High Prevalence of Multidrug-Resistant Bacteria in the Trachea of Intensive Care Units Admitted Patients: Evidence from a Bangladeshi Hospital
Abstract
1. Introduction
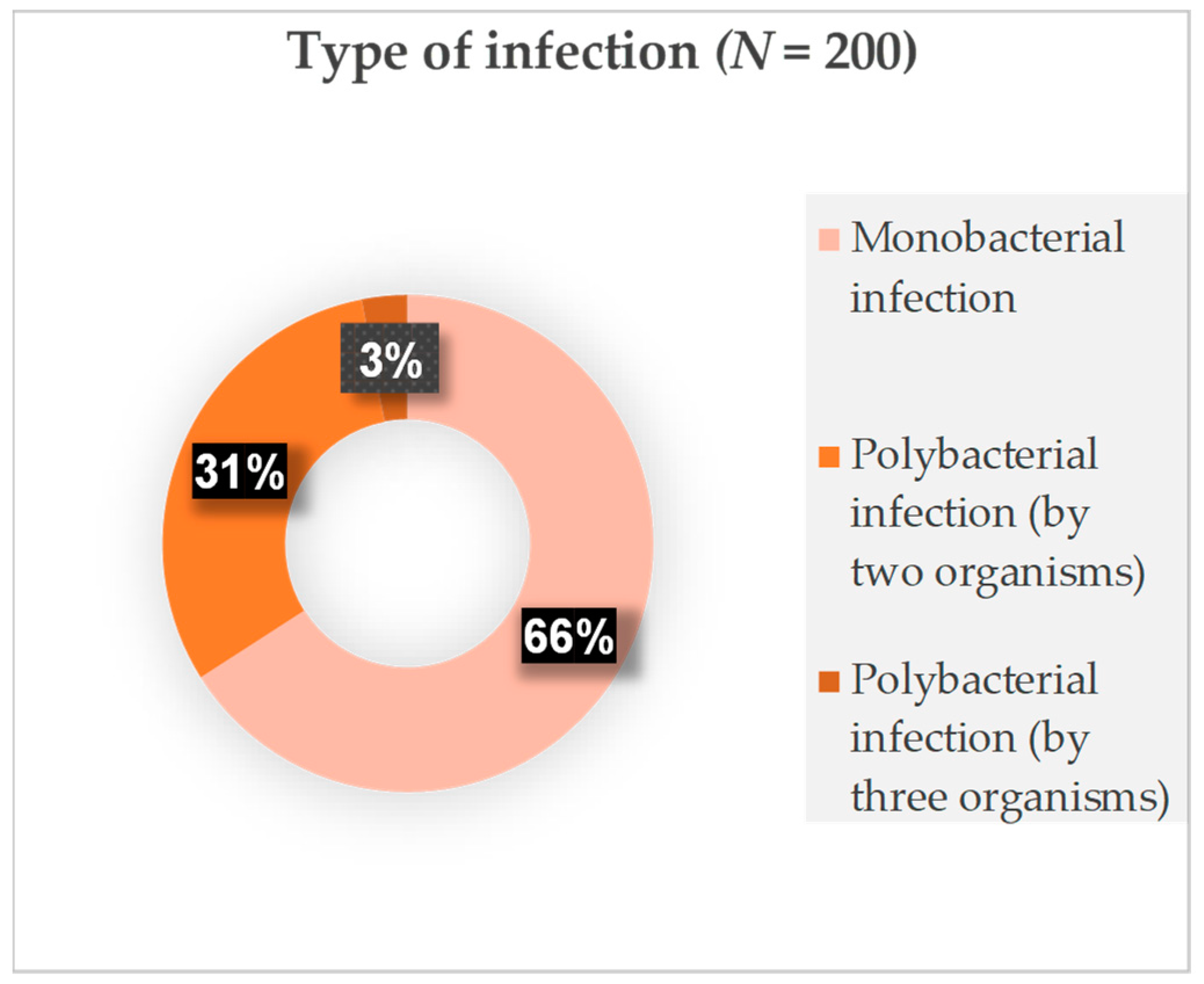
2. Results
3. Discussion
Limitations
4. Materials and Methods
4.1. Study Design and Site
4.2. Study Population and Sample Collection
4.3. Culture and Identification of Organisms
4.4. Antimicrobial Susceptibility Test
4.5. Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zilahi, G.; Artigas, A.; Martin-Loeches, I. What’s new in multidrug-resistant pathogens in the ICU? Ann. Intensive Care 2016, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Heddini, A.; Cars, O.; Qiang, S.; Tomson, G. Antibiotic resistance in China—A major future challenge. Lancet 2009, 373, 30. [Google Scholar] [CrossRef] [PubMed]
- Bonten, M.J.M.; Mascini, E.M. The hidden faces of the epidemiology of antibiotic resistance. Intensive Care Med. 2003, 29, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lo Yan Yam, E.; Hsu, L.Y.; Yap, E.P.H.; Yeo, T.W.; Lee, V.; Schlundt, J.; Lwin, M.O.; Limmathurotsakul, D.; Jit, M.; Dedon, P.C.; et al. Antimicrobial Resistance in the Asia Pacific region: A meeting report. Antimicrob. Resist. Infect. Control 2019, 8, 202. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; A critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Jamil, S.M.T.; Faruq, M.O.; Saleheen, S.; Biswas, P.K.; Hossain, M.S.; Hossain, S.Z.; Arifuzzaman, M.; Basu, B.K.; Kabir, A.; Alam, F.; et al. Microorganisms profile and their antimicrobial resistance pattern isolated from the lower respiratory tract of mechanically ventilated patients in the intensive care unit of a tertiary care hospital in Dhaka. J. Med. 2016, 17, 91–94. [Google Scholar] [CrossRef][Green Version]
- Bhandari, P.; Thapa, G.; Pokhrel, B.M.; Bhatta, D.R.; Devkota, U. Nosocomial Isolates and Their Drug Resistant Pattern in ICU Patients at National Institute of Neurological and Allied Sciences, Nepal. Int. J. Microbiol. 2015, 2015, 572163. [Google Scholar] [CrossRef]
- Frattari, A.; Savini, V.; Polilli, E.; Di Marco, G.; Lucisano, G.; Corridoni, S.; Spina, T.; Costantini, A.; Nicolucci, A.; Fazii, P.; et al. Control of Gram-negative multidrug resistant microorganisms in an Italian ICU: Rapid decline as a result of a multifaceted intervention, including conservative use of antibiotics. Int. J. Infect. Dis. 2019, 84, 153–162. [Google Scholar] [CrossRef]
