5 group II capsule (kpsMII-K5) was found in 10 isolates (58.8%). Notably, the co-occurrence of papGII, cnf1 (cytotoxic necrotizing factor 1), and hlyA (α-hemolysin) was observed in three isolates of virotype E(17.6%) (Table 2).

Table 2.
Phenotypic resistance and virulence profile of the 17 CC131 E. coli isolates.
In Silico Characterization of ST1193 Isolate
The NLF isolate B2-CH14-64 underwent WGS and in silico analysis using bioinformatics tools from the Center for Genomic Epidemiology. Table 3 summarizes its genomic characteristics.

Table 3.
In silico characterization and phenotypic resistance of the ST1193 genome.
Serotyping analysis using SerotypeFinder identified the O75:H5 serotype while CHTyper confirmed its clonotype as CH14-64. Multilocus sequence typing (MLST) based on the seven-gene of the Atchman scheme assigned the sequence type (ST)1193, while the alternative MLST scheme using eight genes (dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA) [34] predicted ST53. Core genome MLST (cgMLST) analysis, based on 2513 loci, assigned the isolate to cgST140226.
Resistance gene profiling using ResFinder confirmed the presence of blaCTX-M-15, along with chromosomal mutations associated with FQR (gyrA p.S83L, gyrA p.D87N, parC p.S80I, and parE p.L416F), consistent with the observed phenotypic resistance. Additionally, acquired resistance genes, such as aac(3)-IIa, aac(6′)-Ib-cr, blaOXA-1, and catB3 were predicted.
Virulence profiling using VirulenceFinder identified different virulence factors, including vat, fyuA, yfcV, chuA, iutA, and kpsMII, confirming its UPEC and ExPEC status. Plasmid analysis revealed the presence of IncI1-I(Alpha) and IncF [F-:A1:B10] plasmids, along with small Col-like plasmids.
To investigate the genomic relationship between the Algerian ST1193 isolate and five ST1193 isolates recently recovered from a hospital in Northwest Spain, we performed a single-nucleotide polymorphism (SNP) comparison of their core genomes, which represented 95.65% of the reference genome LREC-269 (5.4 Mb). The phylogenetic analysis was conducted using CSI Phylogeny 1.4 (Figure 2A; Table S3). The Algerian ST1193 isolate (LREC-468) exhibited a minimum pairwise distance of 78 SNPs from LREC-269 and a maximum distance of 128 SNPs from LREC-270.
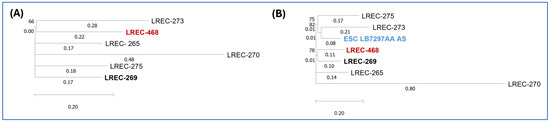
Figure 2.
Phylogenetic dendrogram based on whole-genome SNP analysis. (A) Phylogenetic dendrogram based on SNP counts per substitution within the core genome of the Algerian and Spanish ST1193 isolates. The WGS comparison resulted in a core genome covering 95.65% of the reference genome LREC-269 (5.4 Mb). (B) Phylogenetic dendrogram incorporating the Algerian, Spanish, and ESC_RA5887AA ST1193 isolates, showing SNP counts per substitution. The WGS comparison resulted in a core genome covering 82.6% of the reference genome LREC-269 (5.4 Mb).
Additionally, we accessed Enterobase (https://enterobase.warwick.ac.uk/; accessed on 20 December 2024) to search for ST1193 genomes assigned to cgST140226. We identified and retrieved one publicly available genome, ESC_RA5887AA, from BioProject PRJEB21277 registered in 2020 by the University of Oxford. A subsequent SNP comparison (Figure 2B, Table S4), which included the Algerian (LREC-468), five Spanish ST1193 genomes, and ESC_RA5887AA, showed that 82.6% of nucleotide positions in the reference genome (LREC-269) were conserved across all analysed genomes. The Algerian LREC-468 genome differed by 55 SNPs from ESC_RA5887AA.
2.2. Klebsiella pneumoniae Collection
Seventeen K. pneumoniae isolates were recovered from patients across three healthcare facilities in the Tebessa region of Algeria, comprising five isolates from males and 12 from female patients. Sixteen isolates tested positive for the kp50233 species-specific gene. The single negative isolate was confirmed as K. pneumoniae using the matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF).
2.2.1. AST and Genotypic Characterization of bla Genes
All isolates exhibited resistance to ampicillin, cefuroxime, cefotaxime, and aztreonam. Additionally, the majority (≥60%) were resistant to ceftazidime, ciprofloxacin, fosfomycin, sulfamethoxazole, and amoxicillin-clavulanic acid. Notably, two isolates (11.8%) demonstrated resistance to ertapenem (Figure 3). All isolates were classified as MDR, exhibiting resistance to at least one drug in three or more antimicrobial categories [23], and 94.1% exhibited FQR.
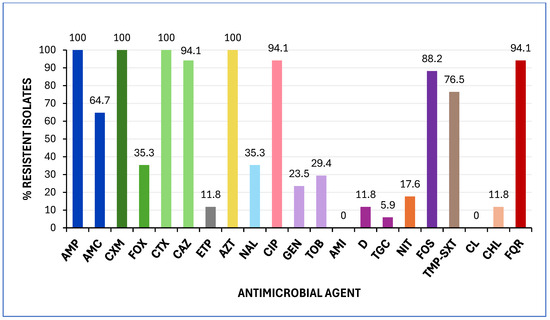
Figure 3.
Prevalence of antimicrobial resistance among the 17 K. pneumoniae analysed in this study. MICs were interpreted according to EUCAST 2025 and CLSI 2024 clinical breakpoints. Abbreviations: AMP, ampicillin; AMC, amoxicillin/clavulanic acid; CXM, cefuroxime; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; AZT, aztreonam; NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin; AMI, amikacin; D, doxycycline; TGC, tigecycline; NIT, nitrofurantoin; FOS, fosfomycin; TMP-SXT, trimethoprim-sulfamethoxazole; CL, colistin; CHL, chloramphenicol; FQR, fluoroquinolone resistance.
PCR analysis was conducted to detect the presence of blaESBL (SHV, CTX-M), blaCMY, and blaCARBA (VIM, IMP, OXA, NDM, KPC) genes, followed by Sanger sequencing. The CTX-M-15 gene was identified in 94.1% of the isolates, with one isolate harbouring the CTX-M-194 variant. Notably, the blaNDM gene was identified in the two K. pneumoniae isolates that exhibited ertapenem resistance, which also carried the CTX-M-15 gene. Additionally, two isolates possessed both the CTX-M-15 and SHV-148 genes. None of the isolates tested positive for mcr (1 to 5) genes or showed polymyxin resistance (Table S2).
2.2.2. Virulence Traits
The hypermucoviscous (HMV) phenotyping of the 17 K. pneumoniae isolates, using the string test described by Shon et al. [35] with modifications, showed that four were classified as HMV-positive in at least two of the tested culture conditions (Table 4 and Table S2). Notably, two isolates exhibited the HMV-positive phenotype across all culture conditions.

Table 4.
Hypervirulent and AMR traits of the four phenotypic HMV K. pneumoniae isolates.
PCR screening was conducted to detect virulence genes commonly associated with hypervirulent K. pneumoniae (hvKp) [36]. All isolates tested negative for iroB, peg-589, peg-1631, and rmpA2 genes. However, more than 50% were positive for terB and rpmA. Notably, the two K. pneumoniae isolates exhibiting the HMV phenotype across all culture conditions tested positive for iucA, peg-344, and rmpA genes, and co-harboured both blaNDM and blaCTX-M-15 (Table 4).