- Japoni, A.; Vazin, A.; Hamedi, M.; Davarpanah, M.A.; Alborzi, A.; Rafaatpour, N. Multidrug-resistant bacteria isolated from intensive-care-unit patient samples. Braz. J. Infect. Dis. 2009, 13, 118–122. [Google Scholar] [CrossRef]
- Jesmin, H.; Ahasan, H.N.; Asaduzzaman, M.; Islam, A.M. Antimicrobial Resistance among Intensive Care Unit Patients in A Tertiary Care Hospital of Bangladesh. Bangladesh J. Med. 2021, 32, 5–11. [Google Scholar] [CrossRef]
- Lim, S.M.; Webb, S. Nosocomial bacterial infections in Intensive Care Units. I: Organisms and mechanisms of antibiotic resistance. Anaesthesia 2005, 60, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; Rello, J.; Marshall, J.; Silva, E.; Anzueto, A.; Martin, C.D.; Moreno, R.; Lipman, J.; Gomersall, C.; Sakr, Y.; et al. International Study of the Prevalence and Outcomes of Infection in Intensive Care Units. JAMA 2009, 302, 2323–2329. [Google Scholar] [CrossRef] [PubMed]
- Gonlugur, U.; Bakici, M.Z.; Akkurt, I.; Efeoglu, T. Antibiotic susceptibility patterns among respiratory isolates of Gram-negative bacilli in a Turkish university hospital. BMC Microbiol. 2004, 4, 32. [Google Scholar] [CrossRef]
- Lagacé-Wiens, P.R.S.; DeCorby, M.R.; Baudry, P.J.; Hoban, D.J.; Karlowsky, J.A.; Zhanel, G.G.; CAN-ICU Study Group. Differences in Antimicrobial Susceptibility in Escherichia coli from Canadian Intensive Care Units Based on Regional and Demographic Variables. Can. J. Infect. Dis. Med. Microbiol. 2008, 19, 568458. [Google Scholar] [CrossRef]
- Mukhopadhyay, C.; Bhargava, A.; Ayyagari, A. Role of mechanical ventilation & development of multidrug resistant organisms in hospital acquired pneumonia. Indian J. Med. Res. 2003, 118, 229–235. [Google Scholar]
- Singh, A.K.; Sen, M.R.; Anupurba, S.; Bhattacharya, P. Antibiotic sensitivity pattern of the bacteria isolated from nosocomial infections in ICU. J. Commun. Dis. 2002, 34, 257–263. [Google Scholar]
- Talbot, G.H.; Bradley, J.; Edwards, J.E., Jr.; Gilbert, D.; Scheld, M.; Bartlett, J.G. Bad Bugs Need Drugs: An Update on the Development Pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 2006, 42, 657–668. [Google Scholar] [CrossRef]