3. Discussion
Urinary tract infections (UTIs) continue to represent a significant public health concern globally, with the pathogens E. coli and K. pneumoniae being the primary causative agents [1,4]. Particularly in Algeria, UTIs and pyelonephritis were reported as the fifth leading cause of death in 2021, accounting for 1,290 fatalities. E. coli and K. pneumoniae were the primary pathogens responsible for 40.5% of the total deaths (370 and 153, respectively). Among these, 84% of deaths were attributed to resistant E. coli and K. pneumoniae (321 and 117, respectively) [37]. Given the significant role of these bacteria in the aetiology of UTIs, their global contribution to the dissemination of MDR, and the limited data on their impact in Algeria, this study aimed to investigate the molecular characteristics of E. coli and K. pneumoniae exhibiting ESC resistance associated with UTIs in the province of Tebessa, where data on such infections remain scarce. Although our study was limited to three healthcare facilities in Tebessa, the selected sites are key referral centers that serve a diverse patient population across the province, making the sample reasonably representative of urban Algeria. The Laboratory of Medical Analysis receives a large and diverse influx of patients from across the province, due to its advanced equipment and strong reputation among local physicians who frequently refer their patients there. The Bouguerra Boulares Hospital and Khaldi Abdelaziz Maternity Hospital are central healthcare institutions in the province, receiving patients from various areas, particularly in complex or high-risk cases. Diagnostic approaches and antimicrobial prescribing practices in these facilities mirror those used in other regions of the country, where antibiotics remain widely accessible without prescription, contributing to similar AMR dynamics.
CTX-M-15 and OXA-48-like enzymes are the most prevalent and widely disseminated ESBLs and carbapenemases, respectively, posing a significant global public health threat [38,39]. In this study, blaCTX-M-15 was detected in 87.5% of E. coli isolates, a prevalence similar to that reported in other regions of Algeria for ESBL-producing E. coli in UTIs [19,20]. The blaOXA-48 carbapenemase gene, endemic in North Africa and highly reported in Algeria [21], was found here in three E. coli isolates.
Notably, we identified two blaCTX-M-27 E. coli isolates, which also harboured blaOXA-48; this would represent the first report of the CTX-M-27 variant in Algeria, whose emergence and global spread have been reported in regions such as Japan and Europe [38,40]. The third OXA-48 E. coli isolate analysed here showed a co-carriage of CMY-59. Furthermore, we observed FQR in 62.5% of ESC-producing E. coli, which aligns with findings from Sétif [19] but is higher than the 30% resistance prevalence reported by Zenati et al. [20] in Tlemcen, Algeria.
The E. coli population is classified into distinct phylogenetic groups, namely A, B1, B2, C, D, F, and G, along with cryptic clades [26,27]. UPEC isolates predominantly belong to phylogroups B2 and D, which are typically associated with the carriage of a higher number of virulence genes in comparison with other phylogroups [41,42]. Accordingly, our ESC-producing E. coli isolates were classified into A, B1, B2, D, and F, with B2 being the most prevalent (52.5%), notably including high-risk clones such as CC131 and ST1193. Phylogroup D was the second, comprising 22.5% of the isolates. Of the 21 B2 isolates, 19 met the criteria for UPEC status, and 20 for ExPEC status. Additionally, all nine phylogroup D isolates fulfilled the ExPEC status.
The CC131 and ST1193 are reported as the most prevalent among FQ and cephalosporin-resistant E. coli isolates globally, commonly linked to MDR UTIs [10,11,43,44]. The CC131 lineage of E. coli has diversified into three clades: A and B, which are susceptible, and C, which exhibits FQ/cephalosporin resistance. Clade C, particularly the fimH30 variant (CH40-30 clonotype), is the most widely distributed. Within clade C, subclade C1 (H30R) includes FQR non-ESBL producers, whereas subclade C2 (H30Rx) is characterized by the presence of both FQR and the blaCTX-M-15 gene [45].
In this study, we used rfbO25b, fliCH4, fliCH5, and the B2 phylogroup as PCR screening markers for CC131, along with clonotyping, as previously described [10]. This approach allowed us to identify 17 O25b:H4-B2 E. coli (42.5%), all assigned to CC131 and exhibiting the CH40-30 clonotype. Moreover, all isolates displayed FQR and carried the blaCTX-M-15 gene, classifying them within subclade C2 [45].
CC131 has been reported in various sources in Algeria, including human clinical samples [46,47], uropathogenic E. coli from non-hospitalized patients [48], fish [49], food [50], and wildlife [51], indicating its widespread distribution across the country. However, detailed molecular characterization of CC131 in Algeria remains limited. Our study focuses on resistance and virulence gene profiles. Thus, we found that 88.2% and 94.1% of CC131 isolates exhibited UPEC and ExPEC status, respectively. Furthermore, analysis of extraintestinal virulence factors following the Dahbi et al. [30] scheme identified two distinct virotypes: F (sat, papG II, kpsM II-K5) in 41.2% of isolates and E (sat, papG II, cnf1, hlyA, kpsM II-K5) in 17.6%. Notably, the remaining seven FQR, CTX-M-15 O25b:H4-B2-CC131 (CH40-30) isolates could not be virotyped.
The ST1193 clone is considered an emerging pathogenic lineage of E. coli [52,53] typically associated with the O75 serotype, fimH64 type 1 pili, and K1 or K5 capsular types. This clone is also characterized by FQR and lactose non-fermentation [11]. Although ST1193 has been reported in Europe [9,10,54,55], Asia [56,57,58], and the United States [59], its detection in Africa remains limited [60,61,62]. In this study, we identified one NLF ST1193 isolate, FQR, CTX-M-15-producer, with genomic key features consistent with globally disseminated ST1193 isolates [9,10,11]. Specifically, the Algerian isolate carried the K1 capsular type and harboured mutations in gyrA (D87N, S83L) and parC (S80I), along with parE (L416F). Plasmidome analysis identified the presence of IncF [F-:A1:B10], IncI1-I, and ColBS512-like plasmids. A large-scale genomic study of ST1193 [52] classified the ST1193 clone into distinct clades based on capsule type (K1 or K5) and the presence of specific IncF plamids: clade A (K5 capsule with F−:A1:B20 plasmids), subclade B1 (K1 capsule with F−:A1:B10 plasmids) and subclade B2 (K1 capsule with F−:A1:B1 plasmids) [11,52]. According to this classification, our ST1193 isolate belongs to subclade B2.
A core genome SNP comparison between our Algerian ST1193 isolate (LREC-468) and previously characterized ST1193 isolates from a Northwest Spanish hospital [9] revealed a minimum distance of 78 SNPs with LREC-269. We then queried Enterobase for genomes assigned to cgST140226 and found a single genome, ESC_RA5887AA, from BioProject PRJEB21277 (University of Oxford, 2020), which differed by only 55 SNPs.
This comparative approach was guided by the core genome sequence typing (cgST) framework proposed by Achtman et al. [63], which offers a robust method for identifying highly related strains based on shared allelic profiles. While hundreds of ST1193 genomes are available in public repositories, few are annotated with cgST identifiers, and fewer still meet the quality criteria and metadata completeness required for SNP-level comparisons. cgMLST, which analyzes 2,513 soft-core genes, offers significantly higher resolution than MLST, making it a powerful tool for tracing transmission dynamics within outbreaks and defining population structures across different levels, including the genus level [63]. Interestingly, the detection of the same cgST in a genome presumably originating from Ireland (based on the metadata provided in the BioProject of this genome) underscores the transcontinental dissemination of this clone and raises public health concerns about the unnoticed spread of high-risk E. coli lineages in North Africa. Our findings, therefore, emphasize the urgent need for continued genomic surveillance of MDR uropathogens in the region, particularly as this is the first genomic report of ST1193 in clinical E. coli isolates from Algeria.