- Khan, F.; Khan, A.; Kazmi, S.U. Prevalence and susceptibility pattern of multi drug resistant clinical isolates of Pseudomonas aeruginosa in Karachi. Pak. J. Med. Sci. 2014, 30, 951. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef]
- Dereli, N.; Ozayar, E.; Degerli, S.; Sahin, S.; Koç, F. Three-Year Evaluation of Nosocomial Infection Rates of the ICU. Braz. J. Anesthesiol. 2013, 63, 73–84. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Islam, Q.T.; Siddiqui, M.M.R.; Raz, F.; Asrafuzzaman, M.; Amin, M.R. Patterns of antimicrobial resistance among intensive care unit patients of a private medical college hospital in Dhaka. Bangladesh J. Med. 2014, 25, 47–51. [Google Scholar] [CrossRef]
- Kumari, H.B.V.; Nagarathna, S.; Chandramuki, A. Antimicrobial resistance pattern among aerobic gram-negative bacilli of lower respiratory tract specimens of intensive care unit patients in a neurocentre. Indian J. Chest Dis. Allied Sci. 2007, 49, 19–22. [Google Scholar] [PubMed]
- Moolchandani, K.; Sastry, A.S.; Deepashree, R.; Sistla, S.; Harish, B.; Mandal, J. Antimicrobial resistance surveillance among intensive care units of a tertiary care hospital in Southern India. J. Clin. Diagn. Res. 2017, 11, DC01. [Google Scholar] [PubMed]
- Lachhab, Z.; Frikh, M.; Maleb, A.; Kasouati, J.; Doghmi, N.; Ben Lahlou, Y.; Belefquih, B.; Lemnouer, A.; Elouennass, M. Bacteraemia in Intensive Care Unit: Clinical, Bacteriological, and Prognostic Prospective Study. Can. J. Infect. Dis. Med. Microbiol. 2017, 2017, 4082938. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.L.; da Silva, B.C.M.; Rezende, G.S.; Nakamura-Silva, R.; Pitondo-Silva, A.; Campanini, E.B.; Brito, M.C.; da Silva, E.M.; Freire, C.C.D.M.; Cunha, A.F.D.; et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2019, 9, 3198. [Google Scholar] [CrossRef]
- Yadav, S.K.; Bhujel, R.; Mishra, S.K.; Sharma, S.; Sherchand, J.B. Emergence of multidrug-resistant non-fermentative gram negative bacterial infection in hospitalized patients in a tertiary care center of Nepal. BMC Res. Notes 2020, 13, 319. [Google Scholar] [CrossRef]
- Anupurba, S.; Sen, M.R. Antimicrobial resistance profile of bacterial isolates from Intensive Care Unit: Changing trends. J. Commun. Dis. 2005, 37, 58–65. [Google Scholar]
- CARA. Canadian Antimicrobial Resistance Alliance (CARA). Available online: https://www.can-r.com/ (accessed on 15 January 2023).
- Biberg, C.A.; Rodrigues, A.C.S.; Carmo, S.F.D.; Chaves, C.E.V.; Gales, A.C.; Chang, M.R. KPC-2-producing Klebsiella pneumoniae in a hospital in the Midwest region of Brazil. Braz. J. Microbiol. 2015, 46, 501–504. [Google Scholar] [CrossRef][Green Version]
- Elgendy, S.G.; Hameed, M.R.A.; El-Mokhtar, M.A. Tigecycline resistance among Klebsiella pneumoniae isolated from febrile neutropenic patients. J. Med. Microbiol. 2018, 67, 972. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Zhang, Y.; Wang, Q.; Zhao, C.; Li, H.; He, W.; Zhang, F.; Wang, Z.; Li, S.; et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: Role of the global regulator RamA and its local repressor RamR. Int. J. Antimicrob. Agents 2015, 45, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Azzab, M.M.; El Sokkary, R.H.; Tawfeek, M.M.; Gebriel, M.G. Multidrug-resistant bacteria among patients with ventilator-associated pneumonia in an emergency intensive care unit, Egypt. EMHJ-East. Mediterr. Health J. 2016, 22, 894–903. [Google Scholar] [CrossRef]
- Tehrani, S.; Saffarfar, V.; Hashemi, A.; Abolghasemi, S. A Survey of Genotype and Resistance Patterns of Ventilator-Associated Pneumonia Organisms in ICU Patients. Tanaffos 2019, 18, 215–222. [Google Scholar] [PubMed]
- Deyno, S.; Fekadu, S.; Astatkie, A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: A meta-analysis. Antimicrob. Resist. Infect. Control 2017, 6, 85. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Karageorgopoulos, D.E.; Leptidis, J.; Korbila, I.P. MRSA in Africa: Filling the global map of antimicrobial resistance. PLoS ONE 2013, 8, e68024. [Google Scholar] [CrossRef]
- Ak, O.; Batirel, A.; Özer, S.; Colakoglu, S. Nosocomial infections and risk factors in the intensive care unit of a teaching and research hospital: A prospective cohort study. Med. Sci. Monit. 2011, 17, PH29–PH34. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Das, S.; Chawan, N.S.; Hazra, A. Nosocomial infections in the intensive care unit: Incidence, risk factors, outcome and associated pathogens in a public tertiary teaching hospital of Eastern India. Indian J. Crit. Care Med. 2015, 19, 14–20. [Google Scholar] [CrossRef]
- Irwin, R.S.; Pratter, M.R. The Clinical Value of Pharmacologic Bronchoprovocation Challenge. Med. Clin. N. Am. 1990, 74, 767–778. [Google Scholar] [CrossRef]
- Whitman, W.B. (Ed.) Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Basavaraju, M.; Gunashree, B. Escherichia coli: An Overview of Main Characteristics; IntechOpen: London, UK, 2023. [Google Scholar] [CrossRef]
- Cetrimide Agar|Principle|Preparation|Results. Available online: https://microbiologie-clinique.com/cetrimide-agar.html (accessed on 1 August 2023).
- Prasad, M.; Shetty, S.K.; Nair, B.; Pal, S.; Madhavan, A. A novel and improved selective media for the isolation and enumeration of Klebsiella species. Appl. Microbiol. Biotechnol. 2022, 106, 8273–8284. [Google Scholar] [CrossRef]
- Public Health England. Identification of Moraxella species and Morphologically Similar Organisms. UK Standards for Microbiology Investigations. ID 11 Issue 3. 2015. Available online: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality-andconsistency-in-clinical-laboratories (accessed on 1 February 2021).
- Libretexts. 3.13: Levine EMB Agar. Biology LibreTexts. 2021. Available online: https://bio.libretexts.org/Learning_Objects/Laboratory_Experiments/Microbiology_Labs/Microbiology_for_Allied_Health_Students%3A_Lab_Manual/3.13%3A_Levine_EMB_Agar#:~:text=Escherichia%20coli%20colonies%20produce%20a,buff%20color%20with%20darker%20centers (accessed on 1 March 2021).