Phylogroup D was the second most prevalent in our E. coli collection, comprising 22.5% of the isolates, all classified as ExPEC. These isolates exhibited either the fumC26 fimH5 or fumC26 fimH-negative clonotypes (no amplification of the 489-bp internal sequence of the clonotype scheme) [31]. According to EnteroBase, all genomes assigned to phylogroup D (Clermont scheme) with fumC26 fimH5/negative are associated with CC38, which includes ST38, a high-risk MDR clone linked to UTI and bloodstream infections, as well as the global spread of OXA-48 [39]. ST38 has also contributed to the emergence of nosocomial and community-acquired OXA-244-producing E. coli ST38 in Europe [64,65]. Additionally, putative inter-host and host-environment transmission events within ST38, where genomes differed by <35 SNPs, underscore its role in maintaining and disseminating AMR genes [66]. In this study, the two D-CH26-Neg isolates carried both OXA-48-like and CTX-M-27, while the one D-CH26-5 isolate harboured OXA-48-like and CMY-59. ST38 OXA-48-producing E. coli has previously been detected in Algeria in white stork (Ciconia ciconia) migratory birds [67], river water [68], human clinical samples [69], and broilers [70]. Our findings further highlight the role of CC38 in the spread of AMR in this region.
All K. pneumoniae isolates, except one, carried the blaCTX-M-15 gene. In addition, two ertapenem-resistant isolates tested positive for the blaNDM gene. The co-occurrence of carbapenem and ESBL resistance in the same isolate significantly complicates treatment options and raises concerns about therapeutic failures. Specifically, K. pneumoniae harbouring blaCTX-M-15 and blaNDM genes was previously reported at Annaba University Hospital (Algeria) in 2014 [71]. Further studies have reported the widespread presence of blaCTX-M-15 in Algeria, including the University Hospital Establishment of Oran, where nearly all ESBL-positive isolates carried blaCTX-M-15 [72]. blaCTX-M K. pneumoniae isolates were also identified in the Regional Military University Hospital of Oran, with a prevalence of 37.5% ESBL producers [17]. Carbapenem-resistant K. pneumoniae strains carrying blaCTX-M-15 have also been documented in Ibn Roched hospital of Annaba, where OXA-48 and KPC-2 carbapenemase-producing isolates were identified in urology patients [16]. These findings underscore the ongoing emergence of multidrug-resistant K. pneumoniae in Algerian hospitals, necessitating enhanced surveillance and infection control measures.
In terms of virulence, K. pneumoniae is categorized into two distinct pathotypes: classical (cKp) and hypervirulent (hvKp). cKp is a common cause of hospital-acquired infections such as pneumonia and UTIs, particularly in elderly or immunocompromised people, and is known for its ability to acquire multiple AMR genes [73]. By contrast, hvKp is more virulent and capable of causing severe infections in healthy people, such as pyogenic liver abscesses, endophthalmitis, meningitis, septic arthritis, and other unusual infections, often leading to significant morbidity and mortality [74,75]. The definition of hvKp has evolved. Initially, the HMV phenotype with a positive string test (>5 mm) was used for identification [35,76], but this method lacks accuracy. Currently, murine infection models remain the gold standard [77]; however, their high cost and ethical constraints limit their practicality. Instead, recent evidence supports the presence of five virulence plasmid-associated genes (peg-344, rmpA, rmpA2, iroB, and iucA) as the most accurate molecular markers for hvKp identification, which offer a feasible diagnostic alternative for clinical and surveillance applications [78,79]. In our K. pneumoniae collection, the HMV phenotype was observed in four isolates, though only two were consistently positive across all test conditions. PCR screening of the virulence markers revealed that these two isolates were the only ones carrying three of the five plasmid-associated genes (peg-344, rmpA, and iucA). Notably, both also harboured the blaNDM and blaCTX-M-15 genes, further highlighting their clinical relevance.
Initially, hvKp isolates were susceptible to common antibiotics, including last generation cephalosporins and carbapenems. However, the emergence of MDR hvKp in recent years has raised significant concerns. Notably, the European Centre for Disease Prevention and Control reported the spread of hvKp ST23 strains carrying carbapenemase genes across multiple EU/EEA countries, highlighting the convergence of hypervirulence and antimicrobial resistance [80]. Although our two HMV blaNDM and blaCTX-M-15-producing K. pneumoniae possessed only three of the five key virulence plasmid-associated genes (peg-344, rmpA, and iucA), they exemplify the concerning trend of MDR K. pneumoniae strains enhancing their clinical virulence potential. This convergence of resistance and virulence factors poses significant challenges for treatment and infection control strategies.
In Algeria, the widespread and often unregulated use of antibiotics, especially beta-lactams, continues to drive the emergence and spread of ESBL-producing bacteria. In Tebessa province, a recent study confirmed the strong correlation between antibiotic misuse and the growing prevalence of resistant pathogens [18]. Similar findings from northeastern [19] and northwestern Algeria [20] linked high resistance rates to inappropriate prescribing practices and over-the-counter antibiotic sales without medical oversight. These trends highlight the urgent need for robust antibiotic stewardship and strict regulation of antimicrobial use nationwide.
AMR data in Algeria remains limited and fragmented. As noted by Touati and Mairi [21], most available evidence stems from isolated academic efforts, often in collaboration with European labs, rather than from coordinated national surveillance. The Algerian Antimicrobial Resistance Network (AARN) [81] has also reported increasingly alarming resistance patterns in E. coli and K. pneumoniae from hospitals across the country, emphasizing challenges such as diagnostic variability and under-resourced microbiology services.
In this context, our study contributes valuable region-specific data by characterizing the virulence and antibiotic resistance profiles of E. coli and K. pneumoniae strains causing UTIs in Tebessa. By exploring a simplified laboratory workflow for AMR monitoring, this work supports the development of cost-effective surveillance tools. A clearer understanding of local epidemiological patterns is essential to guide empirical therapy, inform infection control strategies, and strengthen Algeria’s national AMR response.
4. Materials and Methods
4.1. E. coli and K. pneumoniae Collections
This study analyzed 57 non-duplicate isolates, comprising 40 E. coli and 17 K. pneumoniae, recovered from urine samples of both male (9 E. coli and 5 K. pneumoniae) and female (31 E. coli and 12 K. pneumoniae) patients. The samples were obtained from both outpatients and inpatients at three healthcare facilities in the Tebessa region of northeast Algeria. These facilities were the Bouguerra Boularess-Bekaria Public Hospital Establishment (EPH) (252 beds, 8 wards), the Khaldi Abdelaziz-Tebessa Maternity Hospital (EPH) (166 beds, 4 wards), and the private laboratory Elite. Isolates were collected between July 2022 and January 2024.
Following incubation on MacConkey (ML) agar (Oxoid, Sin El Fil, Beirut, Lebanon) at 37 °C for 18–24 h, bacterial identification was performed using the API 20E system (bioMérieux, Dely Ibrahim, Alger, Algeria) and the Vitek 2 GN system (bioMérieux Inc., Hazelwood, MO, USA). Isolates were selected based on extended-spectrum cephalosporin (ESC) resistance using the Vitek 2 AST system (bioMérieux, Dely Ibrahim, Alger, Algeria). Selected isolates were stored on slanted nutrient agar (Difco, Sin El Fil, Beirut, Lebanon) at room temperature until further molecular analysis at the Reference Laboratory for E. coli (LREC, University of Santiago de Compostela, Spain) (Tables S1 and S2).