- Ajao, A.; Robinson, G.; Lee, M.S.; Ranke, T.D.; Venezia, R.A.; Furuno, J.P.; Harris, A.D.; Johnson, J.K. Comparison of culture media for detection of Acinetobacter baumannii in surveillance cultures of critically ill patients. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1425–1430. [Google Scholar] [CrossRef]
- Tankeshwar, A.; Tankeshwar, A. Proteus species: Properties, Diseases, Identification. Microbe Online. 2022. Available online: https://microbeonline.com/proteus-species-properties-diseases-identification/#:~:text=Proteus%20grows%20on%20the%20Blood,or%20colorless%20(NLF)%20colonies.&text=increase%20in%20agar%20concentration%20in,instead%20of%201%2D2%25 (accessed on 1 April 2022).
- Missiakas, D.M.; Schneewind, O. Growth and laboratory maintenance of Staphylococcus aureus. Curr. Protoc. Microbiol. 2013, 28, 9C-1. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6211185/ (accessed on 1 March 2021). [CrossRef] [PubMed]
- Hossain, Z. Bacteria: Streptococcus. In Encyclopedia of Food Safety; Motarjemi, Y., Ed.; Academic Press: Waltham, MA, USA, 2014; pp. 535–545. [Google Scholar] [CrossRef]
- M-enterococcus (ME) agar. Prog. Ind. Microbiol. 2003, 37, 524–526. [CrossRef]
- Aryal, S. Biochemical Test and Identification of E. coli. Microbiology Info.com. 2022. Available online: https://microbiologyinfo.com/biochemical-test-and-identification-of-e-coli/ (accessed on 1 April 2022).
- Aryal, S. Biochemical Test and Identification of Pseudomonas aeruginosa. Microbiology Info.com. 2022. Available online: https://microbiologyinfo.com/biochemical-test-and-identification-of-pseudomonas-aeruginosa/ (accessed on 1 April 2022).
- Aryal, S. Biochemical Test of Klebsiella pneumoniae. Microbe Notes. 2022. Available online: https://microbenotes.com/biochemical-test-of-klebsiella-pneumoniae/ (accessed on 1 April 2022).
- Sheikh, A.F.; Feghhi, M.; Torabipour, M.; Saki, M.; Veisi, H. Low prevalence of Moraxella catarrhalis in the patients who suffered from conjunctivitis in the southwest of Iran. BMC Res. Notes 2020, 13, 547. [Google Scholar] [CrossRef]
- Aryal, S. Biochemical Test of Enterobacter aerogenes. Microbe Notes. 2022. Available online: https://microbenotes.com/biochemical-test-of-enterobacter-aerogenes/ (accessed on 1 March 2021).
- Aryal, S. Biochemical Test of Acinetobacter baumannii. Microbe Notes. 2022. Available online: https://microbenotes.com/biochemical-test-of-acinetobacter-baumannii/ (accessed on 1 April 2022).
- Aryal, S. Biochemical Test and Identification of Proteus mirabilis. Microbiology Info.com. 2022. Available online: https://microbiologyinfo.com/biochemical-test-and-identification-of-proteus-mirabilis/ (accessed on 1 April 2022).
- Aryal, S. Biochemical Test and Identification of Staphylococcus aureus. Microbiology Info.com. 2022. Available online: https://microbiologyinfo.com/biochemical-test-and-identification-of-staphylococcus-aureus/ (accessed on 1 April 2022).
- Batra, S. Biochemical Tests for Streptococcus Pyogenes. Paramedics World. 2020. Available online: https://paramedicsworld.com/streptococcus-pyogenes/biochemical-tests-for-streptococcus-pyogenes/medical-paramedical-studynotes (accessed on 1 March 2021).
- Aryal, S. Biochemical Test of Enterococcus faecium. Microbe Notes. 2022. Available online: https://microbenotes.com/biochemical-test-of-enterococcus-faecium/ (accessed on 1 April 2022).

| Age Interval (Years) | Percentage Distribution among Age Groups | Number of Patients (N = 200) (%) | Acinetobacter spp. (N = 93) (%) | Klebsiella spp. (N = 60) (%) | E. coli (N = 25) (%) | Pseudomonas spp. (N = 38) (%) | Staphylococcus aureus (N = 16) (%) |
|---|---|---|---|---|---|---|---|
| 13–19 | 1 | 2 (1) | 2 (2.2) | 1 (1.7) | 0 (0) | 1 (2.6) | 0 (0) |
| 20–29 | 3 | 6 (3) | 2 (2.2) | 2 (3.3) | 0 (0) | 1 (2.6) | 0 (0) |
| 30–39 | 9 | 19 (9.5) | 12 (12.9) | 2 (3.3) | 2 (8.0) | 2 (5.3) | 1 (6.3) |
| 40–49 | 7 | 14 (7) | 4 (4.3) | 3 (5.0) | 1 (4.0) | 3 (7.9) | 3 (18.8) |
| 50–59 | 16 | 32 (16) | 16 (17.2) | 10 (16.7) | 4 (16) | 7 (18.4) | 3 (18.8) |
| 60–69 | 27 | 54 (27) | 28 (30.1) | 15 (25) | 9 (36) | 9 (23.7) | 5 (31.3) |
| 70–79 | 19 | 39 (19.5) | 19 (20.4) | 12 (20) | 4 (16) | 5 (13.2) | 3 (18.8) |
| 80–89 | 12 | 23 (11.5) | 7 (7.5) | 9 (15) | 4 (16) | 7 (18.4) | 1 (6.3) |
| 90–99 | 6 | 11 (5.5) | 3 (3.2) | 6 (10) | 1 (4.0) | 3 (7.9) | 0 (0) |
| Gram Stain Type | Isolated Organisms | Number of Isolated Organisms N (%) |
|---|---|---|
| Gram-negative | E. coli | 25 (9.2) |
| Pseudomonas spp. | 38 (14) | |
| Klebsiella spp. | 60 (22) | |
| Moraxella spp. | 2 (0.7) | |
| Enterobacter spp. | 2 (0.7) | |
| Acinetobacter spp. | 93 (34) | |
| Proteus spp. | 2 (0.7) | |
| Gram-positive | Staphylococcus aureus | 16 (5.9) |
| Staphylococcus spp. | 2 (0.7) | |
| Streptococcus spp. | 6 (2.2) | |
| Enterococcus spp. | 4 (1.5) | |
| Fungi | Candida spp. | 20 (7.3) |
| Fungi | 1 (0.4) | |
| Saprophytic fungi | 2 (0.7) | |
| Total | 273 (100) |
| Gram-Negative Microorganisms Number of (%) Isolates | Gram-Positive Microorganisms Number of (%) Isolates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotics | Acinetobacter spp. | Klebsiella spp. | E. coli | Pseudomonas spp. | Staphylococcus aureus | ||||||||||
| (N = 93) | (N = 60) | (N = 25) | (N = 38) | (N = 16) | |||||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |
| Penicillin | |||||||||||||||
| Amoxicillin | 1 (1.1) | 0 | 92 (98.9) | 7 (11.7) | 0 | 53 (88.3) | 1 (4) | 0 | 24 (96) | 1 (2.6) | 0 | 37 (97.4) | 7 (43.8) | 0 | 9 (56.3) |
| Ampicillin | 1 (1.1) | 0 | 92 (98.9) | 2 (3.3) | 0 | 58 (96.7) | 0 | 0 | 25 (100) | 0 | 0 | 38 (100) | 2 (12.5) | 0 | 14 (87.5) |
| Cloxacillin | 3 (3.2) | 0 | 90 (96.8) | 8 (13.3) | 0 | 52 (86.7) | 1 (4) | 0 | 24 (96) | 2 (5.3) | 0 | 36 (94.7) | 8 (50) | 0 | 8 (50) |
| Amoxiclav | 1 (1.1) | 0 | 92 (98.9) | 7 (11.7) | 0 | 53 (88.3) | 1 (4) | 0 | 24 (96) | 1 (2.6) | 0 | 37 (97.4) | 6 (37.5) | 0 | 10 (62.5) |