Species confirmation was conducted by PCR amplification of the β-d-glucuronidase (uidA) gene for E. coli [22] and the putative acyltransferase (kp50233) gene for K. pneumoniae [82] (Table S5). When PCR results were inconclusive, bacterial identification was performed using matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF) (Bruker Daltonik, Bremen, Germany). A species-level identification was considered reliable only if the obtained score exceeded 2.
Non-lactose fermenting (NLF) E. coli were identified phenotypically based on their inability to ferment lactose after overnight incubation on ML agar at 37 °C.
4.2. Antimicrobial Susceptibility Testing (AST)
At the LREC (University of Santiago de Compostela, Spain), antimicrobial susceptibility testing (AST) was performed using the disc diffusion method (Becton Dickinson, Sparks, MD, USA) on Mueller–Hinton (MH) agar (Oxoid, Madrid, Spain). A total of 20 antibiotics were tested: penicillin (ampicillin), penicillin + beta-lactamase inhibitors (amoxicillin-clavulanic acid), non-broad spectrum cephalosporins (cefuroxime); broad-spectrum cephalosporins (cefoxitin, cefotaxime, ceftazidime), carbapenems (ertapenem), monobactams (aztreonam), fluoroquinolones (nalidixic acid, ciprofloxacin), aminoglycosides (amikacin, gentamicin, tobramycin), tetracyclines (doxycycline), glycylcyclines (tigecycline), phosphonic acids (fosfomycin), nitrofurans (nitrofurantoin), folate pathway inhibitors (trimethoprim-sulfamethoxazole), polymyxins (colistin), and amphenicols (chloramphenicol).
AST results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing [83] clinical breakpoints when available, or Clinical & Laboratory Standards Institute [84] as an alternative. Isolates were classified as MDR if they exhibited resistance to at least one drug in three or more antimicrobial categories [23].
4.3. Detection and Typing of Antimicrobial Resistance Genes
The isolates were screened by PCR for clinically relevant blaESC genes using primers specific for SHV, CMY, CTX-M, VIM, IMP, OXA-48, KPC, and NDM. Sanger sequencing was performed when appropriate, in order to confirm gene variants or subtyping, as described elsewhere [85,86,87,88,89,90,91] (Table S6). In addition, mobile colistin resistance genes mcr-1 to mcr-5 were screened by PCR [92,93] (Table S7).
4.4. Molecular Characterization of E. coli: Virulence Traits, Phylogroup, Clonotype, and Virotype Assignment
For the molecular characterization of the E. coli collection, we followed the workflow scheme proposed by García-Meniño et al. [10]. Briefly, specific virulence-associated genes statistically linked to increased efficiency in urinary tract colonization were tested by PCR. Isolates were classified as UPEC if they harbored ≥3 of the following genes: chuA, fyuA, vat, or yfcV [24] (Table S8). The designation of extraintestinal pathogenic E. coli (ExPEC) status was attributed to isolates positive for ≥2 of these five markers: papAH, sfa/focDE, afa/draBC, kpsM II, or iutA [25,33,94] (Table S8).
The clonal structure of the E. coli collection was investigated using the phylogroup assignment method of Clermont et al. [26,27,95] (Table S9), which differentiates eight E. coli phylogroups (A, B1, B2, C, D, E, F, and G). Clonotype (CH) determination was performed by Sanger sequencing a 469-nucleotide (nt) internal region of the fumC gene (allele derived from MLST) and a 489-nt internal fragment of the fimH gene, encoding type 1 fimbrial adhesion [31] (Table S10).
To presumptively identify the pandemic CC131 lineage, we screened for phylogroup B2 along with rfbO25, fliCH4, and fliCH5. In addition, the fliCH5 flagellar-encoding gene is commonly associated with NFL E. coli ST1193 (Table S11) [96]. Isolates confirmed as O25b:H4-B2-CH40-30 were further characterized for their virotypes according to the protocol established by Dahbi et al. [30], based on the presence or absence of specific extraintestinal virulence factors (afa/draBC, afa operon FM955459, iroN, sat, ibeA, papG II, papG III, cnf1, hlyA, cdtB, kpsM II-K1, -K2, and -K5) (Tables S12 and S13) [97,98,99,100,101,102].
4.5. Phenotypic and Genotypic Detection of Hypervirulent K. pneumoniae
Hypervirulent K. pneumoniae (hvKp) isolates were initially screened based on the hypermucoviscous (HMV) phenotype using the string test, as described by Shon et al. [35]. String tests were performed on colonies grown on MH agar, ML agar, trypticase soy (TSA) agar (Oxoid, Madrid, Spain), and Columbia agar (5% sheep blood) (Oxoid, Madrid, Spain) incubated at 37 °C/24 h. A positive string test was defined as the formation of a viscous string ≥ 5 mm in length when a bacteriological inoculation loop was used to stretch bacterial colonies on an agar plate [35].
Additionally, the presence of molecular markers statistically associated with hvKp was assessed by PCR. Isolates were classified as hvKp if they carried all five hvKp virulence plasmid-associated genes: peg-344 (putative transporter), rmpA and rmpA2(regulators of the mucoid phenotype via increased capsule production), iroB (salmochelin siderophore biosynthesis), and iucA (aerobactin siderophore biosynthesis) [36,78] (Table S14).
4.6. Whole Genome Sequencing (WGS) and Bioinformatics Analysis
The E. coli isolate classified as fliCH5-B2 and CH14-64 was further analysed by WGS as described by García et al. [103]. Briefly, DNA was extracted with the DNeasey Blood and Tissue Kit (Qiagen, Hilen, Germany) according to the manufacturer’s instructions. After extraction, the DNA was quantified by an Invitrogen Qubit fluorimeter (Thermo Fisher Scientific, Waltham, MA, USA) and evaluated for purity using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). The DNA sequencing was performed using Illumina technology with a NovaSeq 6000 S4PE150 XP system to obtain 150 bp paired-end reads at Eurofins Genomics (Eurofins Genomics GmbH, Konstanz, Germany), following library preparation using the standard Illumina DNA Prep kit (Illumina Cambridge Ltd., Cambridge, UK). The quality of the paired-end Illumina reads was evaluated using FastQC v0.12.1.
The genome reconstruction and in silico analysis were performed as described elsewhere [104]. Briefly, the raw reads were assembled with the VelvetOptimiser.pl. script implemented in the online version of the PLAsmid Constellation NETwork (PLACNETw). The assembled contigs, with genomic size 5.0 Mbp, were analyzed using the bioinformatics tools of the Center for Genomic Epidemiology (CGE) as specified, and applying the thresholds suggested by default when required (minimum identity of 95% and coverage of 60%) for the presence of acquired genes and or chromosomal mutations mediating antimicrobial resistance (ResFinder 4.6.) [105,106]; for identification of acquired virulence genes (VirulenceFinder 2.0) [107,108]; plasmid replicon types (PlasmidFinder 2.1/pMLST 2.0) [109]; identification of clonotypes (CHTyper 1.0) [110]; and serotypes (SeroTypeFinder 2.0.1) [111]. Two different MLST (2.0.9) schemes were applied for phylogenetic typing [34,112]. Additionally, cgMLSTFinder1.2 was called for the core genome multi-locus typing (cgMLST) [106,113].
To investigate the phylogenetic relationship of the ST1193 isolate sequenced in this study, a comparative analysis was performed using CSI Phylogeny 1.4 with five previously described ST1193 isolates from a hospital in northwest Spain [9].