| Aminoglycoside | |||||||||||||||
| Amikacin | 5 (5.4) | 3 (3.2) | 85 (91.4) | 23 (38.3) | 0 | 37 (61.7) | 8 (32) | 1 (4) | 16 (64) | 12 (31.6) | 5 (13.2) | 21 (55.2) | 1 (6.2) | 9 (56.3) | 6 (37.5) |
| Gentamicin | 7 (7.5) | 0 | 86 (92.5) | 24 (40) | 0 | 36 (60) | 5 (20) | 0 | 20 (80) | 13 (34.2) | 1 (2.6) | 24 (63.2) | 10 (62.5) | 1 (6.3) | 5 (31.2) |
| Netilmicin | 9 (9.7) | 0 | 84 (90.3) | 23 (38.3) | 1 (1.7) | 36 (60) | 11 (44) | 0 | 14 (56) | 9 (23.7) | 1 (2.6) | 28 (73.8) | 13 (81.2) | 0 | 3 (18.8) |
| Cephalosporin | |||||||||||||||
| Cefepime | 2 (2.2) | 1 (1.1) | 90 (96.8) | 7 (11.7) | 0 | 53 (88.3) | 2 (8) | 0 | 23 (92) | 10 (26.3) | 3 (7.9) | 25 (65.8) | 5 (31.3) | 0 | 11 (68.8) |
| Cefixime | 0 | 1 (1.1) | 92 (98.9) | 6 (10) | 0 | 54 (90) | 0 | 0 | 25 (100) | 2 (5.3) | 0 | 36 (94.7) | 2 (12.5) | 0 | 14 (87.5) |
| Cefotaxime | 0 | 1 (1.1) | 92 (98.9) | 7 (11.7) | 1 (1.7) | 52 (86.7) | 1 (4) | 0 | 24 (96) | 1 (2.6) | 0 | 37 (97.4) | 2 (12.5) | 2 (12.5) | 12 (75) |
| Ceftazidime | 1 (1.1) | 0 | 92 (98.9) | 8 (13.3) | 0 | 52 (86.7) | 0 | 1 (4) | 24 (96) | 6 (15.8) | 1 (2.6) | 31 (81.6) | 2 (12.5) | 2 (12.5) | 12 (75) |
| Cefuroxime | 1 (1.1) | 0 | 92 (98.9) | 6 (10) | 0 | 54 (90) | 1 (4) | 0 | 24 (96) | 0 | 0 | 38 (100) | 5 (31.2) | 0 | 11 (68.8) |
| Ceftriaxone | 0 | 1 (1.1) | 92 (98.9) | 7 (11.7) | 0 | 53 (88.3) | 1 (4) | 0 | 24 (96) | 0 | 0 | 38 (100) | 4 (25) | 0 | 12 (75) |
| Cephalexin | 1 (1.1) | 0 | 92 (98.9) | 5 (8.3) | 1 (1.7) | 54 (90) | 1 (4) | 0 | 24 (96) | 1 (2.6) | 0 | 37 (97.4) | 4 (25) | 1 (6.2) | 11 (68.8) |
| Fluoroquinolones | |||||||||||||||
| Ciprofloxacin | 4 (4.3) | 0 | 89 (95.7) | 16 (26.7) | 2 (3.3) | 42 (70) | 3 (12) | 0 | 22 (88) | 6 (15.8) | 1 (2.63) | 31 (81.6) | 6 (37.5) | 1 (6.3) | 9 (56.3) |
| Levofloxacin | 5 (5.4) | 1 (1.1) | 87 (93.6) | 18 (30) | 2 (3.3) | 40 (66.7) | 3 (12) | 0 | 22 (88) | 7 (18.4) | 0 | 31 (81.6) | 5 (31.2) | 0 | 11 (68.8) |
| Carbapenem | |||||||||||||||
| Imipenem | 7 (7.5) | 1 (1.1) | 85 (91.4) | 24 (40) | 6 (10) | 30 (50) | 11 (44) | 0 | 14 (56) | 12 (31.6) | 1 (2.6) | 25 (65.8) | 9 (56.2) | 0 | 7 (43.8) |
| Meropenem | 2 (2.2) | 0 | 91 (97.9) | 20 (33.3) | 1 (1.7) | 39 (65) | 6 (24) | 0 | 19 (76) | 8 (21.1) | 1 (2.6) | 29 (76.3) | 8 (50) | 0 | 8 (50) |
| Polymyxin | |||||||||||||||
| Colistin | 82 (88.2) | 3 (3.2) | 8 (8.6) | 57 (95) | 1 (1.7) | 2 (3.3) | 23 (92) | 0 | 2 (8) | 32 (84.2) | 0 | 6 (15.8) | |||
| Sulfonamide | |||||||||||||||