The pipeline was run, using the genome of LREC-269 as the reference for single-nucleotide polymorphism (SNP) calling, with the following parameters: min. depth at SNP positions ×10; min. relative depth at SNP positions: ×10; min. distance between SNPs (prune): 10 bp; min. SNP quality: 30; min. read mapping quality: 25, a min. Z-score of 1.96 and by ignoring heterozygous SNPs). Branch values represent substitutions per site. Bootstrap support for the consensus phylogenetic tree was calculated using 1000 replicates [114]. The resulting SNP matrix is shown in Table S3.
Finally, EnteroBase (https://EnteroBase.warwick.ac.uk/; accessed on 20 December 2024) was queried for ST1193 genomes based on the Achtman 7-gene MLST scheme. Specific core genome MLST (cgMLST) sequences were retrieved, and their raw reads were used for comparative genomic analysis (Table S4).
5. Conclusions
This study provides important insights into the epidemiology of MDR E. coli and K. pneumoniae in Algeria, highlighting the presence and diversity of E. coli CC131 CH40-30 and reporting, for the first time, E. coli ST1193 carrying CTX-M-15 in this region. The detection of virulent K. pneumoniae co-harbouring carbapenemase and ESBL resistance genes is particularly concerning, as it raises the potential for increased pathogenicity and treatment failure in clinical settings.
The co-occurrence of AMR and virulence factors underscores the need for enhanced surveillance and infection control measures. A simplified surveillance method based on virulence traits in E. coli and K. pneumoniae is proposed for early detection and outbreak monitoring. In addition, monitoring high-risk clones such as CC38 and OXA-48 is crucial for preventing further public health threats. A practical lab workflow for identifying high-risk E. coli clones associated with UTIs is outlined, emphasizing non-lactose fermenting isolates, antimicrobial susceptibility testing, and phylogroup/clonotyping.
This study had certain limitations, as the isolates were collected from only three healthcare facilities within a single province (Tebessa), which may limit the broader geographical representation of the findings. The sample size, while sufficient for exploratory analysis, may not fully capture the genetic and phenotypic diversity of E. coli and K. pneumoniae circulating in other regions of Algeria. Nevertheless, our findings are consistent with previous reports from northern and western Algeria, suggesting that the AMR patterns observed in uropathogens may reflect broader national trends. Furthermore, future studies involving in-depth genomic characterization, such as plasmid profiling and mobility analyses, are needed to understand the dynamics of resistance gene dissemination.
Given the significant resistance profiles observed here, targeted antibiotic optimization programs should focus on rational antibiotic use, improved prescription practices, and enhanced surveillance, especially in LMICs where MDR infections pose a major public health burden.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics14050485/s1, Table S1. Metadata and main traits of the 40 UTI E. coli isolates analysed in this study; Table S2. Metadata and main traits of the 17 UTI K. pneumoniae isolates analysed in this study; Table S3. Pairwise distance matrix calculated from SNP in the Algerian and Spanish ST1193 genomes; Table S4. Pairwise distance matrix calculated from SNP in the Algerian, Spanish and ESC_RA5887AA ST1193 genomes; Table S5. Targets and primers used for Escherichia coli and Klebsiella pneumoniae identification; Table S6. Primers used for the detection and/or sequencing of bla genes; Table S7. Primers used for the detection of mcr genes; Table S8. Targets and primers used to determine the UPEC and ExPEC status; Table S9. Targets and primers used for the phylogroup determination in E. coli; Table S10. Targets and primers to determine clonotypes (CH); Table S11. Primers used for the rbfO25b, H4 (fliCH4), and H5 (fliCH5) screening; Table S12. Targets and primers used in the virotype scheme of CC131 isolates; Table S13. Virotype designation scheme for CC131 E. coli; Table S14. Targets and primers used for virulence genes of Klebsiella pneumoniae.
Author Contributions
Conceptualization, V.G. and A.M.; methodology, H.Z., V.G. and A.M.; formal analysis, H.Z. and L.C.; investigation, H.Z., L.C., F.C., A.C., V.G. and A.M.; resources, A.M.; writing—original draft preparation, H.Z. and V.G.; writing—review and editing, V.G. and A.M.; supervision, V.G. and A.M.; funding acquisition, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants PID2022-143041OB-I00 funded by MICIU/AEI/10.13039/501100011033 and FEDER, UE, and ED431C 2021/11 from the Consellería de Cultura, Educación e Ordenación Universitaria (Xunta de Galicia) and FEDER.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequence of the ST1193 E. coli isolate was deposited in the European Nucleotide Archive (ENA) with the following accession ERS22141751 and BioSample SAMEA117078805 codes, as part of BioProject ID PRJEB82513.
Acknowledgments
Hajer Ziadi’s research at the Laboratorio de Referencia de Escherichia coli (LREC) in Lugo, Spain, under the supervision of Azucena Mora, was supported by an ERASMUS+ KA171 Doctoral Studies grant, in collaboration with the University of Mostaganem (Algeria) and the University of Santiago de Compostela (USC, Spain). The authors appreciate the support of the EPH Bouguerra Boularaas laboratory and GUESSOUM Amina from ELITE private laboratory, for assistance in isolating. Special thanks to Javier Fernández (Hospital Universitario Central de Asturias, HUCA, Oviedo, Spain) for his help with Klebsiella pneumoniae identification when PCR results were inconclusive.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yang, X.; Chen, H.; Zheng, Y.; Qu, S.; Wang, H.; Yi, F. Disease burden and long-term trends of urinary tract infections: A worldwide report. Front. Public. Health 2022, 10, 888205. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, R.; Murt, A. Epidemiology of urological infections: A global burden. World J. Urol. 2020, 38, 2669–2679. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. N. Am. 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- He, Y.; Zhao, J.; Wang, L.; Han, C.; Yan, R.; Zhu, P.; Qian, T.; Yu, S.; Zhu, X.; He, W.H. Epidemiological trends and predictions of urinary tract infections in the global burden of disease study 2021. Sci. Rep. 2025, 15, 4702. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fan, H.; Zi, H.; Hu, H.; Li, B.; Huang, J.; Luo, P.; Zeng, X. Global and Regional Burden of Bacterial Antimicrobial Resistance in Urinary Tract Infections in 2019. J. Clin. Med. 2022, 11, 2817. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; Lumbreras, P.; Lestón, L.; Álvarez-Álvarez, M.; García, V.; Hammerl, J.A.; Fernández, J.; Mora, A. Occurrence and Genomic Characterization of Clone ST1193 Clonotype 14-64 in Uncomplicated Urinary Tract Infections Caused by Escherichia coli in Spain. Microbiol. Spectr. 2022, 10, e0004122. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; García, V.; Lumbreras-Iglesias, P.; Fernández, J.; Mora, A. Fluoroquinolone resistance in complicated urinary tract infections: Association with the increased occurrence and diversity of clonal complex 131, together with ST1193. Front. Cell Infect. Microbiol. 2024, 14, 1351618. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Peirano, G.; Chen, L.; DeVinney, R.; Matsumura, Y. Escherichia coli ST1193: Following in the Footsteps of E. coli ST131. Antimicrob. Agents Chemother. 2022, 66, e0051122. [Google Scholar] [CrossRef]
- Cummins, E.A.; Snaith, A.E.; McNally, A.; Hall, R.J. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- WHO’s List of Medically Important Antimicrobials: A Risk Management Tool for Mitigating Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2024.
- Ait-Mimoune, N.; Hassaine, H.; Boulanoir, M. Bacteriological profile of urinary tract infections and antibiotic susceptibility of Escherichia coli in Algeria. Iran. J. Microbiol. 2022, 14, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Bakour, S.; Sahli, F.; Touati, A.; Rolain, J.M. Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbes New Infect. 2015, 3, 34–36. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brahmia, S.; Lalaoui, R.; Nedjai, S.; Djahmi, N.; Chettibi, S.; Rolain, J.M.; Bakour, S. First Clinical Cases of KPC-2-Producing Klebsiella pneumoniae ST258 in Algeria and Outbreak of Klebsiella pneumoniae ST101 Harboring blaOXA-48 Gene in the Urology Department of Annaba Hospital. Microb. Drug Resist. 2021, 27, 652–659. [Google Scholar] [CrossRef]
- Benbrahim, C.; Barka, M.S.; Benmahdi, L.; Zatout, A.; Khadir, A. Klebsiella pneumoniae producing extended spectrum β-lactamase in Regional Military University Hospital of Oran, Algeria: Antibiotic resistance, biofilm formation, and detection of blaCTX-M and blaTEM genes. Afr. J. Clin. Exper Microbiol. 2021, 22, 28–37. [Google Scholar] [CrossRef]
- Fares, R.; Debabza, M.; Mechai, A. Detection prevalence of extended spectrum β-lactamases production among Enterobacteriaceae isolated from urinary tract infections. Biosyst. Divers. 2023, 31, 163–169. [Google Scholar] [CrossRef]
- Nabti, L.Z.; Sahli, F.; Radji, N.; Mezaghcha, W.; Semara, L.; Aberkane, S.; Lounnas, M.; Solassol, J.; Didelot, M.N.; Jean-Pierre, H.; et al. High Prevalence of Multidrug-Resistant Escherichia coli in Urine Samples from Inpatients and Outpatients at a Tertiary Care Hospital in Sétif, Algeria. Microb. Drug Resist. 2019, 25, 386–393. [Google Scholar] [CrossRef]
- Zenati, F.; Barguigua, A.; Nayme, K.; Benbelaïd, F.; Khadir, A.; Bellahsene, C.; Bendahou, M.; Hafida, H.; Timinouni, M. Characterization of uropathogenic ESBL-producing Escherichia coli isolated from hospitalized patients in western Algeria. J. Infect. Dev. Ctries. 2019, 13, 291–302. [Google Scholar] [CrossRef]
- Touati, A.; Mairi, A. Carbapenemase-Producing Enterobacterales in Algeria: A Systematic Review. Microb. Drug Resist. 2020, 26, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Duarte, O.G.; Arzuza, O.; Urbina, D.; Bai, J.; Guerra, J.; Montes, O.; Puello, M.; Mendoza, K.; Castro, G.Y. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children’s diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog. Dis. 2010, 7, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L.T. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Johnson, J.R.; Murray, A.C.; Gajewski, A.; Sullivan, M.; Snippes, P.; Kuskowski, M.A.; Smith, K.E. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 2003, 47, 2161–2168. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.; Alonso, M.P.; Nicolas-Chanoine, M.H.; Dahbi, G.; Mora, A.; Blanco, J.E.; López, C.; Cortés, P.; Llagostera, M.; Leflon-Guibout, V.; et al. Molecular epidemiology of Escherichia coli producing extended-spectrum {beta}-lactamases in Lugo (Spain): Dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2009, 63, 1135–1141. [Google Scholar] [CrossRef]
- Clermont, O.; Dhanji, H.; Upton, M.; Gibreel, T.; Fox, A.; Boyd, D.; Mulvey, M.R.; Nordmann, P.; Ruppé, E.; Sarthou, J.L.; et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 2009, 64, 274–277. [Google Scholar] [CrossRef]
- Dahbi, G.; Mora, A.; Mamani, R.; López, C.; Alonso, M.P.; Marzoa, J.; Blanco, M.; Herrera, A.; Viso, S.; García-Garrote, F.; et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. 2014, 304, 1247–1257. [Google Scholar] [CrossRef]
- Weissman, S.J.; Johnson, J.R.; Tchesnokova, V.; Billig, M.; Dykhuizen, D.; Riddell, K.; Peggy, R.; Qin, X.; Butler-Wu, S.; Cookson, B.T.; et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Tchesnokova, V.; Johnston, B.; Clabots, C.; Roberts, P.L.; Billig, M.; Riddell, K.; Rogers, P.; Qin, X.; Butler-Wu, S.; et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 2013, 207, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Shon, A.S.; Bajwa, R.P.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Olson, R.; Fang, C.T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Hohnson, J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, e00776-18. [Google Scholar] [CrossRef]
- Institute for Health Metrics and Evaluation (IHME), University of Oxford. MICROBE. Available online: https://vizhub.healthdata.org/microbe (accessed on 23 March 2025).
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Pitout, J.D.D. Rapidly spreading Enterobacterales with OXA-48-like carbapenemases. J. Clin. Microbiol. 2025, 63, e0151524. [Google Scholar] [CrossRef]
- López-Cerero, L.; Salamanca, E.; Delgado-Valverde, M.; Rodríguez-Martínez, J.M.; Rodríguez-Baño, J.; Pascual, Á. Higher prevalence of CTX-M-27-producing Escherichia coli belonging to ST131 clade C1 among residents of two long-term care facilities in Southern Spain. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 335–338. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Mobley, H.L.T. Chapter 9—Uropathogenic Escherichia coli. In Escherichia coli, Pathotypes and Principles of Pathogenesis, 2nd ed.; Donnenberg, M., Ed.; Elsevier Inc.: Amsterdam, The Netherlands; Academic Press: Amsterdam, The Netherlands, 2013; pp. 275–304. [Google Scholar]
- Wang, M.C.; Fan, Y.H.; Zhang, Y.Z.; Bregente, C.J.B.; Lin, W.H.; Chen, C.A.; Lin, T.L.; Kao, C.Y. Characterization of uropathogenic Escherichia coli phylogroups associated with antimicrobial resistance, virulence factor distribution, and virulence-related phenotypes. Infect. Genet. Evol. 2023, 114, 105493. [Google Scholar] [CrossRef] [PubMed]
- Colpan, A.; Johnston, B.; Porter, S.; Clabots, C.; Anway, R.; Thao, L.; Kuskowski, M.A.; Tchesnokova, V.; Solurenko, E.V.; Johnson, J.R. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin. Infect. Dis. 2013, 57, 1256–1265. [Google Scholar] [CrossRef]
- Banerjee, R.; Johnson, J.R. A new clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 2014, 58, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; DeVinney, R. ST131: A multidrug-resistant clone primed for global domination. F1000Research 2017, 6, 195. [Google Scholar] [CrossRef] [PubMed]
- Baba Ahmed-Kazi Tani, Z.; Decré, D.; Genel, N.; Boucherit-Otmani, Z.; Arlet, G.; Drissi, M. Molecular and epidemiological characterization of enterobacterial multidrug-resistant strains in Tlemcen Hospital (Algeria) (2008–2010). Microb. Drug Resist. 2013, 19, 185–190. [Google Scholar] [CrossRef]
- Agabou, A.; Pantel, A.; Ouchenane, Z.; Lezzar, N.; Khemissi, S.; Satta, D.; Lavigne, J.P. First description of OXA-48-producing Escherichia coli and the pandemic clone ST131 from patients hospitalised at a military hospital in Algeria. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1641–1646. [Google Scholar] [CrossRef]