| Cotrimoxazole | 32 (34.4) | 8 (8.6) | 53 (57) | 18 (30) | 1 (1.7) | 41 (68.3) | 4 (16) | 0 | 21 (84) | 5 (13.2) | 0 | 33 (86.8) | 11 (68.7) | 0 | 5 (31.3) |
| Oxazolidinone | |||||||||||||||
| Linezolid | 13 (81.3) | 0 | 3 (18.7) | ||||||||||||
| Glycopeptide | |||||||||||||||
| Vancomycin | 16 (100) | 0 | 0 | ||||||||||||
| Glycylcycline | |||||||||||||||
| Tigecycline | 67 (72.0) | 15 (16.1) | 11 (11.8) | 42 (70) | 12 (20) | 6 (10) | 18 (72) | 4 (16) | 3 (12) | 8 (21.1) | 3 (7.9) | 27 (71) | 12 (75) | 0 | 4 (25) |
| Fusidane | |||||||||||||||
| Fusidic Acid | 12 (75) | 0 | 4 (25) | ||||||||||||
| Monobactam | |||||||||||||||
| Aztreonam | 1 (1.1) | 2 (2.2) | 90 (96.8) | 7 (11.7) | 1 (1.7) | 52 (86.7) | 1 (4) | 0 | 24 (96) | 13 (34.2) | 1 (2.6) | 24 (63.2) | |||
| Beta-lactamase inhibitor + penicillin | |||||||||||||||
| Tazobactam + Piperacillin | 2 (2.2) | 2 (2.2) | 89 (95.6) | 19 (31.7) | 1 (1.7) | 40 (66.7) | 6 (24) | 2 (8) | 17 (68) | 23 (60.5) | 6 (15.8) | 9 (23.7) | 7 (43.7) | 0 | 9 (56.3) |
| Isolated Organism | SDR | MDR |
|---|---|---|
| N (%) | N (%) | |
| Acinetobacter spp. | 1 (1.08) | 92 (98.92) |
| Pseudomonas spp. | 3 (7.89) | 35 (92.09) |
| Klebsiella spp. | 6 (9.99) | 54 (90.01) |
| Staphylococcus aureus | 11 (68.75) | 5(31.25) |
| E. coli | 1 (4.00) | 24 (96.00) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haque, S.; Ahmed, A.; Islam, N.; Haque, F.K.M. High Prevalence of Multidrug-Resistant Bacteria in the Trachea of Intensive Care Units Admitted Patients: Evidence from a Bangladeshi Hospital. Antibiotics 2024, 13, 62. https://doi.org/10.3390/antibiotics13010062
Haque S, Ahmed A, Islam N, Haque FKM. High Prevalence of Multidrug-Resistant Bacteria in the Trachea of Intensive Care Units Admitted Patients: Evidence from a Bangladeshi Hospital. Antibiotics. 2024; 13(1):62. https://doi.org/10.3390/antibiotics13010062
Chicago/Turabian StyleHaque, Sabrina, Akash Ahmed, Nazrul Islam, and Fahim Kabir Monjurul Haque. 2024. "High Prevalence of Multidrug-Resistant Bacteria in the Trachea of Intensive Care Units Admitted Patients: Evidence from a Bangladeshi Hospital" Antibiotics 13, no. 1: 62. https://doi.org/10.3390/antibiotics13010062
APA StyleHaque, S., Ahmed, A., Islam, N., & Haque, F. K. M. (2024). High Prevalence of Multidrug-Resistant Bacteria in the Trachea of Intensive Care Units Admitted Patients: Evidence from a Bangladeshi Hospital. Antibiotics, 13(1), 62. https://doi.org/10.3390/antibiotics13010062