- Yahiaoui, M.; Robin, F.; Bakour, R.; Hamidi, M.; Bonnet, R.; Messai, Y. Antibiotic Resistance, Virulence, and Genetic Background of Community-Acquired Uropathogenic Escherichia coli from Algeria. Microb. Drug Resist. 2015, 21, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Brahmi, S.; Dunyach-Rémy, C.; Touati, A.; Lavigne, J.P. CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin. Microbiol. Infect. 2015, 21, e18–e20. [Google Scholar] [CrossRef] [PubMed]
- Chenouf, N.S.; Carvalho, I.; Messaï, C.R.; Ruiz-Ripa, L.; Mama, O.M.; Titouche, Y.; Zitouni, A.; Hakem, A.; Torres, C. Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Brolier Liver in the Center of Algeria, with Detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli. Microb. Drug Resist. 2021, 27, 268–276. [Google Scholar] [CrossRef]
- Bachiri, T.; Bakour, S.; Ladjouzi, R.; Thongpan, L.; Rolain, J.M.; Touati, A. High rates of CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in wild boars and Barbary macaques in Algeria. J. Glob. Antimicrob. Resist. 2017, 8, 35–40. [Google Scholar] [CrossRef]
- Johnson, T.J.; Elnekave, E.; Miller, E.A.; Munoz-Aguayo, J.; Flores Figueroa, C.; Johnston, B.; Nielson, D.W.; Logue, C.M.; Johnson, J.R. Phylogenomic Analysis of Extraintestinal Pathogenic Escherichia coli Sequence Type 1193, an Emerging Multidrug-Resistant Clonal Group. Antimicrob. Agents Chemother. 2019, 63, e01913-18. [Google Scholar] [CrossRef] [PubMed]
- Wyrsch, E.R.; Bushell, R.N.; Marenda, M.S.; Browning, G.F.; Djordjevic, S.P. Global Phylogeny and F Virulence Plasmid Carriage in Pandemic Escherichia coli ST1193. Microbiol. Spectr. 2022, 10, e0255422. [Google Scholar] [CrossRef]
- Birgy, A.; Madhi, F.; Jung, C.; Levy, C.; Cointe, A.; Bidet, P.; Hobson, C.A.; Bechet, S.; Sobral, E.; Vuthien, H.; et al. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J. Antimicrob. Chemother. 2020, 75, 96–105. [Google Scholar] [CrossRef]
- Valenza, G.; Werner, M.; Eisenberger, D.; Nickel, S.; Lehner-Reindl, V.; Höller, C.; Lehner-Reindl, V.; Höller, C.; Bagdan, C. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J. Glob. Antimicrob. Resist. 2019, 17, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhang, J.; Yao, K.; Gao, W.; Wang, Y. Molecular characteristics of the new emerging global clone ST1193 among clinical isolates of Escherichia coli from neonatal invasive infections in China. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Oh, T.; Nam, Y.S.; Cho, S.Y.; Lee, H.J. Prevalence of ST131 and ST1193 Among Bloodstream Isolates of Escherichia coli not Susceptible to Ciprofloxacin in a Tertiary Care University Hospital in Korea, 2013–2014. Clin. Lab. 2017, 63, 1541–1543. [Google Scholar] [CrossRef]
- Nguyen, Q.; Nguyen, T.T.N.; Pham, P.; Chau, V.; Nguyen, L.P.H.; Nguyen, T.D.; Ha, T.T.; Le, N.T.Q.; Vu, D.T.; Baker, S.; et al. Genomic insights into the circulation of pandemic fluoroquinolone-resistant extra-intestinal pathogenic Escherichia coli ST1193 in Vietnam. Microb. Genom. 2021, 7, 000733. [Google Scholar] [CrossRef]
- Tchesnokova, V.L.; Rechkina, E.; Larson, L.; Ferrier, K.; Weaver, J.L.; Schroeder, D.W.; She, R.; Butler-Wu, S.M.; Aguero-Rosenfeld, M.E.; Zerr, D.; et al. Rapid and Extensive Expansion in the United States of a New Multidrug-resistant Escherichia coli Clonal Group, Sequence Type 1193. Clin. Infect. Dis. 2019, 68, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Yehouenou, C.L.; Bogaerts, B.; De Keersmaecker, S.C.J.; Roosens, N.H.C.; Marchal, K.; Tchiakpe, E.; Affolabi, D.; Simon, A.; Dossou, F.M.; Vanneste, K.; et al. Whole-Genome Sequencing-Based Antimicrobial Resistance Characterization and Phylogenomic Investigation of 19 Multidrug-Resistant and Extended-Spectrum Beta-Lactamase-Positive Escherichia coli Strains Collected From Hospital Patients in Benin in 2019. Front. Microbiol. 2021, 12, 752883. [Google Scholar] [CrossRef] [PubMed]
- Byarugaba, D.K.; Erima, B.; Wokorach, G.; Alafi, S.; Kibuuka, H.; Mworozi, E.; Musinguzi, A.K.; Kiyengo, J.; Najjuka, F.; Wabwire-Mangen, F. Resistome and virulome of high-risk pandemic clones of multidrug-resistant extra-intestinal pathogenic Escherichia coli (ExPEC) isolated from tertiary healthcare settings in Uganda. PLoS ONE 2023, 18, e0294424. [Google Scholar] [CrossRef]
- Gomi, R.; Matsumura, Y.; Yamamoto, M.; Tanaka, M.; Komakech, A.J.; Matsuda, T.; Harada, H. Genomic surveillance of antimicrobial-resistant Escherichia coli in fecal sludge and sewage in Uganda. Water Res. 2024, 248, 120830. [Google Scholar] [CrossRef] [PubMed]
- Achtman, M.; Zhou, Z.; Charlesworth, J.; Baxter, L. EnteroBase: Hierarchical clustering of 100 000s of bacterial genomes into species/subspecies and populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20210240. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Nordmann, P.; Imirzalioglu, C.; Yao, Y.; Falgenhauer, J.; Hauri, A.M.; Heinmüller, P.; Chakraborty, T. Cross-border emergence of clonal lineages of ST38 Escherichia coli producing the OXA-48-like carbapenemase OXA-244 in Germany and Switzerland. Int. J. Antimicrob. Agents 2020, 56, 106157. [Google Scholar] [CrossRef] [PubMed]
- Grevskott, D.H.; Radisic, V.; Salvà-Serra, F.; Moore, E.R.B.; Akervold, K.S.; Victor, M.P.; Marathe, N.P. Emergence and dissemination of epidemic-causing OXA-244 carbapenemase-producing Escherichia coli ST38 through hospital sewage in Norway, 2020–2022. J. Hosp. Infect. 2024, 145, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, P.; Hastak, P.; DeMaere, M.; Wyrsch, E.; Li, D.; Elankumaran, P.; Dolejska, M.; Browning, G.F.; Marenda, M.S.; Gottlieb, T.; et al. Phylogenomic analysis of a global collection of Escherichia coli ST38: Evidence of interspecies and environmental transmission? mSystems 2023, 8, e0123622. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Loucif, L.; Ayachi, A.; Guehaz, K.; Bendjama, E.; Rolain, J.M. Migratory White Stork (Ciconia ciconia): A Potential Vector of the OXA-48-Producing Escherichia coli ST38 Clone in Algeria. Microb. Drug Resist. 2018, 24, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Tafoukt, R.; Touati, A.; Leangapichart, T.; Bakour, S.; Rolain, J.M. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017, 120, 185–189. [Google Scholar] [CrossRef]
- Yagoubat, M.; Ould El-Hadj-Khelil, A.; Malki, A.; Bakour, S.; Touati, A.; Rolain, J.M. Genetic characterisation of carbapenem-resistant Gram-negative bacteria isolated from the University Hospital Mohamed Boudiaf in Ouargla, southern Algeria. J. Glob. Antimicrob. Resist. 2017, 8, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Belmahdi, M.; Bakour, S.; Al Bayssari, C.; Touati, A.; Rolain, J.M. Molecular characterisation of extended-spectrum β-lactamase- and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. J. Glob. Antimicrob. Resist. 2016, 6, 108–112. [Google Scholar] [CrossRef]
- Abderrahim, A.; Djahmi, N.; Pujol, C.; Nedjai, S.; Bentakouk, M.C.; Kirane-Gacemi, D.; Dekhil, M.; Sotto, A.; Lavigne, J.P.; Pantel, A. First Case of NDM-1-Producing Klebsiella pneumoniae in Annaba University Hospital, Algeria. Microb. Drug Resist. 2017, 23, 895–900. [Google Scholar] [CrossRef]
- Zemmour, A.; Dali-Yahia, R.; Maatallah, M.; Saidi-Ouahrani, N.; Rahmani, B.; Benhamouche, N.; Al-Farsi, H.M.; Giske, C.G. High-risk clones of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from the University Hospital Establishment of Oran, Algeria (2011–2012). PLoS ONE 2021, 16, e0254805. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J.; Lu, M.; Shen, N.; Du, P. Dissemination of the mobilised RND efflux pump gene cluster tmexCD-toprJ among Klebsiella pneumoniae. Lancet Microbe 2023, 4, e135. [Google Scholar] [CrossRef]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed]
- Al Ismail, D.; Campos-Madueno, E.I.; Donà, V.; Endimiani, A. Hypervirulent Klebsiella pneumoniae (hvKP): Overview, Epidemiology, and Laboratory Detection. Pathog. Immun. 2025, 10, 80–119. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, G.; Yu, Y.; Li, N.; Chen, M.; Jin, R.; Jiao, Y.; Wu, H. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 2014, 58, 225–232. [Google Scholar] [CrossRef]
- Russo, T.A.; MacDonald, U. The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. mSphere 2020, 5, e00850-19. [Google Scholar] [CrossRef]
- Russo, T.A.; Alvarado, C.L.; Davies, C.J.; Drayer, Z.J.; Carlino-MacDonald, U.; Hutson, A.; Luo, T.L.; Martin, M.J.; Corey, B.W.; Moser, K.A.; et al. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. mBio 2024, 15, e0286723. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Du, P.; Du, C.; Yang, P.; Shen, N.; Russo, T.A.; Liu, C. Genomically defined hypervirulent Klebsiella pneumoniae contributed to early-onset increased mortality. Nat. Commun. 2025, 16, 2096. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Emergence of Hypervirulent Klebsiella pneumoniae ST23 Carrying Carbapenemase Genes in EU/EEA Countries, First Update; ECDC: Stockholm, Sweden, 2024; ISBN 978-92-9498-691-7. [Google Scholar] [CrossRef]
- Institut Pasteur d’Algérie. Surveillance de la Résistance des Bactéries Aux Antibiotiques; 23ème Rapport D’évaluation 2023; Algerian Antimicrobial Resistance Network: Algiers, Algeria, 2023. [Google Scholar]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Nicolas-Chanoine, M.H.; Decré, D.; Brisse, S. Development of a multiplex PCR assay for identification of Klebsiella pneumoniae hypervirulent clones of capsular serotype K2. J. Med. Microbiol. 2014, 63 Pt 12, 1608–1614. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical Breakpoints. 2025. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 10 January 2025).
- CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; CLSI Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- García-Meniño, I.; García, V.; Alonso, M.P.; Blanco, J.E.; Blanco, J.; Mora, A. Clones of enterotoxigenic and Shiga toxin-producing Escherichia coli implicated in swine enteric colibacillosis in Spain and rates of antibiotic resistance. Vet. Microbiol. 2021, 252, 108924. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Viso, S.; López, C.; Alonso, M.P.; García-Garrote, F.; Dabhi, G.; Mamani, R.; Herrera, A.; Marzoa, J.; Blanco, M.; et al. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet Microbiol. 2013, 167, 506–512. [Google Scholar] [CrossRef]
- Saladin, M.; Cao, V.T.; Lambert, T.; Donay, J.L.; Herrmann, J.L.; Ould-Hocine, Z.; Verdet, C.; Delisle, F.; Philippon, A.; Arlet, G. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Simarro, E.; Navarro, F.; Ruiz, J.; Miró, E.; Gómez, J.; Mirelis, B. Salmonella enterica serovar virchow with CTX-M-like beta-lactamase in Spain. J. Clin. Microbiol. 2000, 38, 4676–4678. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine enteric colibacillosis in Spain: Pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 17-00672. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Le Bouguenec, C.; Archambaud, M.; Labigne, A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1189–1193. [Google Scholar] [CrossRef]
- Lescat, M.; Clermont, O.; Woerther, P.L.; Glodt, J.; Dion, S.; Skurnik, D.; Djossou, F.; Dupont, C.; Perroz, G.; Picard, B.; et al. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ. Microbiol. Rep. 2013, 5, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Herrrera, A.; Lopez, C.; Dahbi, G.; Mamani, R.; Pita, J.M.; Alonso, M.P.; Llovo, J.; Bernardez, M.I.; Blanco, J.E.; et al. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 2011, 4, 121–141. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A.; Tarr, P.I.; Carlino, U.; Bilge, S.S.; Vary, J.C.; Stell, A.L. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN (E. coli), among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 2000, 68, 3040–3047. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Gajewski, A.; Lesse, A.J.; Russo, T.A. Extraintestinal pathogenic Escherichia coli as a cause of invasive non urinary infections. J. Clin. Microbiol. 2003, 41, 5798–5802. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Terai, A.; Yuri, K.; Kurazono, H.; Takeda, Y.; Yoshida, O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol. 1995, 12, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Tóth, I.; Hérault, F.; Beutin, L.; Oswald, E. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: Establishment of the existence of a new cdt variant (Type IV). J. Clin. Microbiol. 2003, 41, 4285–4291. [Google Scholar] [CrossRef] [PubMed]
- Moulin-Schouleur, M.; Schouler, C.; Tailliez, P.; Kao, M.-R.; Brée, A.; Germon, P.; Oswald, E.; Mainil, J.; Blanco, M.; Blanco, J. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J Clin. Microbiol. 2006, 44, 3484–3492. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; O’Bryan, T.T. Detection of the Escherichia coli group 2 polysaccharide capsule synthesis Gene kpsM by a rapid and specific PCR-based assay. J. Clin. Microbiol. 2004, 42, 1773–1776. [Google Scholar] [CrossRef]
- García, V.; García-Meniño, I.; Gómez, V.; Jiménez-Orellana, M.; Méndez, A.; Aguarón, A.; Roca, E.; Mora, A. Mobile colistin resistance (MCR), extended-spectrum beta-lactamase (ESBL) and multidrug resistance monitoring in Escherichia coli (commensal and pathogenic) in pig farming: Need of harmonized guidelines and clinical breakpoints. Front. Microbiol. 2022, 13, 1042612. [Google Scholar] [CrossRef]
- García-Meniño, I.; Díaz-Jiménez, D.; García, V.; de Toro, M.; Flament-Simon, S.C.; Blanco, J.; Mora, A. Genomic Characterization of Prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli Within Swine Enteric Colibacillosis in Spain. Front. Microbiol. 2019, 10, 2469. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoer, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesoe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a Web Tool for Subtyping of Extraintestinal Pathogenic Escherichia coli Based on the fumC and fimH alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).